Synthesis and Structural Characterization of Selenium Nanoparticles–Bacillus sp. MKUST-01 Exopolysaccharide (SeNPs–EPS) Conjugate for Biomedical Applications
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation and Screening of EPS-Producing Bacteria
2.2. Identification of EPS-Producing Bacteria
2.3. Production, Extraction, and Purification of EPS
2.4. Synthesis of Selenium Nanoparticles (SeNPs)
2.5. Exopolysaccharide–Nanoparticle Conjugation
2.6. Characterization of SeNPs–EPS
2.7. Biological Activities
2.7.1. Reducing and Scavenging Activity
2.7.2. Hemolytic Activity
2.7.3. Cell Viability using MTT Assay
2.7.4. In Vivo Toxicity Assay
2.8. Statistical Analysis
3. Results and Discussion
3.1. Isolation and Identification of EPS-Producing Bacteria
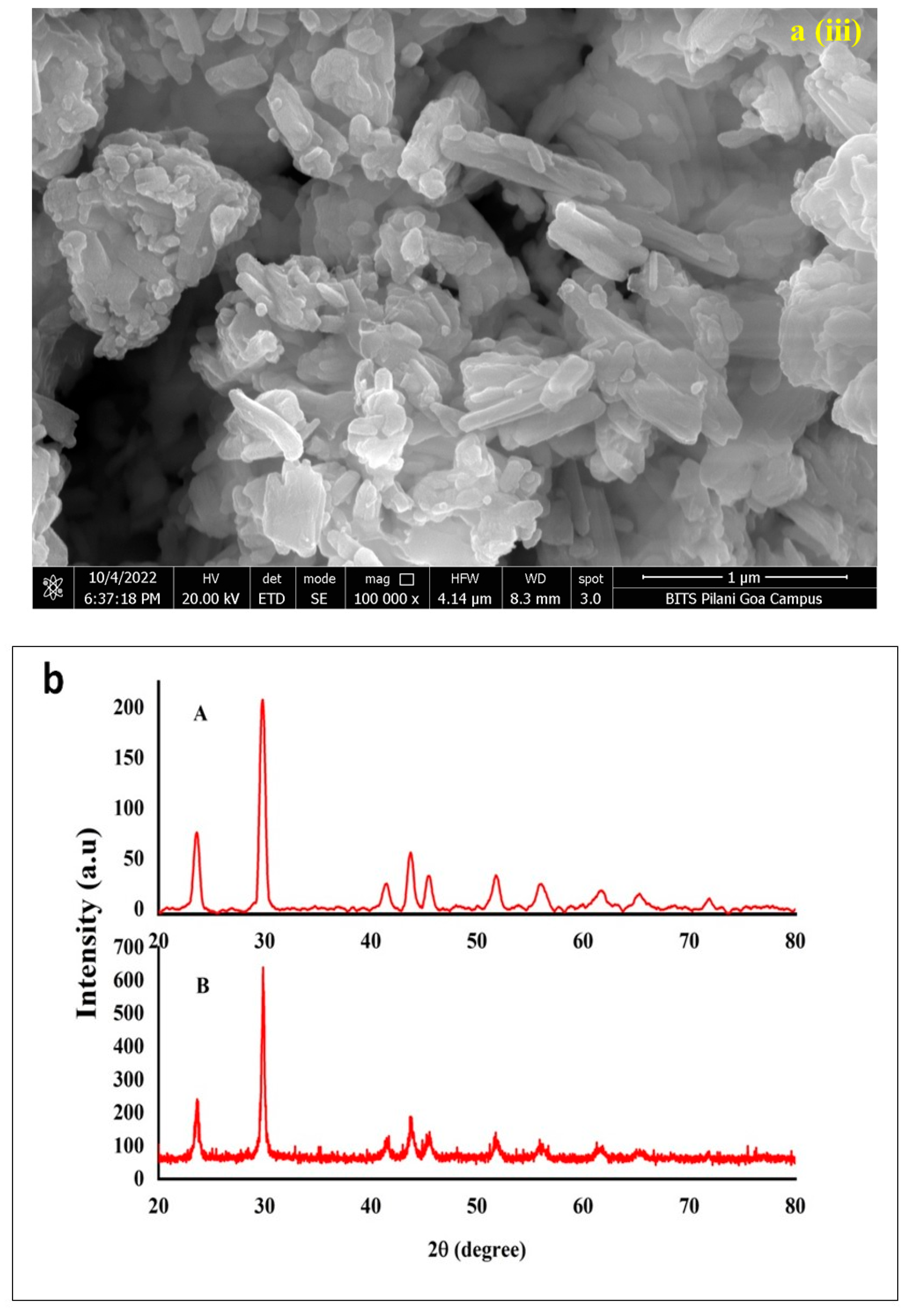
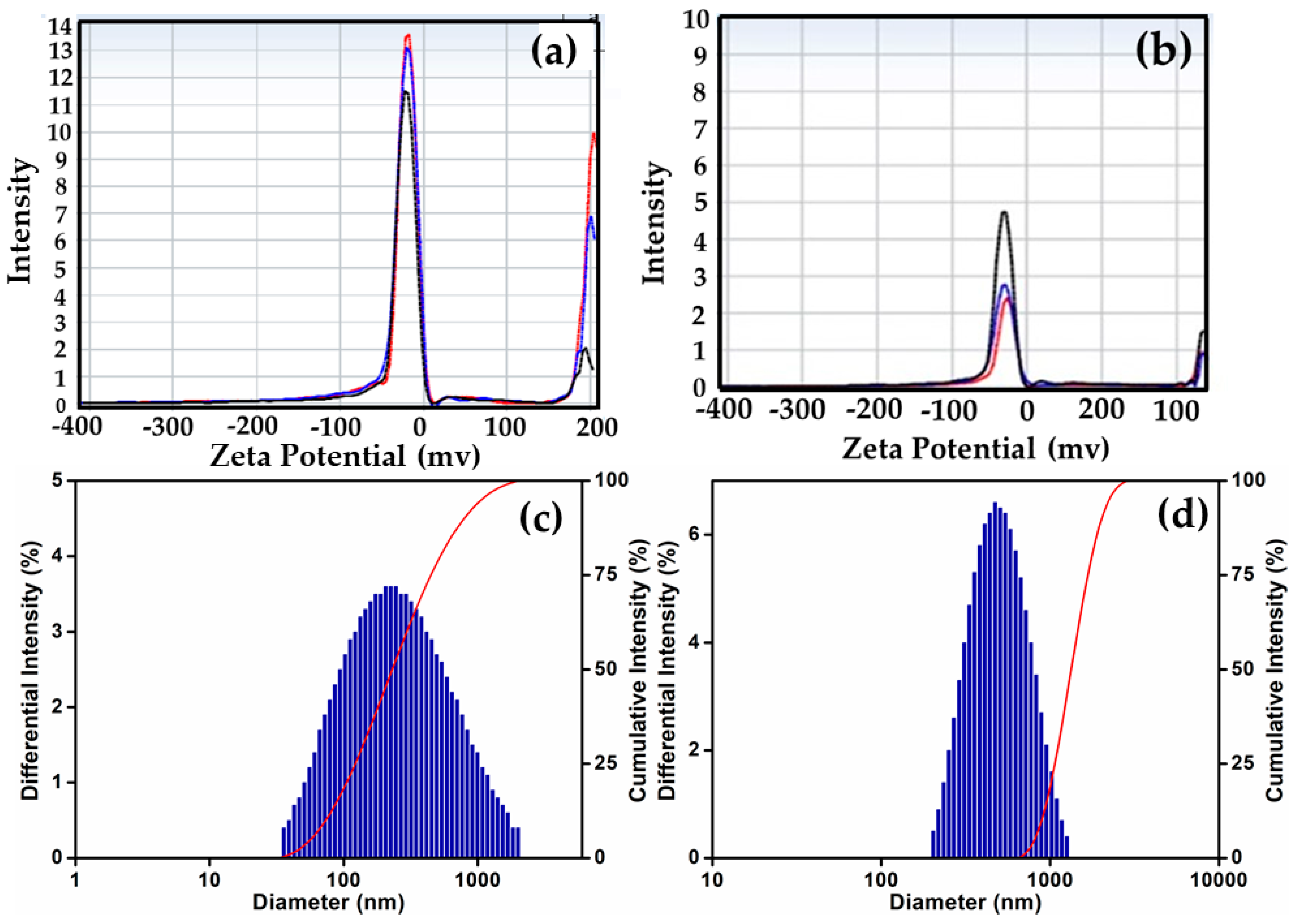
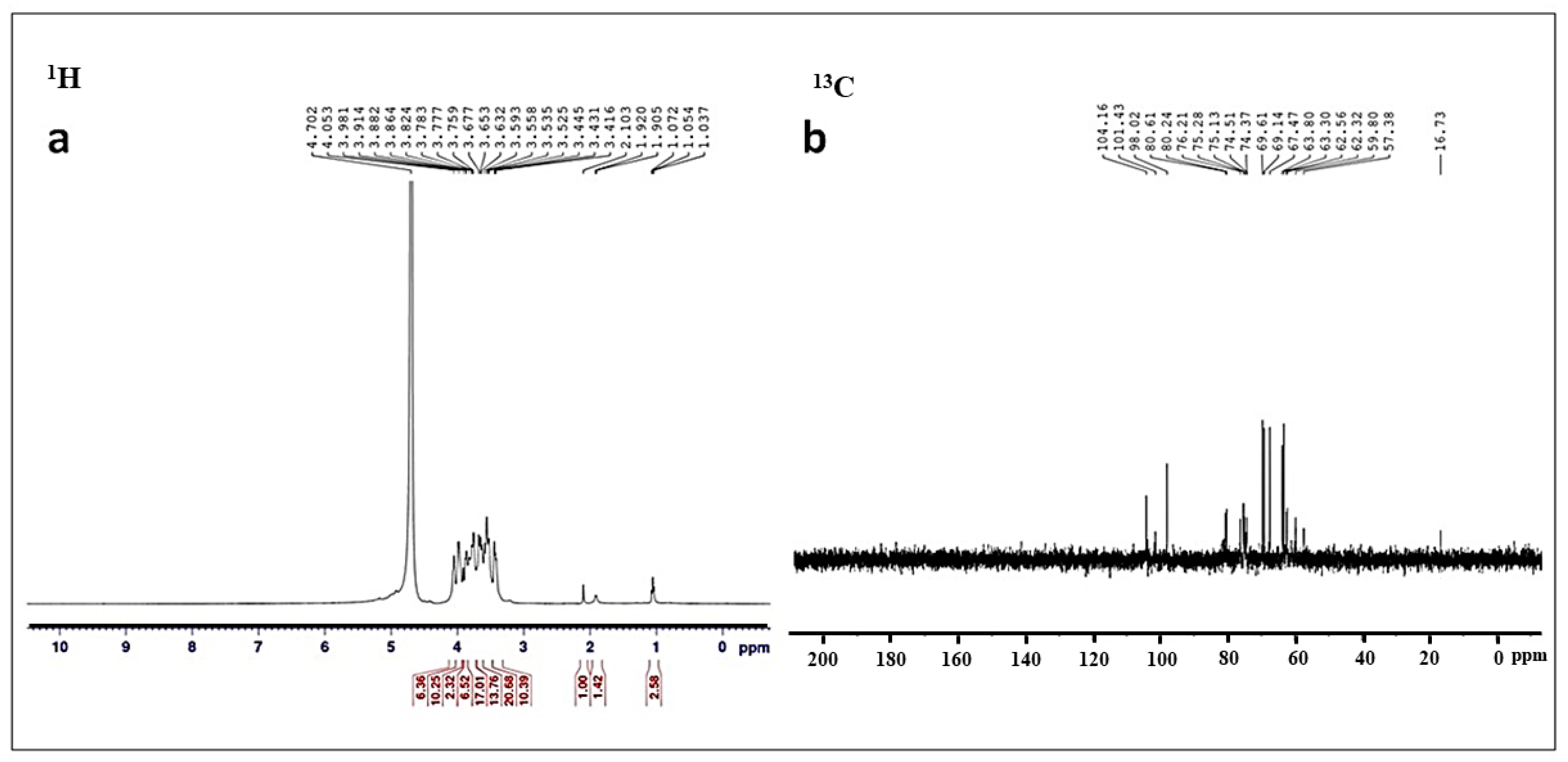
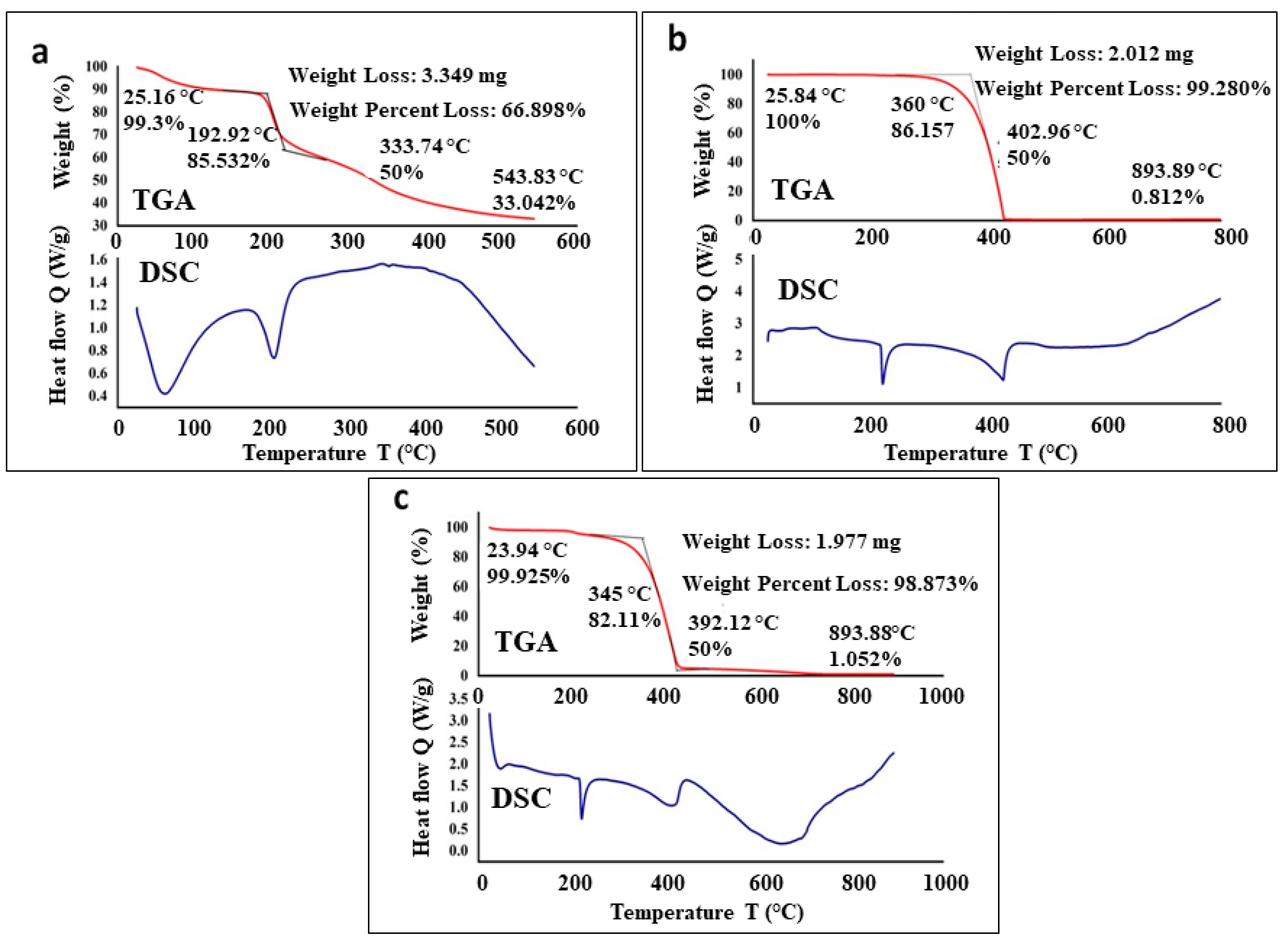
3.2. Characterization of Conjugated SeNPs–EPS
3.3. Biological Activities
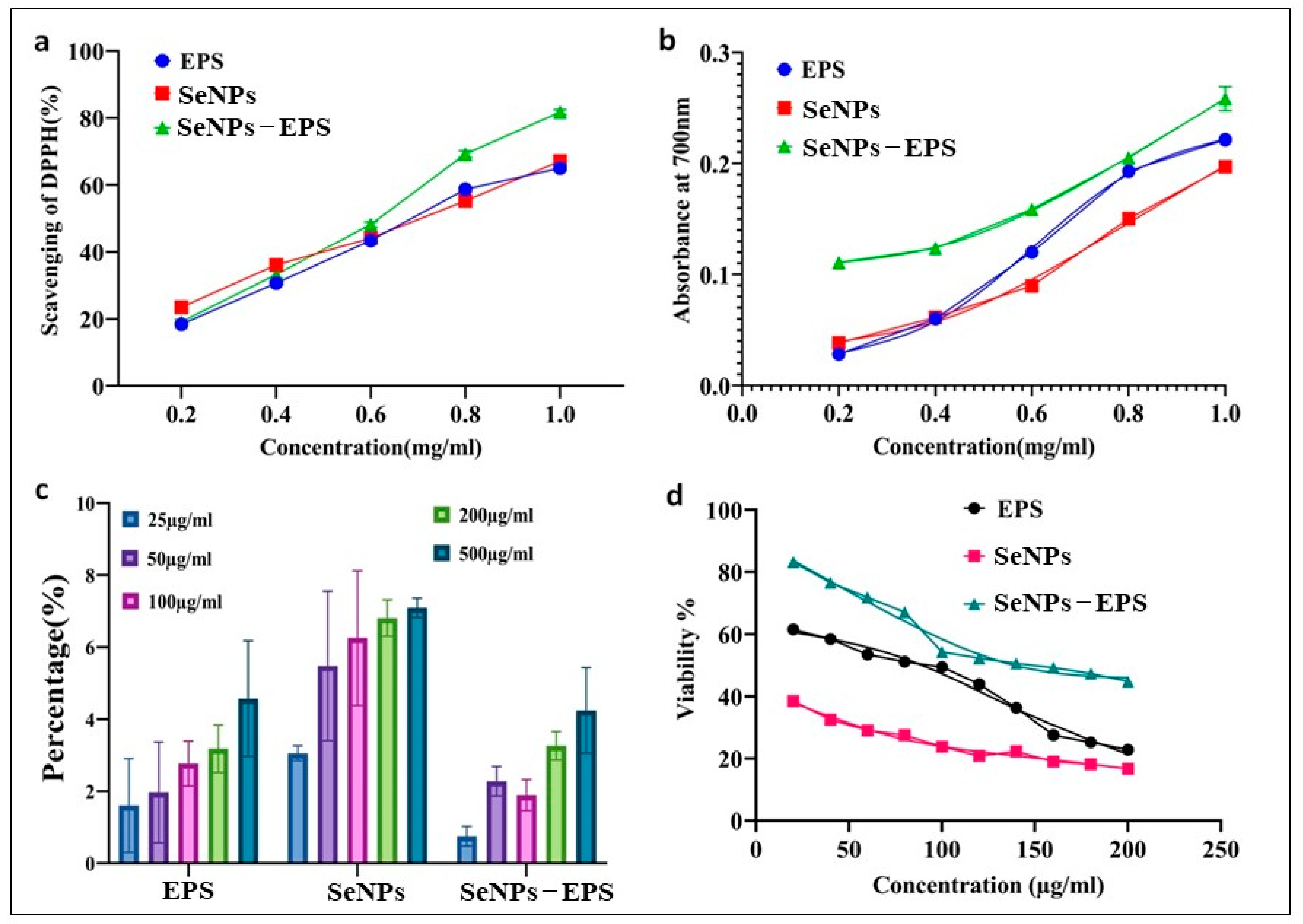
DPPH and Reducing Power Assay
3.4. In Vivo Toxicity Study
Gnotobiotic Brine Shrimp
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yougbaré, S.; Mutalik, C.; Okoro, G.; Lin, I.-H.; Krisnawati, D.I.; Jazidie, A.; Nuh, M.; Chang, C.-C.; Kuo, T.-R. Emerging trends in nanomaterials for antibacterial applications. Int. J. Nanomed. 2021, 16, 5831–5867. [Google Scholar] [CrossRef] [PubMed]
- Lucaci, A.-R.; Bulgariu, D.; Bulgariu, L. Green Synthesis of Gold Nanoparticles Using Marine Red Algae Biomass. In Proceedings of the 2021 International Conference on e-Health and Bioengineering (EHB), Iasi, Romania, 18–19 November 2021; IEEE: Piscataway Township, NJ, USA, 2021; pp. 1–4. [Google Scholar]
- Arif, R.; Uddin, R. A review on recent developments in the biosynthesis of silver nanoparticles and its biomedical applications. Med. Devices Sens. 2021, 4, e10158. [Google Scholar] [CrossRef]
- Zhu, M.; Niu, G.; Tang, J. Elemental Se: Fundamentals and its optoelectronic applications. J. Mater. Chem. C 2019, 7, 2199–2206. [Google Scholar] [CrossRef]
- Nayak, V.; Singh, K.R.; Singh, A.K.; Singh, R.P. Potentialities of selenium nanoparticles in biomedical science. New J. Chem. 2021, 45, 2849–2878. [Google Scholar] [CrossRef]
- Kieliszek, M. Selenium–fascinating microelement, properties and sources in food. Molecules 2019, 24, 1298. [Google Scholar] [CrossRef] [PubMed]
- Skalickova, S.; Milosavljevic, V.; Cihalova, K.; Horky, P.; Richtera, L.; Adam, V. Selenium nanoparticles as a nutritional supplement. Nutrition 2017, 33, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, S.Y.; Xu, J.J.; Chen, H.Y. A new method for the synthesis of selenium nanoparticles and the application to construction of H2O2 biosensor. Chin. Chem. Lett. 2004, 15, 1345–1348. [Google Scholar]
- Constantinescu-Aruxandei, D.; Frîncu, R.M.; Capră, L.; Oancea, F. Selenium analysis and speciation in dietary supplements based on next-generation selenium ingredients. Nutrients 2018, 10, 1466. [Google Scholar] [CrossRef]
- Back, T.G. Investigations of new types of glutathione peroxidase mimetics. In Biochalcogen Chemistry: The Biological Chemistry of Sulfur, Selenium, and Tellurium; ACS Publications: Washiington, DC, USA, 2013; pp. 143–162. [Google Scholar]
- Gupta, M.; Gupta, S. An overview of selenium uptake, metabolism, and toxicity in plants. Front. Plant Sci. 2017, 7, 2074. [Google Scholar] [CrossRef]
- Shoeibi, S.; Mozdziak, P.; Golkar-Narenji, A. Biogenesis of selenium nanoparticles using green chemistry. Top. Curr. Chem. 2017, 375, 88. [Google Scholar] [CrossRef]
- Varlamova, E.G.; Turovsky, E.A.; Blinova, E.V. Therapeutic potential and main methods of obtaining selenium nanoparticles. Int. J. Mol. Sci. 2021, 22, 10808. [Google Scholar] [CrossRef] [PubMed]
- Zong, A.; Cao, H.; Wang, F. Anticancer polysaccharides from natural resources: A review of recent research. Carbohydr. Polym. 2012, 90, 1395–1410. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Monty, J.; Linhardt, R.J. Polysaccharide-based nanocomposites and their applications. Carbohydr. Res. 2015, 405, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Gunti, L.; Dass, R.S.; Kalagatur, N.K. Phytofabrication of selenium nanoparticles from Emblica officinalis fruit extract and exploring its biopotential applications: Antioxidant, antimicrobial, and biocompatibility. Front. Microbiol. 2019, 10, 931. [Google Scholar] [CrossRef]
- Vu, B.; Chen, M.; Crawford, R.J.; Ivanova, E.P. Bacterial extracellular polysaccharides involved in biofilm formation. Molecules 2009, 14, 2535–2554. [Google Scholar] [CrossRef] [PubMed]
- Osemwegie, O.O.; Adetunji, C.O.; Ayeni, E.A.; Adejobi, O.I.; Arise, R.O.; Nwonuma, C.O.; Oghenekaro, A.O. Exopolysaccharides from bacteria and fungi: Current status and perspectives in Africa. Heliyon 2020, 6, e04205. [Google Scholar] [CrossRef]
- Chaisuwan, W.; Jantanasakulwong, K.; Wangtueai, S.; Phimolsiripol, Y.; Chaiyaso, T.; Techapun, C.; Phongthai, S.; You, S.; Regenstein, J.M.; Seesuriyachan, P. Microbial exopolysaccharides for immune enhancement: Fermentation, modifications and bioactivities. Food Biosci. 2020, 35, 100564. [Google Scholar] [CrossRef]
- Yildiz, H.; Karatas, N. Microbial exopolysaccharides: Resources and bioactive properties. Process. Biochem. 2018, 72, 41–46. [Google Scholar] [CrossRef]
- Ikram, M.; Javed, B.; Raja, N.I.; Mashwani, Z.-u.-R. Biomedical potential of plant-based selenium nanoparticles: A comprehensive review on therapeutic and mechanistic aspects. Int. J. Nanomed. 2021, 16, 249–268. [Google Scholar] [CrossRef]
- Roy, R.; Tiwari, M.; Donelli, G.; Tiwari, V. Strategies for combating bacterial biofilms: A focus on anti-biofilm agents and their mechanisms of action. Virulence 2018, 9, 522–554. [Google Scholar] [CrossRef]
- Garrity, G.; Brenner, D.; Krieg, N.; Staley, J.; Manual, B. Systematic Bacteriology: The Proteobacteria, Part C: The Alpha-, Beta-, Delta-, and Epsilonproteobacteria. Bergey’s Man. Trust. Dep. Microbiol. Mol. Genet. 2005, 2. [Google Scholar]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- DuBois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.t.; Smith, F. Colorimetric method for determination of sugars and related substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Yang, Y.; Peng, Q.; Guo, Y.; Han, Y.; Xiao, H.; Zhou, Z. Isolation and characterization of dextran produced by Leuconostoc citreum NM105 from manchurian sauerkraut. Carbohydr. Polym. 2015, 133, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Vahdati, M.; Tohidi Moghadam, T. Synthesis and characterization of selenium nanoparticles-lysozyme nanohybrid system with synergistic antibacterial properties. Sci. Rep. 2020, 10, 510. [Google Scholar] [CrossRef] [PubMed]
- Yeo, L.K.; Olusanya, T.O.; Chaw, C.S.; Elkordy, A.A. Brief effect of a small hydrophobic drug (cinnarizine) on the physicochemical characterisation of niosomes produced by thin-film hydration and microfluidic methods. Pharmaceutics 2018, 10, 185. [Google Scholar] [CrossRef]
- Singh, R.P.; Shukla, M.K.; Mishra, A.; Kumari, P.; Reddy, C.; Jha, B. Isolation and characterization of exopolysaccharides from seaweed associated bacteria Bacillus licheniformis. Carbohydr. Polym. 2011, 84, 1019–1026. [Google Scholar] [CrossRef]
- Akgül, H.; Mohammed, F.S.; Kına, E.; Uysal, İ.; Sevindik, M.; Doğan, M. Total Antioxidant and Oxidant Status and DPPH Free radical activity of Euphorbia eriophora. Turk. J. Agric. Food Sci. Technol. 2022, 10, 272–275. [Google Scholar] [CrossRef]
- Shimada, K.; Fujikawa, K.; Yahara, K.; Nakamura, T. Antioxidative properties of xanthan on the autoxidation of soybean oil in cyclodextrin emulsion. J. Agric. Food Chem. 1992, 40, 945–948. [Google Scholar] [CrossRef]
- Younis, F.A.; Saleh, S.R.; El-Rahman, S.S.A.; Newairy, A.-S.A.; El-Demellawy, M.A.; Ghareeb, D.A. Preparation, physicochemical characterization, and bioactivity evaluation of berberine-entrapped albumin nanoparticles. Sci. Rep. 2022, 12, 17431. [Google Scholar] [CrossRef]
- Mosmann, T.; Fong, T. Specific assays for cytokine production by T cells. J. Immunol. Methods 1989, 116, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Rajabi, S.; Ramazani, A.; Hamidi, M.; Naji, T. Artemia salina as a model organism in toxicity assessment of nanoparticles. DARU J. Pharm. Sci. 2015, 23, 20. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Selvi, S.S.; Praba, A.; Selvaraj, M.; Rani, L.M.; Suganthi, P.; Devi, B.S.; Govindaraju, M. Antibacterial activity and in-vitro cytotoxicity assay against brine shrimp using silver nanoparticles synthesized from Sargassum ilicifolium. Dig. J. Nanomater. Biostructures 2012, 7, 1447–1455. [Google Scholar]
- Ammar, S.; Fiévet, F. Polyol Synthesis: A Versatile Wet-Chemistry Route for the Design and Production of Functional Inorganic Nanoparticles. Nanomaterials 2020, 10, 1217. [Google Scholar] [CrossRef] [PubMed]
- Nikam, A.V.; Prasad, B.L.V.; Kulkarni, A.A. Wet chemical synthesis of metal oxide nanoparticles: A review. CrystEngComm 2018, 20, 5091–5107. [Google Scholar] [CrossRef]
- Tan, C.; Zhang, H. Wet-chemical synthesis and applications of non-layer structured two-dimensional nanomaterials. Nat. Commun. 2015, 6, 7873. [Google Scholar] [CrossRef]
- Shubharani, R.; Mahesh, M.; Yogananda Murthy, V. Biosynthesis and characterization, antioxidant and antimicrobial activities of selenium nanoparticles from ethanol extract of Bee Propolis. J. Nanomed. Nanotechnol. 2019, 10, 1–7. [Google Scholar]
- Fritea, L.; Laslo, V.; Cavalu, S.; Costea, T.; Vicas, S.I. Green biosynthesis of selenium nanoparticles using parsley (Petroselinum crispum) leaves extract. Stud. Univ. “Vasile Goldis” Arad. Ser. Stiintele Vietii (Life Sci. Ser.) 2017, 27, 203–208. [Google Scholar]
- Ashrafpour, S.; Moghadam, T.T. Interaction of silver nanoparticles with Lysozyme: Functional and structural investigations. Surf. Interfaces 2018, 10, 216–221. [Google Scholar] [CrossRef]
- Moghadam, T.T.; Ranjbar, B.; Khajeh, K.; Etezad, S.M.; Khalifeh, K.; Ganjalikhany, M.R. Interaction of lysozyme with gold nanorods: Conformation and activity investigations. Int. J. Biol. Macromol. 2011, 49, 629–636. [Google Scholar] [CrossRef]
- Nel, A.E.; Mädler, L.; Velegol, D.; Xia, T.; Hoek, E.M.; Somasundaran, P.; Klaessig, F.; Castranova, V.; Thompson, M. Understanding biophysicochemical interactions at the nano–bio interface. Nat. Mater. 2009, 8, 543–557. [Google Scholar] [CrossRef] [PubMed]
- Cai, P.; Zhang, X.; Wang, M.; Wu, Y.-L.; Chen, X. Combinatorial nano–bio interfaces. ACS Nano 2018, 12, 5078–5084. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, J.; Zhang, L. Creation of highly stable selenium nanoparticles capped with hyperbranched polysaccharide in water. Langmuir 2010, 26, 17617–17623. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Tang, Q.; Zhong, X.; Bai, Y.; Chen, T.; Zhang, Y.; Li, Y.; Zheng, W. Surface decoration by Spirulina polysaccharide enhances the cellular uptake and anticancer efficacy of selenium nanoparticles. Int. J. Nanomed. 2012, 7, 835–844. [Google Scholar]
- Kannan, S.; Mohanraj, K.; Prabhu, K.; Barathan, S.; Sivakumar, G. Synthesis of selenium nanorods with assistance of biomolecule. Bull. Mater. Sci. 2014, 37, 1631–1635. [Google Scholar] [CrossRef]
- Cittrarasu, V.; Kaliannan, D.; Dharman, K.; Maluventhen, V.; Easwaran, M.; Liu, W.C.; Balasubramanian, B.; Arumugam, M. Green synthesis of selenium nanoparticles mediated from Ceropegia bulbosa Roxb extract and its cytotoxicity, antimicrobial, mosquitocidal and photocatalytic activities. Sci. Rep. 2021, 11, 1032. [Google Scholar] [CrossRef]
- Zhang, J.; Cao, Y.; Wang, J.; Guo, X.; Zheng, Y.; Zhao, W.; Mei, X.; Guo, T.; Yang, Z. Physicochemical characteristics and bioactivities of the exopolysaccharide and its sulphated polymer from Streptococcus thermophilus GST-6. Carbohydr. Polym. 2016, 146, 368–375. [Google Scholar] [CrossRef]
- Wang, J.; Zhao, X.; Tian, Z.; Yang, Y.; Yang, Z. Characterization of an exopolysaccharide produced by Lactobacillus plantarum YW11 isolated from Tibet Kefir. Carbohydr. Polym. 2015, 125, 16–25. [Google Scholar] [CrossRef]
- Zhang, J.; Ji, T.; Yang, X.; Liu, G.; Liang, L.; Liu, X.; Wen, C.; Ye, Z.; Wu, M.; Xu, X. Properties of selenium nanoparticles stabilized by Lycium barbarum polysaccharide-protein conjugates obtained with subcritical water. Int. J. Biol. Macromol. 2022, 205, 672–681. [Google Scholar] [CrossRef]
- Kokila, K.; Elavarasan, N.; Sujatha, V. Diospyros montana leaf extract-mediated synthesis of selenium nanoparticles and their biological applications. New J. Chem. 2017, 41, 7481–7490. [Google Scholar] [CrossRef]
- Srivastava, N.; Mukhopadhyay, M. Biosynthesis and structural characterization of selenium nanoparticles mediated by Zooglea ramigera. Powder Technol. 2013, 244, 26–29. [Google Scholar] [CrossRef]
- Xiao, Y.; Huang, Q.; Zheng, Z.; Guan, H.; Liu, S. Construction of a Cordyceps sinensis exopolysaccharide-conjugated selenium nanoparticles and enhancement of their antioxidant activities. Int. J. Biol. Macromol. 2017, 99, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Carneiro-da-Cunha, M.G.; Cerqueira, M.A.; Souza, B.W.; Teixeira, J.A.; Vicente, A.A. Influence of concentration, ionic strength and pH on zeta potential and mean hydrodynamic diameter of edible polysaccharide solutions envisaged for multinanolayered films production. Carbohydr. Polym. 2011, 85, 522–528. [Google Scholar] [CrossRef]
- Khan, M.; Shaik, M.R.; Khan, S.T.; Adil, S.F.; Kuniyil, M.; Khan, M.; Al-Warthan, A.A.; Siddiqui, M.R.H.; Nawaz Tahir, M. Enhanced antimicrobial activity of biofunctionalized zirconia nanoparticles. ACS Omega 2020, 5, 1987–1996. [Google Scholar] [CrossRef] [PubMed]
- Sathishkumar, R.; Kannan, R.; Jinendiran, S.; Sivakumar, N.; Selvakumar, G.; Shyamkumar, R. Production and characterization of exopolysaccharide from the sponge-associated Bacillus subtilis MKU SERB2 and its in-vitro biological properties. Int. J. Biol. Macromol. 2021, 166, 1471–1479. [Google Scholar] [CrossRef]
- Ahmed, Z.; Wang, Y.; Anjum, N.; Ahmad, H.; Ahmad, A.; Raza, M. Characterization of new exopolysaccharides produced by coculturing of L. kefiranofaciens with yoghurt strains. Int. J. Biol. Macromol. 2013, 59, 377–383. [Google Scholar] [CrossRef]
- Chen, W.; Li, Y.; Yang, S.; Yue, L.; Jiang, Q.; Xia, W. Synthesis and antioxidant properties of chitosan and carboxymethyl chitosan-stabilized selenium nanoparticles. Carbohydr. Polym. 2015, 132, 574–581. [Google Scholar] [CrossRef]
- Peng, D.; Zhang, J.; Liu, Q.; Taylor, E.W. Size effect of elemental selenium nanoparticles (Nano-Se) at supranutritional levels on selenium accumulation and glutathione S-transferase activity. J. Inorg. Biochem. 2007, 101, 1457–1463. [Google Scholar] [CrossRef]
- Mishra, R.R.; Prajapati, S.; Das, J.; Dangar, T.K.; Das, N.; Thatoi, H. Reduction of selenite to red elemental selenium by moderately halotolerant Bacillus megaterium strains isolated from Bhitarkanika mangrove soil and characterization of reduced product. Chemosphere 2011, 84, 1231–1237. [Google Scholar] [CrossRef]
- Forootanfar, H.; Adeli-Sardou, M.; Nikkhoo, M.; Mehrabani, M.; Amir-Heidari, B.; Shahverdi, A.R.; Shakibaie, M. Antioxidant and cytotoxic effect of biologically synthesized selenium nanoparticles in comparison to selenium dioxide. J. Trace Elem. Med. Biol. 2014, 28, 75–79. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, H.; Yan, X.; Zhang, L. Comparison of short-term toxicity between Nano-Se and selenite in mice. Life Sci. 2005, 76, 1099–1109. [Google Scholar] [CrossRef] [PubMed]
- Jinendiran, S.; Boopathi, S.; Sivakumar, N.; Selvakumar, G. Functional characterization of probiotic potential of novel pigmented bacterial strains for aquaculture applications. Probiotics Antimicrob. Proteins 2019, 11, 186–197. [Google Scholar] [CrossRef] [PubMed]




| Isolates (Strains) | Biomass (gm/L) | Crude Preparation | Partially Purified | |||
|---|---|---|---|---|---|---|
| EPS (g/L) | Carbohydrate (mg/g) | Protein (mg/g) | EPS (g/L) | Carbohydrate (%) | ||
| MKUST03 | 1.73 ± 0.10 | 3.03 ± 0.34 | 60.66 ± 1.52 | 58.00 ± 0.98 | 0.891 ± 0.52 | 89.00 ± 1.14 |
| MKUST04 | 5.9 ± 0.39 | 2.78 ± 0.20 | 43.00 ± 2.64 | 23.00 ± 0.22 | 0.826 ± 0.11 | 71.00 ± 0.96 |
| MKUST15 | 7.01 ± 0.34 | 1.41 ± 0.27 | 86.33 ± 2.51 | 72.00 ± 0.15 | 0.502 ± 0.01 | 82.00 ± 0.22 |
| MKUST19 | 1.63 ± 0.07 | 1.38 ± 0.17 | 71.33 ± 2.30 | 64.00 ± 0.78 | 0.317 ± 0.058 | 87.00 ± 0.05 |
| MKUST25 | 0.51 ± 0.24 | 1.52 ± 0.37 | 59.23 ± 8.02 | 72.00 ± 0.241 | 0.243 ± 0.021 | 82.00 ± 1.06 |
| MKUST29 | 0.93 ± 0.045 | 1.35 ± 0.16 | 29.45 ± 3.56 | 76.00 ± 1.02 | 0.14 ± 0.06 | 72.00 ± 0.09 |
| MKUST30 | 1.24 ± 0.10 | 1.42 ± 0.39 | 52.34 ± 2.08 | 56.00 ± 1.18 | 0.252 ± 0.12 | 70.00 ± 1.0 |
| Sampling Day | Survival Rates (%) * | Individual Length (µm) | ||||||
|---|---|---|---|---|---|---|---|---|
| Control | EPS | SeNPs | SeNPs–EPS | Control | EPS | SeNPs | SeNPs–EPS | |
| 1 | 94 ± 0.4 | 96 ± 1.2 | 87 ± 1.0 | 95 ± 1 | 85 ± 0.4 | 82.5 ± 6.6 | 57.5 ± 3.6 | 74.2 ± 1.02 |
| 2 | 75 ± 0.9 | 83 ± 0.8 | 78 ± 0.2 | 94 ± 0 | 86 ± 0.26 | 92.5 ± 0.2 | 85. ± 9.2 | 95. ± 2.6 |
| 3 | 52 ± 1.2 | 76 ± 1 | 58 ± 0 | 77 ± 5 | 92 ± 0.71 | 97.5 ± 1.6 | 92.5 ± 1.9 | 100 ± 4.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramachandran, T.; Manoharan, D.; Natesan, S.; Rajaram, S.K.; Karuppiah, P.; Shaik, M.R.; Khan, M.; Shaik, B. Synthesis and Structural Characterization of Selenium Nanoparticles–Bacillus sp. MKUST-01 Exopolysaccharide (SeNPs–EPS) Conjugate for Biomedical Applications. Biomedicines 2023, 11, 2520. https://doi.org/10.3390/biomedicines11092520
Ramachandran T, Manoharan D, Natesan S, Rajaram SK, Karuppiah P, Shaik MR, Khan M, Shaik B. Synthesis and Structural Characterization of Selenium Nanoparticles–Bacillus sp. MKUST-01 Exopolysaccharide (SeNPs–EPS) Conjugate for Biomedical Applications. Biomedicines. 2023; 11(9):2520. https://doi.org/10.3390/biomedicines11092520
Chicago/Turabian StyleRamachandran, Thirumalaivasan, Devaprakash Manoharan, Sivakumar Natesan, Shyam Kumar Rajaram, Ponmurugan Karuppiah, Mohammed Rafi Shaik, Mujeeb Khan, and Baji Shaik. 2023. "Synthesis and Structural Characterization of Selenium Nanoparticles–Bacillus sp. MKUST-01 Exopolysaccharide (SeNPs–EPS) Conjugate for Biomedical Applications" Biomedicines 11, no. 9: 2520. https://doi.org/10.3390/biomedicines11092520









