OCT Imaging in Murine Models of Alzheimer’s Disease in a Systematic Review: Findings, Methodology and Future Perspectives
Abstract
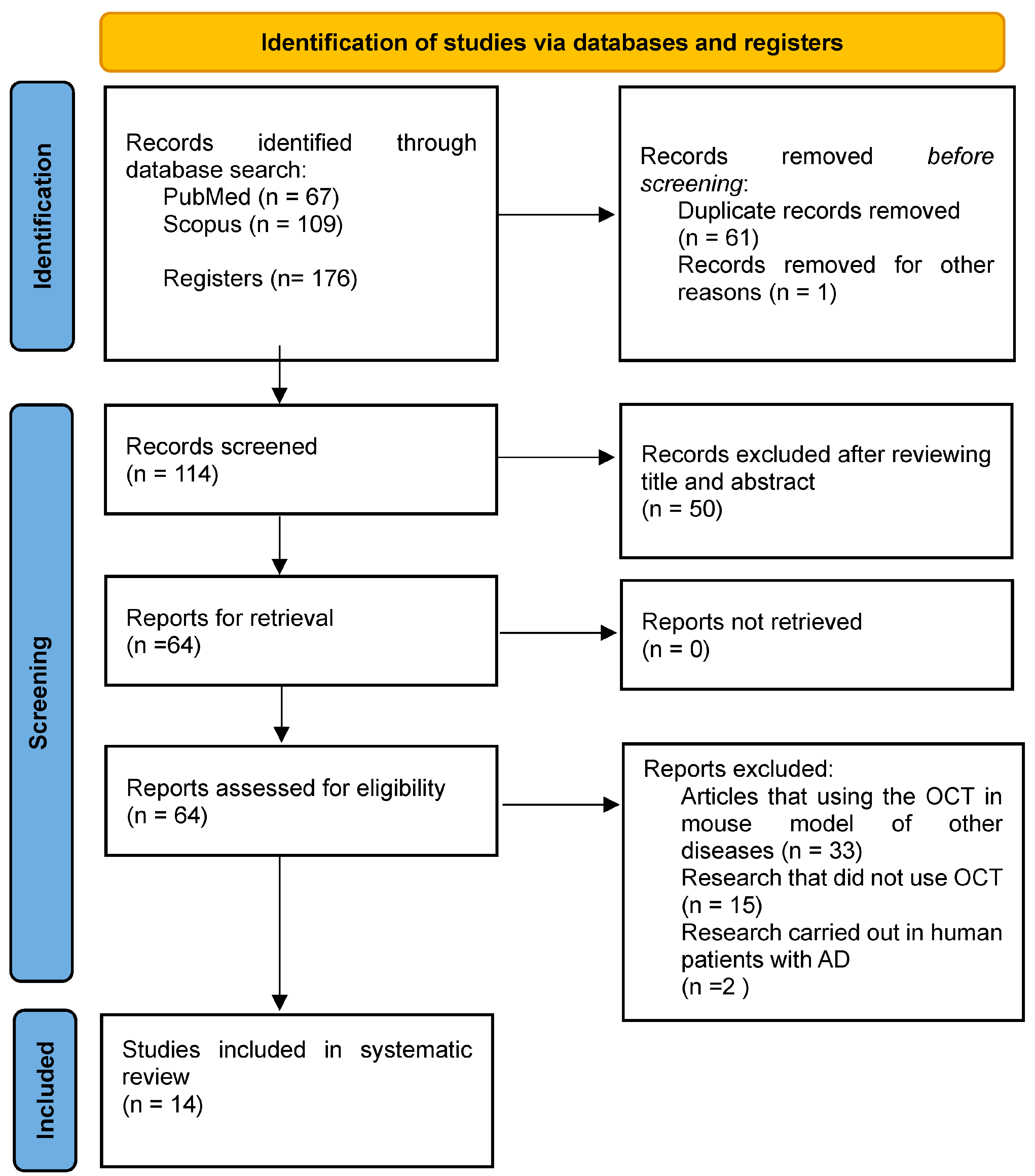
:1. Introduction
2. Materials and Methods
2.1. Search Strategy
2.2. Inclusion and Exclusion Criteria
3. Results
3.1. Mouse Model Analysed
- (i)
- Chiquita et al. [44] conducted a longitudinal analysis, examining male mice at 4, 8, 12, and 16 months of age and comparing them with age-matched WT mice (C57BL/6J);
- (ii)
- (iii)
- Gardner et al. [46] performed a cross-sectional study with 20 3xTg-AD mice (16 males and 4 females) at different ages (2, 4, 7 and 10 months), comparing them with 12 C57BL/6J mice (only males) as the control group;
- (iv)
- Ferreira et al. [47] utilised the 3xTg-AD model to compare the effects of disease and aging with a normative database of the C57BL6/129S model, using male mice at various ages (1, 2, 3, and 4 months);
- (v)
- Guimarães et al. [48] analysed 60 male 3xTg-AD mice and 57 male C57BL6/129S mice at different ages (1, 2, 3, 4, 8, 12, and 16 months) in a longitudinal study;
- (vi)
- Batista et al. [49] examined both eyes of 144 male mice, including 57 3xTg-AD mice and 57 WT mice, at various ages (1, 2, 3, 4, 8, 12, and 16 months).
- (i)
- Lim et al. [51] examined 32 5xFAD mice and 38 non-transgenic (WT) littermates with a C57BL/6J genetic background as controls at different ages (6, 12, and 17 months);
- (ii)
- Kim et al. [22] used five transgenic mice and six B6SJLF1/J WT controls, all 6 months of age;
- (iii)
- Matei et al. [52] employed 16 male transgenic mice and 16 male C57BL/6J WT controls, all of them 3 months old.
| AD Murine Model | WT | Laboratory | Age | Gender | N (Number) | Eye Selected | |
|---|---|---|---|---|---|---|---|
| Buccarello et al., 2017 [35] | TgCRND8 | 129/Sv | Jackson Laboratories, USA | 8 months | Male | 4 AD/4 WT | Both |
| Salobrar-García et al., 2021 [37] | APPNL-F/NL-F | C57BL/6J | RIKEN Brain Science Institute, Saitama, Japan | 6, 9, 12, 15, 17, and 20 months | Male | 55 AD/41 WT | Left eye |
| Vandenabeele et al., 2021 [39] | APPNL-G-F | Not specify | Jackson Laboratories, USA | 3, 6, 9, 12, and 18 months | Male (18 months) | Not specify | Not specify |
| Female (all time points) | |||||||
| Georgevsky et al., 2019 [41] | APP/PS1 | Littermates | Australian phonemics facility (ANU Canberra) | 3, 6, 9, and 12 months | Male and female (50:50) | 70 (no more data) | Both |
| Harper et al., 2020 [42] | APP/PS1 | Littermates | Jackson Laboratories, USA | 10 to 24 months | AD (17F/7M)/WT (9F/6M) | 24 AD/15 WT | Both |
| Chiquita et al., 2019 [44] | 3xTg-AD | C57BL/6J | Not specify | 4, 8, 12, and 16 months | Male | 21 AD/22 WT | Both |
| Song et al., 2020 [45] | 3xTg-AD | B6129SF2/J | Jackson Laboratories, USA | 15–16 months | Female | Not specify | Ex vivo |
| Gardner et al., 2020 [46] | 3xTg-AD | C57BL/6J | Not specify | 2, 4, 7, and 10 months | AD (4F/16M)/WT (12M) | 20 AD/12 WT | Left eye |
| Ferreira et al., 2021 [47] | 3xTg-AD | C57BL6/129S | Not specify | 1, 2, 3, 4 months | Male | 57 AD/57 WT | Both |
| Guimarães et al., 2022 [48] | 3xTg-AD | C57BL6/129S | Not specify | 1, 2, 3, 4, 8, 12, and 16 months | Male | 60 AD/57 WT | Both |
| Batista et al., 2023 [49] | 3xTg-AD | C57BL6/129S | Not specify | 1, 2, 3, 4, 8, 12, and 16 months | Male | 57 AD/57 WT | Both |
| Lim et al., 2020 [51] | 5xFAD | Littermates | Jackson Laboratories, USA | 6, 12, and 17 months | Not specify | 32 AD/38 WT | Not specify |
| Kim et al., 2021 [22] | 5xFAD | B6SJLF1/J | Jackson Laboratories, USA | 6 months | Female | 5 AD/6 WT | One random |
| Matei et al., 2022 [52] | 5xFAD | C57BL/6J | Jackson Laboratories, USA | 3 months | Male | 16 AD/16 WT | One random |
3.2. Selection of the Analysed Eye
3.3. OCT Model
3.4. Image Acquisition Protocol and Retinal Sector Analysed
- (i)
- In the study by Buccarello et al., a two-dimensional scanning protocol (B-scan) with a circular diameter of 550 μm around the optic nerve head (ONH) was utilised [35].
- (ii)
- Chiquita and colleagues employed two scanning protocols. One was a circular scanning protocol around the ONH, and the other was a linear scanning protocol consisting of one scan centred in the middle of the optic nerve head, along with six superior and six inferior scans [44].
- (iii)
- Ferreira and collaborators used a scanning protocol with 512 B-scans, each containing 512 A-scans of 1024 pixels in length. All scans were performed horizontally centred with the optical disc and vertically positioned above it [47].
- (iv)
- Guimarães et al. performed their analysis in a region located above the optic nerve. Their protocol involved 512 B-scans, with regularly spaced B-scans selected at intervals of five B-scans. This resulted in a total of 14,730 B-scans (512 × 1024 pixels). Each selected B-scan was cut out to obtain a region of 512 × 512 pixels, centred on the retina, with its location determined automatically [48].
- (v)
- In the work by Batista et al., each data volume consisted of 512 B-scans, with each B-scan composed of 512 discrete samples of 1024 A-scans. The optic disc served as the reference point to select the retinal region in the image, which was aligned horizontally just above it [49].
- (i)
- Salobrar-García et al. analysed the retina by centring the optic nerve head, placing it in the centre of the scans with 61 horizontal scans. The thickness of the nerve fibre layer was analysed using an axonal ring scan around the optic nerve head. Additionally, these authors placed a +25 dp lens in front of the OCT camera and a contact lens that covered the mouse cornea, creating a uniform refractive surface [37].
- (ii)
- Matei et al. analysed the retina by focusing on two adjacent regions to the optic nerve head: the nasal and temporal regions. In each region, 13 scans were performed, including one in the middle of the optic nerve head, six superior scans, and six inferior scans [52].
| Authors and Year | OCT Device | Retina Protocol Scans | Software Segmentation and Retinal Layer/Complex Analysed | Retinal Findings | |
|---|---|---|---|---|---|
| Retinal Thinning | Retinal Thickening | ||||
| Buccarello et al., 2017 [35] | Phoenix Micron IV Image-guided OCT | Circular scan (diameter of 550 μm) around the ONH. | Insight software (Phoenix Research Laboratories). NFL/GCL, IRL, ORL, total retinal thickness. | RNFL-GCC (4 months) | - |
| Salobrar-García et al., 2021 [37] | SD-OCT Spectralis | 61 horizontal scans centred in the ONH. RNFL Circular scan around the ONH | Heidelberg Eye Explorer software v6.13 Total retinal thickness and RNFL | Total retinal thickness (6, 9, 12, 15, and 20 months), RNFL (6, 20 months) | Total retinal thickness and RNFL (17 months) |
| Vandenabeele et al., 2021 [39] | Envisu R2200 spectral domain OCT | 16 equidistant points from the optic nerve. | InVivoVue Diver 3.0.8 Total retinal thickness | 12 until 18 months ONL | - |
| Georgevsky et al., 2019 [41] | OCT from Wasatch Photonics | 3 × 3 mm square centred in the ONH. Five scans (spaced 125 μm apart), centred in the ONH and two superior and two inferior parallel scans. | A modified segmentation algorithm based on graph theory Inner retina (GCL-INL) Outer retina (OPL-RPE) | Inner retinal thickness at 9 months and outer retinal thickness at 12 months | - |
| Harper et al., 2020 [42] | High-resolution polarisation-sensitive OCT (PS-OCT) system | Five B-scans. Ring scans centred in the ONH at 500 µm and 900 µm of diameter. | Algorithm by Augustin et al., 2018 [55]. Total retinal thickness, inner retinal thickness, and outer retinal thickness of the ring surrounding the ONH | No changes | - |
| Chiquita et al., 2019 [44] | Phoenix Micron IV Image-guided OCT | Circular scan (diameter of 550 μm) around the ONH. One scan centred in ONH, six superior and six inferior scans | Insight software (Phoenix Research Laboratories). GCL + IPL, INL + OPL ONL, IS + OS, total retinal thickness | Total retinal thickness 4–16 months GCL + IPL, INL + OPL, at 4, 8, 12, and 16 months | ONL |
| Song et al., 2020 [45] | Angle-resolved low-coherence interferometry (a/LCI) | Ex vivo acquisition. Aligned using the ONH. Eight scans, spaced 500 µm apart along the vertical and horizontal axis. | Definition described by Srinivasan et al., 2014 [56] RNFL, OPL, and RPE | RNFL (15–16 months) | - |
| Gardner et al., 2020 [46] | Specific subtype of angular resolution OCT, termed scatter angular resolution OCT (SAR-OCT) | Four 1.3 × 1.3 mm2 (512 × 512 A-scans) square sections (nasal, superior, temporal, and inferior), with the ONH at one corner of the frontal view | Previously established algorithm Two regions: the superficial layers consisting of NFL, GCL, and IPL, and the ONL | Total retinal thickness (3, 5, 8 and 11 months) Inner layers (NFL + GCL + IPL) at 2 months and 7 months and ONL at 2 months | |
| Ferreira et al., 2021 [47] | Phoenix Micron IV Image-guided OCT | 512 B-scans, each containing 512 A-scans of 1024 pixels in length. Centred in the ONH | Convolutional neural network (FCNN) following a U-type architecture RNFL-GCL, IPL, INL, OPL, ONL, IS, OS, and RPE. | Total retinal thickness (1, 2, 3 and 4 months) | RNFL-GCC |
| Guimarães et al., 2022 [48] | Phoenix Micron IV Image-guided OCT | 512 B-scans, each containing 512 A-scans of 1024 pixels in length. Centred in the ONH | Not segmentation software Total retinal thickness | Not specified | Not specified |
| Batista et al., 2023 [49] | Phoenix Micron IV Image-guided OCT | 512 B-scans, each containing 512 A-scans of 1024 pixels in length. Centred in the ONH | Deep learning approach based in Convolutional neural network (FCNN) following a U-type architecture RNFL-GCL, IPL, INL, OPL, ONL, IS, OS, and RPE. | Total retinal thickness (1, 2, 3, 4, 12 and 16 months) RNFL-GCL, IPL, ONL OS | OPL, IS and RPE |
| Lim et al., 2020 [51] | Envisu R2200 spectral domain OCT | Retinal volumes (1.4 × 1.4 × 1.57 mm) centred ONH, acquiring them with 200 horizontal B-scans, each composed of 1000 A-scans. | FIJI analysis software GCC (from RNFL to the IPL), total retinal thickness. | RNFL-GCC (6, 12 and 17 months) | IPL (6 months) |
| Kim et al., 2021 [22] | Custom-built spectral-domain OCT | In four quadrants of the retina, starting from ONH to fixate the central point (first dorsal and ventral, followed by nasal and temporal). Each OCT volume 4 × 600 × 600 A-scans | Manual segmentation RNFL, inner retina, outer retina, and total retinal thickness | Total retinal thickness, NFL, Innes retina thickness and Outer Retina thickness (6 months) | - |
| Matei et al., 2022 [52] | OCT SD-OCT Spectralis | 13 scans, one centred in ONH, six superior and six inferior scans. | Heidelberg Eye Explorer software (1.9.10.0) Total retinal thickness | No changes | No changes |
3.5. Layer Segmentation and Software Employed
3.6. Retinal Findings in the Retina of Different Murine Models of AD
- (i)
- The work by Chiquita et al. reported a statistically significant reduction in retinal thickness with respect to the WT group in all layers analysed except for the ONL, in which the authors showed a statistically significant increase in thickness in the 3xTgAD group, with respect to the WT group [44].
- (ii)
- Song et al. also found a statistically significant thinning of RNFL in AD mice compared to WT group. However, no statistically significant differences were observed in either OPL or RPE [45].
- (iii)
- Gardner and colleagues observed statistically significant differences in total retinal thickness, as well as in the thickness of the most inner layers (NFL + GCL + IPL) and the ONL. Furthermore, the authors noted that the most consistent changes in thickness between the AD and control groups were observed in the central regions of the retina [46].
- (iv)
- Ferreira et al. conducted a study on the aging 3xTgAD, comparing it with a normative database of the C57BL6/129S model aged 1 to 4 months. The authors concluded that there were no statistically significant differences between the right and left eyes. However, when comparing the groups, a significant decrease in retinal thickness was observed, except in the RNFL-GCL complex, where the 3xTg-AD mice showed thickening compared to the WT group. Additionally, no statistically significant differences were observed in the ONL. The authors further described that the most significant differences in thickness were found in the IPL, ONL, OPL, IS, RPE, and the total retina [47].
- (v)
- Although Guimarães et al. did not provide detailed information on the specific alterations in retinal thickness, their findings clearly indicated that retinal aging exhibits distinct patterns between the AD group and the WT group [48].
- (vi)
- Batista et al. conducted a longitudinal study to analyse retinal thickness in the 3xTg-AD transgenic model compared to the WT group. The mean values of total retinal thickness showed no statistically significant differences between the left and right eyes, except for 3xTg-AD mice at 1 month of age. Additionally, they found an increase in the total retinal thickness near the optic disc (from superior to inferior) in both eyes [49].
4. Discussion
4.1. Differences among Animal Models of AD Used in OCT Studies
4.2. Differences among the Variety of OCT Devices Used in the Studies of AD Animal Models
4.3. Layer Segmentation and Software Employed in OCT Studies
4.4. Retinal Findings in OCT Studies
4.5. Future Perspectives Needed for the Use of OCT Retinal Analysis in Murine Models of AD
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organisation Dementia. Available online: https://www.who.int/news-room/fact-sheets/detail/dementia (accessed on 11 August 2021).
- Selkoe, D.J. Normal and Abnormal Biology of the Beta-Amyloid Precursor Protein. Annu. Rev. Neurosci. 2003, 17, 489–517. [Google Scholar] [CrossRef]
- Walker, L.C. Aβ Plaques. Free Neuropathol. 2020, 1, 31. [Google Scholar] [CrossRef]
- Pimplikar, S.W. Reassessing the Amyloid Cascade Hypothesis of Alzheimer’s Disease. Int. J. Biochem. Cell Biol. 2009, 41, 1261–1268. [Google Scholar] [CrossRef]
- Dubois, B.; Hampel, H.; Feldman, H.H.; Scheltens, P.; Aisen, P.; Andrieu, S.; Bakardjian, H.; Benali, H.; Bertram, L.; Blennow, K.; et al. Preclinical Alzheimer’s Disease: Definition, Natural History, and Diagnostic Criteria. Alzheimer’s Dement. 2016, 12, 292–323. [Google Scholar] [CrossRef]
- Villemagne, V.L.; Burnham, S.; Bourgeat, P.; Brown, B.; Ellis, K.A.; Salvado, O.; Szoeke, C.; Macaulay, S.L.; Martins, R.; Maruff, P.; et al. Amyloid β Deposition, Neurodegeneration, and Cognitive Decline in Sporadic Alzheimer’s Disease: A Prospective Cohort Study. Lancet Neurol. 2013, 12, 357–367. [Google Scholar] [CrossRef] [PubMed]
- Peters, L.L.; Robledo, R.F.; Bult, C.J.; Churchill, G.A.; Paigen, B.J.; Svenson, K.L. The Mouse as a Model for Human Biology: A Resource Guide for Complex Trait Analysis. Nat. Rev. Genet. 2007, 8, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Sasaguri, H.; Hashimoto, S.; Watamura, N.; Sato, K.; Takamura, R.; Nagata, K.; Tsubuki, S.; Ohshima, T.; Yoshiki, A.; Sato, K.; et al. Recent Advances in the Modeling of Alzheimer’s Disease. Front. Neurosci. 2022, 16, 807473. [Google Scholar] [CrossRef] [PubMed]
- Volland, S.; Esteve-Rudd, J.; Hoo, J.; Yee, C.; Williams, D.S. A Comparison of Some Organizational Characteristics of the Mouse Central Retina and the Human Macula. PLoS ONE 2015, 10, e0125631. [Google Scholar] [CrossRef]
- Sterratt, D.C.; Lyngholm, D.; Willshaw, D.J.; Thompson, I.D. Standard Anatomical and Visual Space for the Mouse Retina: Computational Reconstruction and Transformation of Flattened Retinae with the Retistruct Package. PLoS Comput. Biol. 2013, 9, e1002921. [Google Scholar] [CrossRef]
- Carter-Dawson, L.D.; Lavail, M.M. Rods and Cones in the Mouse Retina. I. Structural Analysis Using Light and Electron Microscopy. J. Comp. Neurol. 1979, 188, 245–262. [Google Scholar] [CrossRef]
- Jeon, C.-J.; Strettoi, E.; Masland, R.H. The Major Cell Populations of the Mouse Retina. J. Neurosci. 1998, 18, 8936–8946. [Google Scholar] [CrossRef]
- Zhang, J.; Shi, L.; Shen, Y. The Retina: A Window in Which to View the Pathogenesis of Alzheimer’s Disease. Ageing Res. Rev. 2022, 77, 101590. [Google Scholar] [CrossRef] [PubMed]
- Salobrar-García, E.; de Hoz, R.; Ramírez, A.I.; López-Cuenca, I.; Rojas, P.; Vazirani, R.; Amarante, C.; Yubero, R.; Gil, P.; Pinazo-Durán, M.D.; et al. Changes in Visual Function and Retinal Structure in the Progression of Alzheimer’s Disease. PLoS ONE 2019, 14, e0220535. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Martin, E.S.; Rojas, B.; Ramirez, A.I.; de Hoz, R.; Salazar, J.J.; Yubero, R.; Gil, P.; Triviño, A.; Ramirez, J.M. Macular Thickness as a Potential Biomarker of Mild Alzheimer’s Disease. Ophthalmology 2014, 121, 1149–1151.e3. [Google Scholar] [CrossRef] [PubMed]
- Lad, E.M.; Mukherjee, D.; Stinnett, S.S.; Cousins, S.W.; Potter, G.G.; Burke, J.R.; Farsiu, S.; Whitson, H.E. Evaluation of Inner Retinal Layers as Biomarkers in Mild Cognitive Impairment to Moderate Alzheimer’s Disease. PLoS ONE 2018, 13, e0192646. [Google Scholar] [CrossRef]
- Trebbastoni, A.; D’Antonio, F.; Bruscolini, A.; Marcelli, M.; Cecere, M.; Campanelli, A.; Imbriano, L.; de Lena, C.; Gharbiya, M. Retinal Nerve Fibre Layer Thickness Changes in Alzheimer’s Disease: Results from a 12-Month Prospective Case Series. Neurosci. Lett. 2016, 629, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Majeed, A.; Marwick, B.; Yu, H.; Fadavi, H.; Tavakoli, M. Ophthalmic Biomarkers for Alzheimer’s Disease: A Review. Front. Aging Neurosci. 2021, 13, 574. [Google Scholar] [CrossRef]
- Chan, V.T.T.; Sun, Z.; Tang, S.; Chen, L.J.; Wong, A.; Tham, C.C.; Wong, T.Y.; Chen, C.; Ikram, M.K.; Whitson, H.E.; et al. Spectral-Domain OCT Measurements in Alzheimer’s Disease: A Systematic Review and Meta-Analysis. Ophthalmology 2019, 126, 497–510. [Google Scholar] [CrossRef]
- Ngolab, J.; Honma, P.; Rissman, R.A. Reflections on the Utility of the Retina as a Biomarker for Alzheimer’s Disease: A Literature Review. Neurol. Ther. 2019, 8, 57–72. [Google Scholar] [CrossRef]
- Gupta, V.B.; Chitranshi, N.; den Haan, J.; Mirzaei, M.; You, Y.; Lim, J.K.H.; Basavarajappa, D.; Godinez, A.; Di Angelantonio, S.; Sachdev, P.; et al. Retinal Changes in Alzheimer’s Disease— Integrated Prospects of Imaging, Functional and Molecular Advances. Prog. Retin. Eye Res. 2021, 82, 100899. [Google Scholar] [CrossRef]
- Kim, T.H.; Son, T.; Klatt, D.; Yao, X. Concurrent OCT and OCT Angiography of Retinal Neurovascular Degeneration in the 5XFAD Alzheimer’s Disease Mice. Neurophotonics 2021, 8, 035002. [Google Scholar] [CrossRef]
- Cabrera DeBuc, D.; Somfai, G.M.; Ranganathan, S.; Tátrai, E.; Ferencz, M.; Puliafito, C.A. Reliability and Reproducibility of Macular Segmentation Using a Custom-Built Optical Coherence Tomography Retinal Image Analysis Software. J. Biomed. Opt. 2009, 14, 064023. [Google Scholar] [CrossRef]
- Byun, M.S.; Park, S.W.; Lee, J.H.; Yi, D.; Jeon, S.Y.; Choi, H.J.; Joung, H.; Ghim, U.H.; Park, U.C.; Kim, Y.K.; et al. Association of Retinal Changes with Alzheimer Disease Neuroimaging Biomarkers in Cognitively Normal Individuals. JAMA Ophthalmol. 2021, 139, 548. [Google Scholar] [CrossRef] [PubMed]
- Suh, A.; Ong, J.; Kamran, S.A.; Waisberg, E.; Paladugu, P.; Zaman, N.; Sarker, P.; Tavakkoli, A.; Lee, A.G. Retina Oculomics in Neurodegenerative Disease. Ann. Biomed. Eng. 2023, 51, 2708–2721. [Google Scholar] [CrossRef]
- Kim, B.R.; Yoo, T.K.; Kim, H.K.; Ryu, I.H.; Kim, J.K.; Lee, I.S.; Kim, J.S.; Shin, D.H.; Kim, Y.S.; Kim, B.T. Oculomics for Sarcopenia Prediction: A Machine Learning Approach toward Predictive, Preventive, and Personalized Medicine. EPMA J. 2022, 13, 367–382. [Google Scholar] [CrossRef] [PubMed]
- Jankowsky, J.L.; Zheng, H. Practical Considerations for Choosing a Mouse Model of Alzheimer’s Disease. Mol. Neurodegener. 2017, 12, 89. [Google Scholar] [CrossRef]
- Ferguson, L.R.; Grover, S.; Dominguez, J.M.; Balaiya, S.; Chalam, K.V. Retinal Thickness Measurement Obtained with Spectral Domain Optical Coherence Tomography Assisted Optical Biopsy Accurately Correlates with Ex Vivo Histology. PLoS ONE 2014, 9, e111203. [Google Scholar] [CrossRef]
- Fischer, M.D.; Huber, G.; Beck, S.C.; Tanimoto, N.; Muehlfriedel, R.; Fahl, E.; Grimm, C.; Wenzel, A.; Remé, C.E.; van de Pavert, S.A.; et al. Noninvasive, in Vivo Assessment of Mouse Retinal Structure Using Optical Coherence Tomography. PLoS ONE 2009, 4, e7507. [Google Scholar] [CrossRef]
- Gloesmann, M.; Hermann, B.; Schubert, C.; Sattmann, H.; Ahnelt, P.K.; Drexler, W. Histologic Correlation of Pig Retina Radial Stratification with Ultrahigh-Resolution Optical Coherence Tomography. Investig. Ophthalmol. Vis. Sci. 2003, 44, 1696–1703. [Google Scholar] [CrossRef] [PubMed]
- Strouthidis, N.G.; Grimm, J.; Williams, G.A.; Cull, G.A.; Wilson, D.J.; Burgoyne, C.F. A Comparison of Optic Nerve Head Morphology Viewed by Spectral Domain Optical Coherence Tomography and by Serial Histology. Investig. Ophthalmol. Vis. Sci. 2010, 51, 1464. [Google Scholar] [CrossRef] [PubMed]
- Ochakovski, G.A.; Fischer, M.D. Phenotyping of Mouse Models with OCT. In Methods in Molecular Biology; Humana, New York, NY, USA, 2019; Volume 1834, pp. 285–291, ISBN 9781493986699.
- Yokoyama, M.; Kobayashi, H.; Tatsumi, L.; Tomita, T. Mouse Models of Alzheimer’s Disease. Front. Mol. Neurosci. 2022, 15, 912995. [Google Scholar] [CrossRef]
- Chishti, M.A.; Yang, D.S.; Janus, C.; Phinney, A.L.; Horne, P.; Pearson, J.; Strome, R.; Zuker, N.; Loukides, J.; French, J.; et al. Early-Onset Amyloid Deposition and Cognitive Deficits in Transgenic Mice Expressing a Double Mutant Form of Amyloid Precursor Protein 695. J. Biol. Chem. 2001, 276, 21562–21570. [Google Scholar] [CrossRef]
- Buccarello, L.; Sclip, A.; Sacchi, M.; Castaldo, A.M.; Bertani, I.; ReCecconi, A.; Maestroni, S.; Zerbini, G.; Nucci, P.; Borsello, T. The C-Jun N-Terminal Kinase Plays a Key Role in Ocular Degenerative Changes in a Mouse Model of Alzheimer Disease Suggesting a Correlation between Ocular and Brain Pathologies. Oncotarget 2017, 8, 83038–83051. [Google Scholar] [CrossRef]
- Saito, T.; Saido, T.C. Neuroinflammation in Mouse Models of Alzheimer’s Disease. Clin. Exp. Neuroimmunol. 2018, 9, 211–218. [Google Scholar] [CrossRef]
- Salobrar-García, E.; López-Cuenca, I.; Sánchez-Puebla, L.; de Hoz, R.; Fernández-Albarral, J.A.; Ramírez, A.I.; Bravo-Ferrer, I.; Medina, V.; Moro, M.A.; Saido, T.C.; et al. Retinal Thickness Changes Over Time in a Murine AD Model APPNL-F/NL-F. Front. Aging Neurosci. 2021, 12, 625642. [Google Scholar] [CrossRef]
- Saito, T.; Matsuba, Y.; Mihira, N.; Takano, J.; Nilsson, P.; Itohara, S.; Iwata, N.; Saido, T.C. Single App Knock-in Mouse Models of Alzheimer’s Disease. Nat. Neurosci. 2014, 17, 661–663. [Google Scholar] [CrossRef]
- Vandenabeele, M.; Veys, L.; Lemmens, S.; Hadoux, X.; Gelders, G.; Masin, L.; Serneels, L.; Theunis, J.; Saito, T.; Saido, T.C.; et al. The AppNL-G-F Mouse Retina Is a Site for Preclinical Alzheimer’s Disease Diagnosis and Research. Acta Neuropathol. Commun. 2021, 9, 6. [Google Scholar] [CrossRef] [PubMed]
- Jankowsky, J.L.; Fadale, D.J.; Anderson, J.; Xu, G.M.; Gonzales, V.; Jenkins, N.A.; Copeland, N.G.; Lee, M.K.; Younkin, L.H.; Wagner, S.L.; et al. Mutant Presenilins Specifically Elevate the Levels of the 42 Residue β-Amyloid Peptide In Vivo: Evidence for Augmentation of a 42-Specific γ Secretase. Hum. Mol. Genet. 2004, 13, 159–170. [Google Scholar] [CrossRef]
- Georgevsky, D.; Retsas, S.; Raoufi, N.; Shimoni, O.; Golzan, S.M. A Longitudinal Assessment of Retinal Function and Structure in the APP/PS1 Transgenic Mouse Model of Alzheimer’s Disease. Transl. Neurodegener. 2019, 8, 30. [Google Scholar] [CrossRef] [PubMed]
- Harper, D.J.; Augustin, M.; Lichtenegger, A.; Gesperger, J.; Himmel, T.; Muck, M.; Merkle, C.W.; Eugui, P.; Kummer, S.; Woehrer, A.; et al. Retinal Analysis of a Mouse Model of Alzheimer’s Disease with Multicontrast Optical Coherence Tomography. Neurophotonics 2020, 7, 015006. [Google Scholar] [CrossRef] [PubMed]
- Oddo, S.; Caccamo, A.; Shepherd, J.D.; Murphy, M.P.P.; Golde, T.E.; Kayed, R.; Metherate, R.; Mattson, M.P.; Akbari, Y.; LaFerla, F.M. Triple-Transgenic Model of Alzheimer’s Disease with Plaques and Tangles: Intracellular Aβ and Synaptic Dysfunction. Neuron 2003, 39, 409–421. [Google Scholar] [CrossRef]
- Chiquita, S.; Campos, E.J.; Castelhano, J.; Ribeiro, M.; Sereno, J.; Moreira, P.I.; Castelo-Branco, M.; Ambrósio, A.F. Retinal Thinning of Inner Sub-Layers Is Associated with Cortical Atrophy in a Mouse Model of Alzheimer’s Disease: A Longitudinal Multimodal in Vivo Study. Alzheimer’s Res. Ther. 2019, 11, 90. [Google Scholar] [CrossRef] [PubMed]
- Song, G.; Steelman, Z.A.; Finkelstein, S.; Yang, Z.; Martin, L.; Chu, K.K.; Farsiu, S.; Arshavsky, V.Y.; Wax, A. Multimodal Coherent Imaging of Retinal Biomarkers of Alzheimer’s Disease in a Mouse Model. Sci. Rep. 2020, 10, 7912. [Google Scholar] [CrossRef] [PubMed]
- Gardner, M.R.; Baruah, V.; Vargas, G.; Motamedi, M.; Milner, T.E.; Rylander, H.G. Scattering Angle Resolved Optical Coherence Tomography Detects Early Changes in 3xTg Alzheimer’s Disease Mouse Model. Transl. Vis. Sci. Technol. 2020, 9, 18. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, H.; Martins, J.; Moreira, P.I.; Ambrósio, A.F.; Castelo-Branco, M.; Serranho, P.; Bernardes, R. Longitudinal Normative OCT Retinal Thickness Data for Wild-Type Mice, and Characterization of Changes in the 3×Tg-AD Mice Model of Alzheimer’s Disease. Aging 2021, 13, 9433. [Google Scholar] [CrossRef]
- Guimarães, P.; Serranho, P.; Martins, J.; Moreira, P.I.; Ambrósio, A.F.; Castelo-Branco, M.; Bernardes, R. Retinal Aging in 3× Tg-AD Mice Model of Alzheimer’s Disease. Front. Aging Neurosci. 2022, 14, 832195. [Google Scholar] [CrossRef] [PubMed]
- Batista, A.; Guimarães, P.; Martins, J.; Moreira, P.I.; Ambrósio, A.F.; Castelo-Branco, M.; Serranho, P.; Bernardes, R. Normative Mice Retinal Thickness: 16-Month Longitudinal Characterization of Wild-Type Mice and Changes in a Model of Alzheimer’s Disease. Front. Aging Neurosci. 2023, 15, 232. [Google Scholar] [CrossRef]
- Oakley, H.; Cole, S.L.; Logan, S.; Maus, E.; Shao, P.; Craft, J.; Guillozet-Bongaarts, A.; Ohno, M.; Disterhoft, J.; Van Eldik, L.; et al. Intraneuronal β-Amyloid Aggregates, Neurodegeneration, and Neuron Loss in Transgenic Mice with Five Familial Alzheimer’s Disease Mutations: Potential Factors in Amyloid Plaque Formation. J. Neurosci. 2006, 26, 10129. [Google Scholar] [CrossRef]
- Lim, J.K.H.; Li, Q.X.; He, Z.; Vingrys, A.J.; Chinnery, H.R.; Mullen, J.; Bui, B.V.; Nguyen, C.T.O. Retinal Functional and Structural Changes in the 5xFAD Mouse Model of Alzheimer’s Disease. Front. Neurosci. 2020, 14, 862. [Google Scholar] [CrossRef]
- Matei, N.; Leahy, S.; Blair, N.P.; Burford, J.; Rahimi, M.; Shahidi, M. Retinal Vascular Physiology Biomarkers in a 5XFAD Mouse Model of Alzheimer’s Disease. Cells 2022, 11, 2413. [Google Scholar] [CrossRef]
- Fialová, S.; Augustin, M.; Glösmann, M.; Himmel, T.; Rauscher, S.; Gröger, M.; Pircher, M.; Hitzenberger, C.K.; Baumann, B. Polarization Properties of Single Layers in the Posterior Eyes of Mice and Rats Investigated Using High Resolution Polarization Sensitive Optical Coherence Tomography. Biomed. Opt. Express 2016, 7, 1479. [Google Scholar] [CrossRef]
- Gardner, M.R.; Katta, N.; Rahman, A.S.; Rylander, H.G.; Milner, T.E. Design Considerations for Murine Retinal Imaging Using Scattering Angle Resolved Optical Coherence Tomography. Appl. Sci. 2018, 8, 2159. [Google Scholar] [CrossRef]
- Augustin, M.; Harper, D.J.; Merkle, C.W.; Hitzenberger, C.K.; Baumann, B. Segmentation of Retinal Layers in OCT Images of the Mouse Eye Utilizing Polarization Contrast. In Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Cham, Switzerland, 2018; Volume 11039 LNCS, pp. 310–318. [Google Scholar] [CrossRef]
- Srinivasan, P.P.; Heflin, S.J.; Izatt, J.A.; Arshavsky, V.Y.; Farsiu, S. Automatic Segmentation of up to Ten Layer Boundaries in SD-OCT Images of the Mouse Retina with and without Missing Layers Due to Pathology. Biomed. Opt. Express 2014, 5, 348. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Cham, Switzerland, 2015; Volume 9351, pp. 234–241. [Google Scholar] [CrossRef]
- Chiu, S.J.; Li, X.T.; Nicholas, P.; Toth, C.A.; Izatt, J.A.; Farsiu, S. Automatic Segmentation of Seven Retinal Layers in SDOCT Images Congruent with Expert Manual Segmentation. Opt. Express 2010, 18, 19413. [Google Scholar] [CrossRef]
- Mitchell, S.J.; Scheibye-Knudsen, M.; Longo, D.L.; De Cabo, R. Animal Models of Aging Research: Implications for Human Aging and Age-Related Diseases. Annu. Rev. Anim. Biosci. 2015, 3, 283–303. [Google Scholar] [CrossRef]
- Dutta, S.; Sengupta, P. Men and Mice: Relating Their Ages. Life Sci. 2016, 152, 244–248. [Google Scholar] [CrossRef]
- Sasaguri, H.; Nilsson, P.; Hashimoto, S.; Nagata, K.; Saito, T.; De Strooper, B.; Hardy, J.; Vassar, R.; Winblad, B.; Saido, T.C. APP Mouse Models for Alzheimer’s Disease Preclinical Studies. EMBO J. 2017, 36, 2473–2487. [Google Scholar] [CrossRef]
- Kwart, D.; Gregg, A.; Scheckel, C.; Murphy, E.; Paquet, D.; Duffield, M.; Fak, J.; Olsen, O.; Darnell, R.; Tessier-Lavigne, M. A Large Panel of Isogenic APP and PSEN1 Mutant Human IPSC Neurons Reveals Shared Endosomal Abnormalities Mediated by APP β-CTFs, Not Aβ. Neuron 2019, 104, 256–270.e5. [Google Scholar] [CrossRef]
- Oddo, S.; Caccamo, A.; Kitazawa, M.; Tseng, B.P.; LaFerla, F.M. Amyloid Deposition Precedes Tangle Formation in a Triple Transgenic Model of Alzheimer’s Disease. Neurobiol. Aging 2003, 24, 1063–1070. [Google Scholar] [CrossRef]
- Jawhar, S.; Trawicka, A.; Jenneckens, C.; Bayer, T.A.; Wirths, O. Motor Deficits, Neuron Loss, and Reduced Anxiety Coinciding with Axonal Degeneration and Intraneuronal Aβ Aggregation in the 5XFAD Mouse Model of Alzheimer’s Disease. Neurobiol. Aging 2012, 33, 196.e29–196.e40. [Google Scholar] [CrossRef]
- Eimer, W.A.; Vassar, R. Neuron Loss in the 5XFAD Mouse Model of Alzheimer’s Disease Correlates with Intraneuronal Aβ42 Accumulation and Caspase-3 Activation. Mol. Neurodegener. 2013, 8, 2. [Google Scholar] [CrossRef]
- Cruz Hernández, J.C.; Bracko, O.; Kersbergen, C.J.; Muse, V.; Haft-Javaherian, M.; Berg, M.; Park, L.; Vinarcsik, L.K.; Ivasyk, I.; Rivera, D.A.; et al. Neutrophil Adhesion in Brain Capillaries Reduces Cortical Blood Flow and Impairs Memory Function in Alzheimer’s Disease Mouse Models. Nat. Neurosci. 2019, 22, 413–420. [Google Scholar] [CrossRef]
- Majumdar, S.; Klatt, D. Longitudinal Study of Sub-Regional Cerebral Viscoelastic Properties of 5XFAD Alzheimer’s Disease Mice Using Multifrequency MR Elastography. Magn. Reson. Med. 2021, 86, 405–414. [Google Scholar] [CrossRef]
- Lanoiselée, H.M.; Nicolas, G.; Wallon, D.; Rovelet-Lecrux, A.; Lacour, M.; Rousseau, S.; Richard, A.C.; Pasquier, F.; Rollin-Sillaire, A.; Martinaud, O.; et al. APP, PSEN1, and PSEN2 Mutations in Early-Onset Alzheimer Disease: A Genetic Screening Study of Familial and Sporadic Cases. PLoS Med. 2017, 14, e1002270. [Google Scholar] [CrossRef]
- Citron, M.; Oltersdorf, T.; Haass, C.; McConlogue, L.; Hung, A.Y.; Seubert, P.; Vigo-Pelfrey, C.; Lieberburg, I.; Selkoe, D.J. Mutation of the Beta-Amyloid Precursor Protein in Familial Alzheimer’s Disease Increases Beta-Protein Production. Nature 1992, 360, 672–674. [Google Scholar] [CrossRef]
- Lichtenthaler, S.F.; Wang, R.; Grimm, H.; Uljon, S.N.; Masters, C.L.; Beyreuther, K. Mechanism of the Cleavage Specificity of Alzheimer’s Disease Gamma-Secretase Identified by Phenylalanine-Scanning Mutagenesis of the Transmembrane Domain of the Amyloid Precursor Protein. Proc. Natl. Acad. Sci. USA 1999, 96, 3053–3058. [Google Scholar] [CrossRef]
- Nilsberth, C.; Westlind-Danielsson, A.; Eckman, C.B.; Condron, M.M.; Axelman, K.; Forsell, C.; Stenh, C.; Luthman, J.; Teplow, D.B.; Younkin, S.G.; et al. The “Arctic” APP Mutation (E693G) Causes Alzheimer’s Disease by Enhanced Abeta Protofibril Formation. Nat. Neurosci. 2001, 4, 887–893. [Google Scholar] [CrossRef]
- Liu, X.; Shen, M.; Huang, S.; Leng, L.; Zhu, D.; Lu, F. Repeatability and Reproducibility of Eight Macular Intra-Retinal Layer Thicknesses Determined by an Automated Segmentation Algorithm Using Two SD-OCT Instruments. PLoS ONE 2014, 9, e87996. [Google Scholar] [CrossRef] [PubMed]
- Terry, L.; Cassels, N.; Lu, K.; Acton, J.H.; Margrain, T.H.; North, R.V.; Fergusson, J.; White, N.; Wood, A. Automated Retinal Layer Segmentation Using Spectral Domain Optical Coherence Tomography: Evaluation of Inter-Session Repeatability and Agreement between Devices. PLoS ONE 2016, 11, e162001. [Google Scholar] [CrossRef] [PubMed]
- Matlach, J.; Wagner, M.; Malzahn, U.; Göbel, W. Repeatability of Peripapillary Retinal Nerve Fiber Layer and Inner Retinal Thickness Among Two Spectral Domain Optical Coherence Tomography Devices. Investig. Ophthalmol. Vis. Sci. 2014, 55, 6536–6546. [Google Scholar] [CrossRef] [PubMed]
- Töteberg-Harms, M.; Sturm, V.; Knecht, P.B.; Funk, J.; Menke, M.N. Repeatability of Nerve Fiber Layer Thickness Measurements in Patients with Glaucoma and without Glaucoma Using Spectral-Domain and Time-Domain OCT. Graefes Arch. Clin. Exp. Ophthalmol. 2012, 250, 279–287. [Google Scholar] [CrossRef]
- Domínguez-Vicent, A.; Brautaset, R.; Venkataraman, A.P. Repeatability of Quantitative Measurements of Retinal Layers with SD-OCT and Agreement between Vertical and Horizontal Scan Protocols in Healthy Eyes. PLoS ONE 2019, 14, e221466. [Google Scholar] [CrossRef] [PubMed]
- Ctori, I.; Huntjens, B. Repeatability of Foveal Measurements Using Spectralis Optical Coherence Tomography Segmentation Software. PLoS ONE 2015, 10, e0129005. [Google Scholar] [CrossRef]
- Sohn, E.H.; Van Dijk, H.W.; Jiao, C.; Kok, P.H.B.; Jeong, W.; Demirkaya, N.; Garmager, A.; Wit, F.; Kucukevcilioglu, M.; Van Velthoven, M.E.J.; et al. Retinal Neurodegeneration May Precede Microvascular Changes Characteristic of Diabetic Retinopathy in Diabetes Mellitus. Proc. Natl. Acad. Sci. USA 2016, 113, E2655–E2664. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, H.; Serranho, P.; Guimarães, P.; Trindade, R.; Martins, J.; Moreira, P.I.; Ambrósio, A.F.; Castelo-Branco, M.; Bernardes, R. Stage-Independent Biomarkers for Alzheimer’s Disease from the Living Retina: An Animal Study. Sci. Rep. 2022, 12, 13667. [Google Scholar] [CrossRef] [PubMed]
- Son, T.; Alam, M.; Toslak, D.; Wang, B.; Lu, Y.; Yao, X. Functional Optical Coherence Tomography of Neurovascular Coupling Interactions in the Retina. J. Biophotonics 2018, 11, e201800089. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.H.; Son, T.; Le, D.; Yao, X. Longitudinal OCT and OCTA Monitoring Reveals Accelerated Regression of Hyaloid Vessels in Retinal Degeneration 10 (Rd10) Mice. Sci. Rep. 2019, 9, 16685. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.; Son, T.; Kim, T.H.; Yao, X. In Vivo Optoretinography of Phototransduction Activation and Energy Metabolism in Retinal Photoreceptors. J. Biophotonics 2021, 14, e202000462. [Google Scholar] [CrossRef] [PubMed]
- Dysli, C.; Enzmann, V.; Sznitman, R.; Zinkernagel, M.S. Quantitative Analysis of Mouse Retinal Layers Using Automated Segmentation of Spectral Domain Optical Coherence Tomography Images. Transl. Vis. Sci. Technol. 2015, 4, 9. [Google Scholar] [CrossRef]
- Gende, M.; Mallen, V.; de Moura, J.; Cordón, B.; Garcia-Martin, E.; Sánchez, C.I.; Novo, J.; Ortega, M. Automatic Segmentation of Retinal Layers in Multiple Neurodegenerative Disorder Scenarios. IEEE J. Biomed. Health Inf. 2023, 27, 5483–5494. [Google Scholar] [CrossRef]
- Alber, J.; Bouwman, F.; den Haan, J.; Rissman, R.A.; De Groef, L.; Koronyo-Hamaoui, M.; Lengyel, I.; Thal, D.R. Retina Pathology as a Target for Biomarkers for Alzheimer’s Disease: Current Status, Ophthalmopathological Background, Challenges, and Future Directions. Alzheimer’s Dement. 2023, 20, 728–740. [Google Scholar] [CrossRef]
- Lindovsky, J.; Palkova, M.; Symkina, V.; Raishbrook, M.J.; Prochazka, J.; Sedlacek, R. OCT and ERG Techniques in High-Throughput Phenotyping of Mouse Vision. Genes 2023, 14, 294. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, L.R.; Dominguez, J.M.; Balaiya, S.; Grover, S.; Chalam, K.V. Retinal Thickness Normative Data in Wild-Type Mice Using Customized Miniature SD-OCT. PLoS ONE 2013, 8, e67265. [Google Scholar] [CrossRef] [PubMed]
- Veitch, D.P.; Weiner, M.W.; Aisen, P.S.; Beckett, L.A.; DeCarli, C.; Green, R.C.; Harvey, D.; Jack, C.R.; Jagust, W.; Landau, S.M.; et al. Using the Alzheimer’s Disease Neuroimaging Initiative to Improve Early Detection, Diagnosis, and Treatment of Alzheimer’s Disease. Alzheimer’s Dement. 2021, 18, 824–857. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez-Puebla, L.; López-Cuenca, I.; Salobrar-García, E.; Ramírez, A.I.; Fernández-Albarral, J.A.; Matamoros, J.A.; Elvira-Hurtado, L.; Salazar, J.J.; Ramírez, J.M.; de Hoz, R. OCT Imaging in Murine Models of Alzheimer’s Disease in a Systematic Review: Findings, Methodology and Future Perspectives. Biomedicines 2024, 12, 528. https://doi.org/10.3390/biomedicines12030528
Sánchez-Puebla L, López-Cuenca I, Salobrar-García E, Ramírez AI, Fernández-Albarral JA, Matamoros JA, Elvira-Hurtado L, Salazar JJ, Ramírez JM, de Hoz R. OCT Imaging in Murine Models of Alzheimer’s Disease in a Systematic Review: Findings, Methodology and Future Perspectives. Biomedicines. 2024; 12(3):528. https://doi.org/10.3390/biomedicines12030528
Chicago/Turabian StyleSánchez-Puebla, Lidia, Inés López-Cuenca, Elena Salobrar-García, Ana I. Ramírez, José A. Fernández-Albarral, José A. Matamoros, Lorena Elvira-Hurtado, Juan J. Salazar, José M. Ramírez, and Rosa de Hoz. 2024. "OCT Imaging in Murine Models of Alzheimer’s Disease in a Systematic Review: Findings, Methodology and Future Perspectives" Biomedicines 12, no. 3: 528. https://doi.org/10.3390/biomedicines12030528














