CFH (rs1061170, rs1410996), KDR (rs2071559, rs1870377) and KDR and CFH Serum Levels in AMD Development and Treatment Efficacy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design and Structure
- Early AMD: Defined by the presence of numerous small (<63 microns, “hard”) or intermediate (≥63 microns but <125 microns, “soft”) drusen.
- Intermediate AMD: Macular disease characterized by either extensive drusen of small or intermediate size, or any drusen of large size (≥125 microns).
- Advanced AMD: Defined by the presence of either geographic atrophy or choroidal neovascular membrane (along with its sequelae, such as subretinal or sub-RPE hemorrhage or serous fluid, and subretinal fibrosis).
- Unrelated eye disorders; e.g., high refractive error, cloudy cornea, lens opacity (nuclear, cortical, or posterior subcapsular cataract) except minor opacities, keratitis, acute or chronic uveitis;
- Systemic illnesses; e.g., diabetes mellitus, malignant tumors, systemic connective tissue disorders, chronic infectious and non-infectious diseases, coronary artery disease, stroke, or conditions following organ or tissue transplantation;
- Ungraded color fundus photographs resulting from obscuring the ocular optic system or because of fundus photograph quality;
- Use of antiepileptic or sedative drugs.
- Older than 18 years;
- Patients after senile cataract surgeries (without any other ocular comorbidities);
- Signed informed consent form.
- Unrelated eye disorders; e.g., high refractive error, cloudy cornea, lens opacity (nuclear, cortical, or posterior subcapsular cataract) except minor opacities, keratitis, acute or chronic uveitis, glaucoma, or diseases of the optic nerve;
- Systemic illnesses; e.g., diabetes mellitus, malignant tumors, systemic connective tissue disorders, chronic infectious and non-infectious diseases, hypertension, coronary artery disease, stroke, or conditions following organ or tissue transplantation;
- Ungraded color fundus photographs resulting from obscuring the ocular optic system or because of fundus photograph quality;
- Use of antiepileptic or sedative drugs.
2.2. SNP Selection
2.3. Deoxyribonucleic Acid Extraction from Peripheral Venous Blood and Genotyping
2.4. Serum Protein Concentration Measurement
2.5. Statistical Analysis
3. Results
3.1. Hardy–Weinberg Equilibrium Analysis
3.2. Analysis of KDR (rs2071559, rs1870377), CFH (rs1061170, rs1410996) in Early and Exudative AMD
3.3. Analysis of KDR (rs2071559, rs1870377) and CFH (rs1061170, rs1410996) in Early and Exudative AMD in Male and Female Subgroups
3.4. KDR and CFH Haplotype Associations with AMD
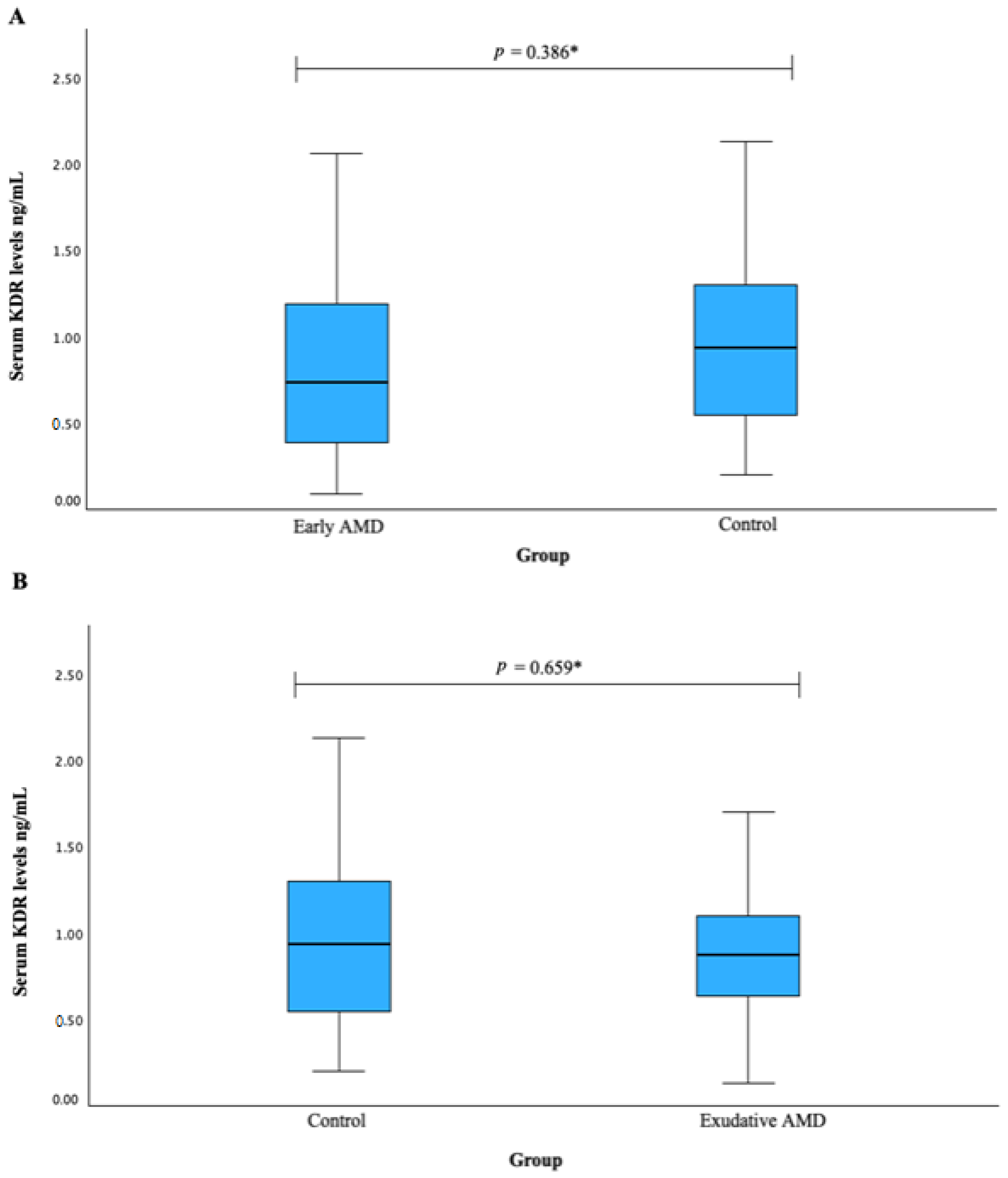
3.5. Serum KDR and CFH Associations with AMD
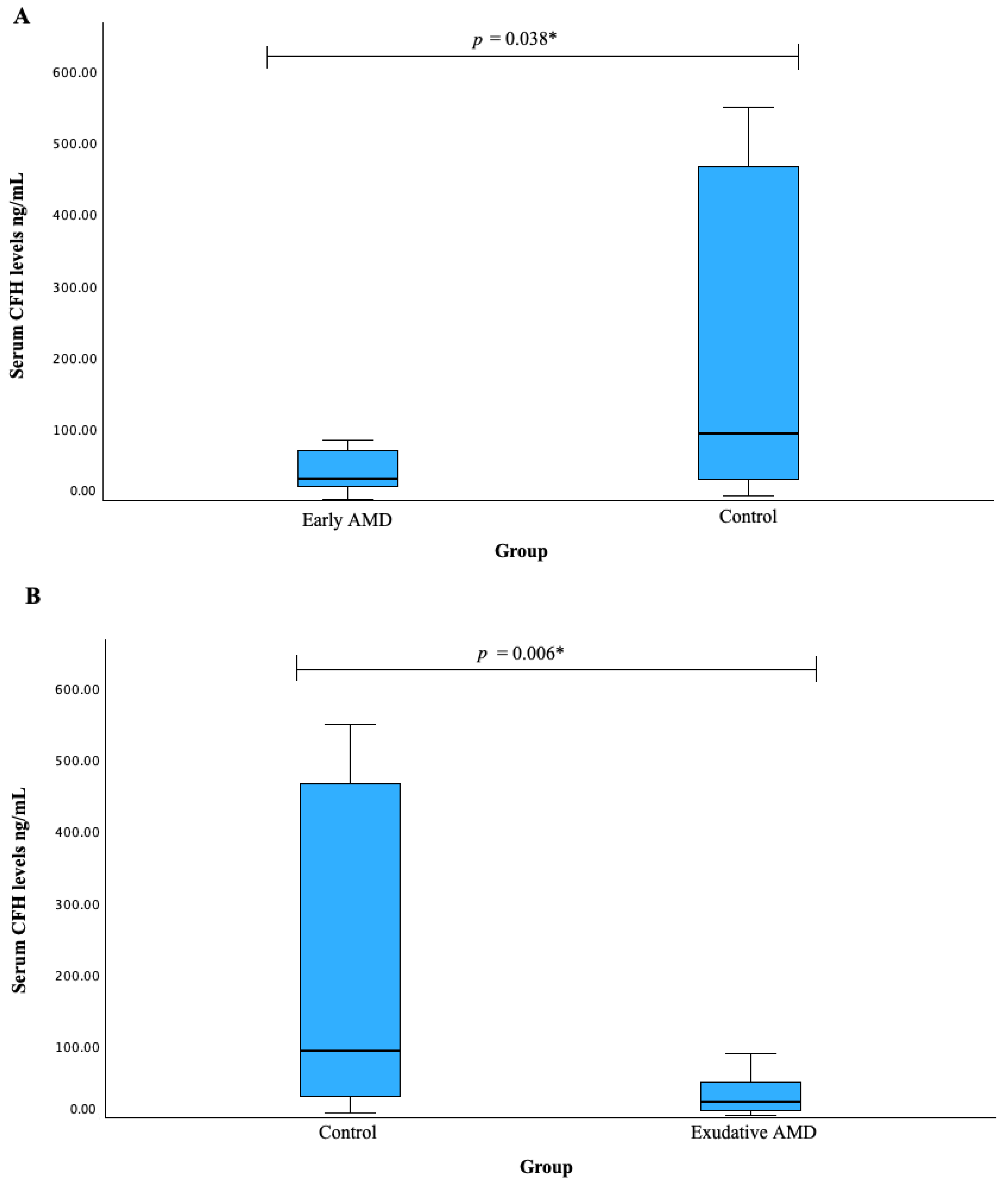
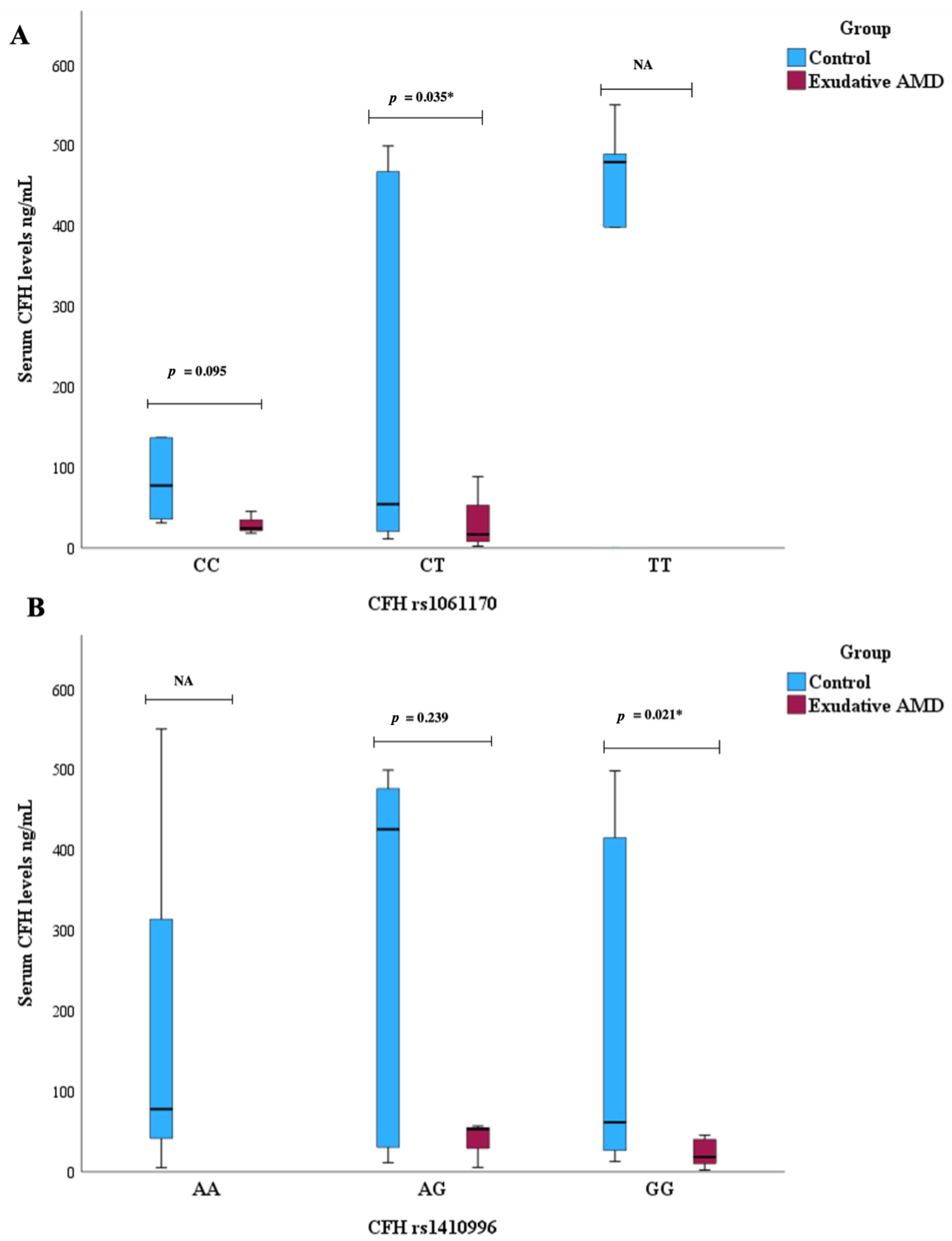
3.6. Serum CFH Levels and CFH SNPs Associations with AMD
3.7. Response to Exudative AMD Treatment with Anti-VEGF Therapy
3.8. Genetic Associations with Exudative AMD Response
3.9. Serum KDR and CFH Associations with Exudative AMD Response
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Klein, R.; Klein, B.E.K.; Linton, K.L.P. Prevalence of age-related maculopathy, The Beaver Dam Eye Study. Ophthalmology 1992, 99, 933–943. [Google Scholar] [CrossRef]
- Mitchell, P.; Smith, W.; Attebo, K.; Wang, J.J. Prevalence of age-related maculopathy in Australia. Ophthalmology 1995, 102, 1450–1460. [Google Scholar] [CrossRef]
- Vingerling, J.R.; Dielemans, I.; Hofman, A.; Grobbee, D.E.; Hijmering, M.; Kramer, C.F.; de Jong, P.T. The prevalence of age-related maculopathy in the Rotterdam study. Ophthalmology 1995, 102, 205–210. [Google Scholar] [CrossRef]
- Wong, W.L.; Su, X.; Li, X.; Cheung, C.M.G.; Klein, R.; Cheng, C.Y.; Wong, T.Y. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: A systematic review and meta-analysis. Lancet Glob. Health 2014, 2, e106–e116. [Google Scholar] [CrossRef]
- Age-Related Eye Disease Study Research Group. A randomized, placebo-controlled, clinical trial of high-dose supplementation with vitamins C and E, beta carotene, and zinc for age-related macular degeneration and vision loss: AREDS report no. 8. Arch. Ophthalmol. 2001, 119, 1417–1436. [Google Scholar] [CrossRef] [PubMed]
- Brandl, C.; Zimmermann, M.E.; Günther, F.; Barth, T.; Olden, M.; Schelter, S.C.; Kronenberg, F.; Loss, J.; Küchenhoff, H.; Helbig, H.; et al. On the impact of different approaches to classify age-related macular degeneration: Results from the German AugUR study. Sci. Rep. 2018, 8, 8675. [Google Scholar] [CrossRef]
- Korb, C.A.; Kottler, U.B.; Wolfram, C.; Hoehn, R.; Schulz, A.; Zwiener, I.; Wild, P.S.; Pfeiffer, N.; Mirshahi, A. Prevalence of age-related macular degeneration in a large European cohort: Results from the population-based Gutenberg Health Study. Graefe’s Arch. Clin. Exp. Ophthalmol. 2014, 252, 1403–1411. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, A.R.; Zielińska, A.; Sanchez-Lopez, E.; dos Santos, T.; Garcia, M.L.; Silva, A.M.; Karczewski, J.; Souto, E.B. Exudative versus nonexudative age-related macular degeneration: Physiopathology and treatment options. Int. J. Mol. Sci. 2022, 23, 2592. [Google Scholar] [CrossRef] [PubMed]
- Fleckenstein, M.; Mitchell, P.; Freund, K.B.; Sadda, S.; Holz, F.G.; Brittain, C.; Henry, E.C.; Ferrara, D. The Progression of Geographic Atrophy Secondary to Age-Related Macular De-generation. Ophthalmology 2018, 125, 369–390. [Google Scholar] [CrossRef]
- Owen, C.G.; Jarrar, Z.; Wormald, R.; Cook, D.G.; Fletcher, A.E.; Rudnicka, A.R. The estimated prevalence and incidence of late stage age related macular degeneration in the UK. Br. J. Ophthalmol. 2012, 96, 752–756. [Google Scholar] [CrossRef]
- Gass, J.D. Pathogenesis of disciform detachment of the neuroepithelium. Am. J. Ophthalmol. 1967, 63, 1–139. [Google Scholar]
- Bakri, S.J.; Thorne, J.E.; Ho, A.C.; Ehlers, J.P.; Schoenberger, S.D.; Yeh, S.; Kim, S.J. Safety and Efficacy of Anti-Vascular Endothelial Growth Factor Therapies for Neovascular Age-Related Macular Degeneration: A Report by the American Academy of Ophthalmology. Ophthalmology 2019, 126, 55–63. [Google Scholar] [CrossRef]
- Rosenfeld, P.J.; Brown, D.M.; Heier, J.S.; Boyer, D.S.; Kaiser, P.K.; Chung, C.Y.; Kim, R.Y. Ranibizumab for neovascular age-related macular degeneration. N. Engl. J. Med. 2006, 355, 1419–1431. [Google Scholar] [CrossRef]
- Heier, J.S.; Brown, D.M.; Chong, V.; Korobelnik, J.-F.; Kaiser, P.K.; Nguyen, Q.D.; Kirchhof, B.; Ho, A.; Ogura, Y.; Yancopoulos, G.D.; et al. Intravitreal aflibercept (VEGF trap-eye) in wet age-related macular degeneration. Ophthalmology 2012, 119, 2537–2548. [Google Scholar] [CrossRef]
- Colijn, J.M.; Buitendijk, G.H.S.; Prokofyeva, E.; Alves, D.; Cachulo, M.L.; Khawaja, A.P.; Cougnard-Gregoire, A.; Merle, B.M.J.; Korb, C.; Erke, M.G.; et al. Prevalence of age-related macular degeneration in Europe. Ophthalmology 2017, 124, 1753–1763. [Google Scholar] [CrossRef]
- Leung, E.H.; Oh, D.J.; Alderson, S.E.; Bracy, J.; McLeod, M.; Perez, L.I.; Bottini, A.; Yee, D.C.; Mukkamala, K. Initial Real-World Experience with Faricimab in Treatment-Resistant Neovascular Age-Related Macular Degeneration. Clin. Ophthalmol. 2023, 17, 1287–1293. [Google Scholar] [CrossRef] [PubMed]
- Khachigian, L.M.; Liew, G.; Teo, K.Y.C.; Wong, T.Y.; Mitchell, P. Emerging therapeutic strategies for unmet need in neovascular age-related macular degeneration. J. Transl. Med. 2023, 21, 133. [Google Scholar] [CrossRef]
- Fisher, S.A.; Abecasis, G.R.; Yashar, B.M.; Zareparsi, S.; Swaroop, A.; Iyengar, S.K.; Klein, B.E.; Klein, R.; Lee, K.E.; Majewski, J.; et al. Meta-analysis of genome scans of age-related macular degeneration. Hum. Mol. Genet. 2005, 14, 2257–2264. [Google Scholar] [CrossRef] [PubMed]
- Klein, M.L.; Schultz, D.W.; Edwards, A.; Matise, T.C.; Rust, K.; Berselli, C.B.; Trzupek, K.; Weleber, R.G.; Ott, J.; Wirtz, M.K. Age-related macular degeneration. Clinical features in a large family and linkage to chromosome 1q. Arch. Ophthalmol. 1998, 116, 1082–1088. [Google Scholar] [CrossRef]
- Majewski, J.; Schultz, D.W.; Weleber, R.G.; Schain, M.B.; Edwards, A.O.; Matise, T.C.; Acott, T.S.; Ott, J.; Klein, M.L. Age-related macular degeneration—A genome scan in extended families. Am. J. Hum. Genet. 2003, 73, 540–550. [Google Scholar] [CrossRef]
- Schick, J.H.; Iyengar, S.K.; Klein, B.E.; Klein, R.; Reading, K.; Liptak, R.; Millard, C.; Lee, K.E.; Tomany, S.C.; Moore, E.L.; et al. A whole-genome screen of a quantitative trait of age-related maculopathy in sibships from the Beaver Dam Eye Study. Am. J. Hum. Genet. 2003, 72, 1412–1424. [Google Scholar] [CrossRef]
- Abecasis, G.R.; Yashar, B.M.; Zhao, Y.; Ghiasvand, N.M.; Zareparsi, S.; Branham, K.E.; Reddick, A.C.; Trager, E.H.; Yoshida, S.; Bahling, J.; et al. Age-related macular degeneration: A high-resolution genome scan for susceptibility loci in a population enriched for late-stage disease. Am. J. Hum. Genet. 2004, 74, 482–494. [Google Scholar] [CrossRef]
- Iyengar, S.K.; Song, D.; Klein, B.E.; Klein, R.; Schick, J.H.; Humphrey, J.; Millard, C.; Liptak, R.; Russo, K.; Jun, G.; et al. Dissection of genomewide-scan data in extended families reveals a major locus and oligogenic susceptibility for age-related macular degeneration. Am. J. Hum. Genet. 2004, 74, 20–39. [Google Scholar] [CrossRef]
- Schmidt, S.; Scott, W.K.; Postel, E.A.; Agarwal, A.; Hauser, E.R.; De La Paz, M.A.; Gilbert, J.R.; Weeks, D.E.; Gorin, M.B.; Haines, J.L.; et al. Ordered subset linkage analysis supports a susceptibility locus for age-related macular degeneration on chromosome 16p12. BMC Genet. 2004, 5, 18. [Google Scholar] [CrossRef] [PubMed]
- Kenealy, S.J.; Schmidt, S.; Agarwal, A.; Postel, E.A.; De La Paz, M.A.; Pericak-Vance, M.A.; Haines, J.L. Linkage analysis for age-related macular degeneration supports a gene on chromosome 10q26. Mol. Vis. 2004, 10, 57–61. [Google Scholar] [CrossRef]
- Jun, G.; Klein, B.E.K.; Klein, R.; Fox, K.; Millard, C.; Capriotti, J.; Russo, K.; Lee, K.E.; Elston, R.C.; Iyengar, S.K. Genome-wide analyses demonstrate novel loci that predispose to drusen formation. Investig. Opthalmol. Vis. Sci. 2005, 46, 3081–3088. [Google Scholar] [CrossRef] [PubMed]
- Silveira, A.C.; Morrison, M.A.; Ji, F.; Xu, H.; Reinecke, J.B.; Adams, S.M.; Arneberg, T.M.; Janssian, M.; Lee, J.-E.; Yuan, Y.; et al. Convergence of linkage, gene expression, and association data demonstrates the influence of the RAR-related orphan receptor alpha (RORA) gene on neovascular AMD: A systems biology-based approach. Vis. Res. 2010, 50, 698–715. [Google Scholar] [CrossRef] [PubMed]
- Fritsche, L.G.; Igl, W.; Bailey, J.N.C.; Grassmann, F.; Sengupta, S.; Bragg-Gresham, J.L.; Burdon, K.P.; Hebbring, S.J.; Wen, C.; Gorski, M.; et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat. Genet. 2016, 48, 134–143. [Google Scholar] [CrossRef]
- Lazzeri, S.; Orlandi, P.; Piaggi, P.; Sartini, M.S.; Casini, G.; Guidi, G.; Figus, M.; Fioravanti, A.; Di Desidero, T.; Ripandelli, G.; et al. IL-8 and VEGFR-2 polymorphisms modulate long-term functional response to intravitreal ranibizumab in exudative age-related macular degeneration. Pharmacogenomics 2016, 17, 35–39. [Google Scholar] [CrossRef]
- Hermann, M.M.; van Asten, F.; Muether, P.S.; Smailhodzic, D.; Lichtner, P.; Hoyng, C.B.; Kirchhof, B.; Grefkes, C.; den Hollander, A.I.; Fauser, S. Polymorphisms in vascular endothelial growth factor receptor 2 are associated with better response rates to ranibizumab treatment in age-related macular degeneration. Ophthalmology 2013, 121, 905–910. [Google Scholar] [CrossRef]
- Blánquez-Martínez, D.; Díaz-Villamarín, X.; Antúnez-Rodríguez, A.; Pozo-Agundo, A.; Muñoz-Ávila, J.I.; Martínez-González, L.J.; Dávila-Fajardo, C.L. Genetic Polymorphisms Affecting Ranibizumab Response in High Myopia Patients. Pharmaceutics 2021, 13, 1973. [Google Scholar] [CrossRef] [PubMed]
- Haines, J.L.; Hauser, M.A.; Schmidt, S.; Scott, W.K.; Olson, L.M.; Gallins, P.; Spencer, K.L.; Kwan, S.Y.; Noureddine, M.; Gilbert, J.R.; et al. Complement factor H variant increases the risk of age-related macular degeneration. Science 2005, 308, 419–421. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Guo, Y.; Ma, Y.; Zheng, Z.; Wang, Q.; Zhou, X. Association of Two Polymorphisms, rs1061170 and rs1410996, in Complement Factor H with Age-Related Macular Degeneration in an Asian Population: A Meta-Analysis. Ophthalmic Res. 2016, 55, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Acar, I.E.; Galesloot, T.E.; Luhmann, U.F.O.; Fauser, S.; Gayán, J.; den Hollander, A.I.; Nogoceke, E. Whole Genome Sequencing Identifies Novel Common and Low-Frequency Variants Associated with Age-Related Macular Degeneration. Investig. Opthalmol. Vis. Sci. 2023, 64, 24. [Google Scholar] [CrossRef] [PubMed]
- Hong, N.; Shen, Y.; Yu, C.-Y.; Wang, S.-Q.; Tong, J.-P. Association of the polymorphism Y402H in the CFH gene with response to anti-VEGF treatment in age-related macular degeneration: A systematic review and meta-analysis. Acta Ophthalmol. 2016, 94, 334–345. [Google Scholar] [CrossRef] [PubMed]
- Coltelli, L.; Allegrini, G.; Orlandi, P.; Finale, C.; Fontana, A.; Masini, L.C.; Scalese, M.; Arrighi, G.; Barletta, M.T.; De Maio, E.; et al. A pharmacogenetic interaction analysis of bevacizumab with paclitaxel in advanced breast cancer patients. NPJ Breast Cancer 2022, 8, 33. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.-B.; Zhan, M.-X.; Zhao, W.; Liu, B.; Huang, J.-W.; He, X.; Fu, S.-R.; Zhao, Y.; Li, Y.; Hu, B.-S.; et al. The relationship of kinase insert domain receptor gene polymorphisms and clinical outcome in advanced hepatocellular carcinoma patients treated with sorafenib. Med. Oncol. 2014, 31, 209. [Google Scholar] [CrossRef] [PubMed]
- Stahl, A. The Diagnosis and Treatment of Age-Related Macular Degeneration. Dtsch. Aerzteblatt Online 2020, 117, 513–520. [Google Scholar] [CrossRef]
- Xu, X.; Wu, J.; Yu, X.; Tang, Y.; Tang, X.; Shentu, X. Regional differences in the global burden of age-related macular degeneration. BMC Public Health 2020, 20, 410. [Google Scholar] [CrossRef]
- Xu, K.; Gupta, V.; Bae, S.; Sharma, S. Metamorphopsia and vision-related quality of life among patients with age-related macular degeneration. Can. J. Ophthalmol. 2018, 53, 168–172. [Google Scholar] [CrossRef]
- Kuroda, Y.; Yamashiro, K.; Miyake, M.; Yoshikawa, M.; Nakanishi, H.; Oishi, A.; Tamura, H.; Ooto, S.; Tsujikawa, A.; Yoshimura, N. Factors associated with recurrence of age-related macular degeneration after an-ti-vascular endothelial growth factor treatment: A retrospective cohort study. Ophthalmology 2015, 122, 2303–2310. [Google Scholar] [CrossRef] [PubMed]
- Hara, C.; Wakabayashi, T.; Fukushima, Y.; Sayanagi, K.; Kawasaki, R.; Sato, S.; Sakaguchi, H.; Nishida, K. Tachyphylaxis during treatment of exudative age-related macular degeneration with aflibercept. Graefe’s Arch. Clin. Exp. Ophthalmol. 2019, 257, 2559–2569. [Google Scholar] [CrossRef] [PubMed]
- Mousavi, M.; Armstrong, R.A. Genetic risk factors and age-related macular degeneration (AMD). J. Optom. 2013, 6, 176–184. [Google Scholar] [CrossRef]
- Yoshida, A.; Yoshida, M.; Yoshida, S.; Shiose, S.; Hiroishi, G.; Ishibashi, T. Familial cases with age-related macular degeneration. Jpn. J. Ophthalmol. 2000, 44, 290–295. [Google Scholar] [CrossRef]
- Seddon, J.M.; Reynolds, R.; Maller, J.; Fagerness, J.A.; Daly, M.J.; Rosner, B. Prediction model for prevalence and incidence of advanced age-related macular degeneration based on genetic, demographic, and environmental variables. Investig. Ophthalmol. Vis. Sci. 2009, 50, 2044–2053. [Google Scholar] [CrossRef] [PubMed]
- Klein, M.L.; Francis, P.J.; Ferris, F.L.; Hamon, S.C.; Clemons, T.E. Risk assessment model for the development of advanced age-related macular degeneration. Arch. Ophthalmol. 2011, 129, 1543–1550. [Google Scholar] [CrossRef] [PubMed]
- Dietzel, M.; Farwick, A.; Hense, H.W. Genetic and risk factors for exudative AMD. Ophthalmology 2010, 107, 1103–1108. [Google Scholar]
- Chan, D. Cigarette smoking and age-related macular degeneration. Optom. Vis. Sci. 1998, 75, 476–484. [Google Scholar] [CrossRef] [PubMed]
- Bauer, P.; Barthelmes, D.; Kurz, M.; Fleischhauer, J.C.; Sutter, F.K. The potential effect of population development, smoking and antioxidant supplementation on the future epidemiology of age-related macular degeneration in Switzerland. Klin. Monatsblätter Augenheilkd. 2008, 225, 376–379. [Google Scholar] [CrossRef]
- Cackett, P.; Yeo, I.; Cheung, C.M.G. Relationship of smoking and cardiovascular risk factors with polypoidal choroidal vascu-lopathy and age-related macular degeneration in Chinese persons. Ophthalmology 2011, 118, 846–852. [Google Scholar] [CrossRef]
- Coleman, A.L.; Seitzman, R.L.; Cummings, S.R. The association of smoking and alcohol use with age-related macular degenera-tion in the oldest old: The study of osteoporotic fractures. Am. J. Ophthalmol. 2010, 149, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.; Hung, S.; Willett, W.C. Prospective study of dietary fat and the risk of age-related macular degeneration. Am. J. Clin. Nutr. 2001, 73, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Chua, B.; Flood, V.; Rochtchina, E.; Wang, J.J.; Smith, W.; Mitchell, P. Dietary fatty acids and the 5-year incidence of age-related maculopathy. Arch. Ophthalmol. 2006, 124, 981–986. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.J.; Milton, R.C.; Klein, R.; Gensler, G.; Taylor, A. Dietary carbohydrate and the progression of age-related macular de-generation: A prospective study from the age-related eye disease study. Am. J. Clin. Nutr. 2007, 86, 1210–1218. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.-J.; Milton, R.C.; Gensler, G.; Taylor, A. Association between dietary glycemic index and age-related macular degeneration in nondiabetic participants in the age-related eye disease study. Am. J. Clin. Nutr. 2007, 86, 180–188. [Google Scholar] [CrossRef]
- Chiu, C.J.; Klein, R.; Milton, R.C.; Gensler, G.; Taylor, A. Does eating particular diets alter the risk of age-related macular degen-eration in users of the age-related eye disease study supplements. Br. J. Ophthalmol. 2009, 93, 1241–1246. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.K.M.; Simpson, J.A.; Aung, K.Z.; Makeyeva, G.A.; Giles, G.G.; English, D.R.; Hopper, J.; Guymer, R.H.; Baird, P.N.; Robman, L.D. Abdominal obesity and age-related macular degeneration. Am. J. Epidemiol. 2011, 173, 1246–1255. [Google Scholar] [CrossRef] [PubMed]
- Delcourt, C.; Carrière, I.; Ponton-Sanchez, A.; Fourrey, S.; Lacroux, A.; Papoz, L. Light exposure and the risk of age-related macular degeneration. Arch. Ophthalmol. 2001, 119, 1463–1468. [Google Scholar] [CrossRef] [PubMed]
- Chalam, K.V.; Khetpal, V.; Rusovici, R.; Balaiya, S. A review: Role of ultraviolet radiation in age-related macular degeneration. Eye Contact Lens 2011, 37, 225–232. [Google Scholar] [CrossRef]
- Ratnayake, K.; Payton, J.L.; Lakmal, O.H.; Karunarathne, A. Blue light excited retinal intercepts cellular signaling. Sci. Rep. 2018, 8, 10207. [Google Scholar] [CrossRef]
- Simader, C.; Ritter, M.; Bolz, M.; Deák, G.G.; Mayr-Sponer, U.; Golbaz, I.; Kundi, M.; Schmidt-Erfurth, U.M. Morphologic Parameters Relevant for Visual Outcome During Anti-Angiogenic Therapy of Neovascular Age-Related Macular Degeneration. Ophthalmology 2014, 121, 1237–1245. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, V.M.; Matonti, F.; Zarranz-Ventura, J.M.; Stewart, M.W. Impact of fluid compartments on functional outcomes for patients with neovascular age-related macular degeneration. Retina 2021, 42, 589–606. [Google Scholar] [CrossRef] [PubMed]
- Tsilimbaris, M.K.; López-Gálvez, M.I.; Gallego-Pinazo, R.; Margaron, P.; Lambrou, G.N. Epidemiological and Clinical Baseline Characteristics as Predictive Biomarkers of Response to Anti-VEGF Treatment in Patients with Neovascular AMD. J. Ophthalmol. 2016, 2016, 4367631. [Google Scholar] [CrossRef] [PubMed]
- Ying, G.-S.; Kim, B.J.; Maguire, M.G.; Huang, J.; Daniel, E.; Jaffe, G.J.; Grunwald, J.E.; Blinder, K.J.; Flaxel, C.J.; Rahhal, F.; et al. Sustained Visual Acuity Loss in the Comparison of Age-Related Macular Degeneration Treatments Trials. JAMA Ophthalmol. 2014, 132, 915–921. [Google Scholar] [CrossRef] [PubMed]
- Amoaku, W.M.; Chakravarthy, U.; Gale, R.; Gavin, M.; Ghanchi, F.; Gibson, J.; Harding, S.; Johnston, R.L.; Kelly, S.P.; Lotery, A.; et al. Defining response to anti-VEGF therapies in neovascular AMD. Eye 2015, 29, 721–731. [Google Scholar] [CrossRef] [PubMed]
- Brantley, M.A.; Fang, A.M.; King, J.M.; Tewari, A.; Kymes, S.M.; Shiels, A. Association of Complement Factor H and LOC387715 Genotypes with Response of Exudative Age-Related Macular Degeneration to Intravitreal Bevacizumab. Ophthalmology 2007, 114, 2168–2173. [Google Scholar] [CrossRef]
- Cruz-González, F.; Cieza-Borrella, C.; Valverde, G.L.; Lorenzo-Pérez, R.; Hernández-Galilea, E.; González-Sarmiento, R. CFH (rs1410996), HTRA1 (rs112000638) and ARMS2 (rs10490923) gene polymorphisms are associated with AMD risk in Spanish patients. Ophthalmic Genet. 2013, 35, 68–73. [Google Scholar] [CrossRef]
- Liao, X.; Lan, C.-J.; Cheuk, I.-W.; Tan, Q.-Q. Four complement factor H gene polymorphisms in association with AMD: A meta-analysis. Arch. Gerontol. Geriatr. 2016, 64, 123–129. [Google Scholar] [CrossRef]
- Fu, Y.; Sun, W.; Xu, C.; Gu, S.; Li, Y.; Liu, Z.; Chen, J. Genetic variants in KDR transcriptional regulatory region affect promoter activity and intramuscular fat deposition in Erhualian pigs. Anim. Genet. 2014, 45, 373–380. [Google Scholar] [CrossRef]
- Hagstrom, S.A.; Ying, G.-S.; Pauer, G.J.; Sturgill-Short, G.M.; Huang, J.; Callanan, D.G.; Kim, I.K.; Klein, M.L.; Maguire, M.G.; Martin, D.F.; et al. Pharmacogenetics for Genes Associated with Age-related Macular Degeneration in the Comparison of AMD Treatments Trials (CATT). Ophthalmology 2013, 120, 593–599. [Google Scholar] [CrossRef]
- Riaz, M.; Lorés-Motta, L.; Richardson, A.J.; Lu, Y.; Montgomery, G.; Omar, A.; Koenekoop, R.K.; Chen, J.; Muether, P.; Altay, L.; et al. GWAS study using DNA pooling strategy identifies association of variant rs4910623 in OR52B4 gene with anti-VEGF treatment response in age-related macular degeneration. Sci. Rep. 2016, 6, 37924. [Google Scholar] [CrossRef] [PubMed]
- Yamashiro, K.; Mori, K.; Honda, S.; Kano, M.; Yanagi, Y.; Obana, A.; Sakurada, Y.; Sato, T.; Nagai, Y.; Hikichi, T.; et al. A prospective multicenter study on genome wide associations to ranibizumab treatment outcome for age-related macular degeneration. Sci. Rep. 2017, 7, 9196. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, M.; Takahashi, A.; Momozawa, Y.; Arakawa, S.; Miya, F.; Tsunoda, T.; Ashikawa, K.; Oshima, Y.; Yasuda, M.; Yoshida, S.; et al. Genome-wide association study suggests four variants influencing outcomes with ranibizumab therapy in exudative age-related macular degeneration. J. Hum. Genet. 2018, 63, 1083–1091. [Google Scholar] [CrossRef] [PubMed]
- Lorés-Motta, L.; Riaz, M.; Grunin, M.; Corominas, J.; van Asten, F.; Pauper, M.; Leenders, M.; Richardson, A.J.; Muether, P.; Cree, A.J.; et al. Association of genetic variants with response to anti-vascular endothelial growth factor therapy in age-related macular degeneration. JAMA Ophthalmol. 2018, 136, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Grunin, M.; Beykin, G.; Rahmani, E.; Schweiger, R.; Barel, G.; Hagbi-Levi, S.; Elbaz-Hayoun, S.; Rinsky, B.; Ganiel, M.; Carmi, S.; et al. Association of a variant in VWA3A with response to anti-vascular endothelial growth factor treatment in neovascular AMD. Investig. Ophthalmol. Vis. Sci. 2020, 61, 48. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zou, M.; Chen, A.; Liu, Z.; Young, C.A.; Wang, S.B.; Zheng, D.; Jin, G. Genetic associations of anti-vascular endothelial growth factor therapy response in age-related macular degeneration: A systematic review and meta-analysis. Acta Ophthalmol. 2021, 100, e669–e680. [Google Scholar] [CrossRef]
- Yang, S.; Zhao, J.; Sun, X. Resistance to anti-VEGF therapy in neovascular age-related macular degeneration: A comprehensive review. Drug Des. Dev. Ther. 2016, 10, 1857–1867. [Google Scholar] [CrossRef]
| Characteristic | Early AMD n = 255 | Exudative AMD n = 252 | Control n = 349 | p-Value |
|---|---|---|---|---|
| Gender Males, n (%) Females, n (%) | 81 (31.8) 174 (68.2) | 94 (37.3) 158 (62.7) | 121 (34.7) 228 (65.3) | 0.455 * 0.507 ** |
| Age years; median (IQR) | 73 (12) | 77 (10) | 72 (11) | 0.119 * <0.001 ** |
| Reagents | 1 Sample | 96 Samples |
|---|---|---|
| TaqMan Universal Master Mix II, no UNG (“Applied Biosystems”, Vilnius, Lithuania) “Applied Biosystems” genotyping assay C___8355565_10 C___2530294_10 C__15869271_10 C__11895315_20 H2O (“ZYMO RESEARCH”, Lithuania) | 5 µL | 480 µL |
| 0.5 µL | 48 µL | |
| 3.5 µL | 336 µL |
| Gene, SNP | RT-PGR Reaction Conditions |
|---|---|
| CFH rs1061170 rs1410996 | 95 °C 10 min 45 cycles: 92 °C 15 s 60 °C 60 s |
| KDR rs2071559 | |
| KDR rs1870377 |
| Gener/Marker | Genotype/ Allele | Group | p-Value * | p-Value ** | ||
|---|---|---|---|---|---|---|
| Early AMD (n = 255) n (%) | Exudative AMD (n = 252) n (%) | Control (n = 349) n (%) | ||||
| KDR rs2071559 | GG GA AA G A | 60 (23.2) 118 (46.3) 77 (30.2) 238 (46.7) 272 (53.3) | 49 (19.4) 134 (53.2) 69 (27.4) 232 (46.0) 272 (54.0) | 77 (22.1) 188 (53.9) 84 (24.1) 342 (49.0) 356 (51.0) | 0.143 0.423 | 0.571 0.310 |
| KDR rs1870377 | TT TA AA T A | 127 (49.7) 101 (39.6) 27 (10.6) 355 (69.6) 155 (30.4) | 125 (49.6) 99 (39.3) 28 (11.1) 349 (69.2) 155 (30.8) | 172 (49.3) 145 (41.5) 32 (9.2) 489 (70.1) 209 (29.9) | 0.799 0.866 | 0.691 0.763 |
| CFH rs1061170 | TT TC CC T C | 80 (31.4) 130 (51) 45 (17.6) 290 (56.9) 220 (43.1) | 38 (15.1) 130 (51.6) 84 (33.3) 206 (40.9) 298 (59.1) | 137 (39.3) 158 (45.3) 54 (15.5) 432 (61.9) 266 (38.1) | 0.137 0.078 | <0.001 <0.001 |
| CFH rs1410996 | GG GA AA G A | 114 (44.7) 108 (42.4) 33 (12.9) 336 (65.9) 174 (34.1) | 162 (64.3) 82 (32.5) 8 (3.2) 406 (80.6) 98 (19.4) | 132 (37.8) 168 (48.1) 49 (14) 432 (61.9) 266 (38.1) | 0.232 0.155 | <0.001 <0.001 |
| Genetic Model | Genotype/Allele | OR * (95 %CI) | p-Value | AIC |
|---|---|---|---|---|
| Early AMD | ||||
| CFH rs1061170 | ||||
| Dominant | TC + CC vs. TT | 1.414 (1.005; 1.988) | 0.046 | 820.629 |
| Exudative AMD | ||||
| CFH rs1061170 | ||||
| Codominant | TC vs. TT CC vs. TT | 2.961 (1.894; 4.630) 5.578 (3.319; 9.377) | <0.001 <0.001 | 715.997 |
| Dominant | TC + CC vs. TT | 3.629 (2.372; 5.552) | <0.001 | 722.380 |
| Recessive | CC vs. TT + TC | 2.704 (1.796; 4.071) | <0.001 | 738.372 |
| Additive | C | 2.354 (1.820; 3.046) | <0.001 | 715.559 |
| CFH rs1410996 | ||||
| Codominant | GA vs. GG AA vs. GG | 0.380 (0.262; 0.550) 0.119 (0.053; 0.266) | <0.001 <0.001 | 712.575 |
| Dominant | GA + AA vs. GG | 0.318 (0.223; 0.453) | <0.001 | 719.925 |
| Recessive | AA vs. GG + GA | 0.184 (0.084; 0.403) | <0.001 | 737.824 |
| Overdominant | GA vs. GG + AA | 0.509 (0.357; 0.725) | <0.001 | 747.483 |
| Additive | A | 0.363 (0.270; 0.488) | <0.001 | 710.725 |
| Genetic Model | Genotype/Allele | OR * (95 %CI) | p-Value | AIC |
|---|---|---|---|---|
| Males | ||||
| Early AMD | ||||
| KDR rs2071559 | ||||
| Codominant | GA AA | 0.491 (0.254; 0.946) 0.657 (0.295; 1.463) | 0.033 0.304 | 271.499 |
| Dominant | GA + AA vs. TT | 0.536 (0.290; 0.991) | 0.047 | 270.093 |
| Exudative AMD | ||||
| KDR rs2071559 | ||||
| Codominant | GA AA | 0.500 (0.266; 0.940) 0.531 (0.239; 1.181) | 0.031 0.121 | 292.937 |
| Dominant | GA + AA vs. TT | 0.508 (0.279; 0.925) | 0.027 | 290.964 |
| CFH rs1061170 | ||||
| Codominant | TC vs. TT CC vs. TT | 3.698 (1.775; 7.704) 8.457 (3.442; 20.780) | <0.001 <0.001 | 271.865 |
| Dominant | TC + CC vs. TT | 4.620 (2.278; 9.369) | <0.001 | 274.871 |
| Recessive | CC vs. TT + TC | 3.432 (1.687; 6.983) | <0.001 | 283.553 |
| Additive | C | 2.354 (1.820; 3.046) | <0.001 | 270.515 |
| CFH rs1410996 | ||||
| Codominant | GA vs. GG AA vs. GG | 0.400 (0.223; 0.719) 0.036 (0.036; 0.278) | 0.002 0.001 | 270.037 |
| Dominant | GA + AA vs. GG | 0.302 (0.171; 0.532) | <0.001 | 277.719 |
| Recessive | AA vs. GG + GA | 0.053 (0.007; 0.406) | 0.005 | 277.689 |
| Overdominant | GA vs. GG + AA | 0.566 (0.322; 0.995) | 0.048 | 281.94\ |
| Additive | A | 0.314 (0.193; 0.510) | <0.001 | 270.282 |
| Females | ||||
| Exudative AMD | ||||
| CFH rs1061170 | ||||
| Codominant | TC vs. TT CC vs. TT | 2.608 (1.470; 4.629) 4.476 (2.346; 8.539) | 0.001 <0.001 | 438.402 |
| Dominant | TC + CC vs. TT | 3.163 (1.843; 5.427) | <0.001 | 440.214 |
| Recessive | CC vs. TT + TC | 2.399 (1.440; 3.996) | <0.001 | 447.718 |
| Additive | C | 2.102 (1.562; 2.895) | <0.001 | 437.210 |
| CFH rs1410996 | ||||
| Codominant | GA vs. GG AA vs. GG | 0.359 (0.221; 0.583) 0.182 (0.072; 0.460) | <0.001 <0.001 | 434.805 |
| Dominant | GA + AA vs. GG | 0.321 (0.202; 0.511) | <0.001 | 434.974 |
| Recessive | AA vs. GG + GA | 0.291 (0.119; 0.713) | 0.007 | 450.722 |
| Overdominant | GA vs. GG + AA | 0.464 (0.292; 0.736) | <0.001 | 448.285 |
| Additive | A | 0.392 (0.269; 0.573) | <0.001 | 433.132 |
| SNPs | D’ | r2 |
|---|---|---|
| Early AMD vs. Controls | ||
| rs2071550-rs1870377 | 0.2282 | 0.0207 |
| rs1061170-rs1410996 | 0.8068 | 0.2510 |
| Exudative AMD vs. Controls | ||
| rs2071550-rs1870377 | 0.2341 | 0.0216 |
| rs1061170-rs1410996 | 0.7954 | 0.2430 |
| Haplotype | rs2071559 | rs1870377 | rs1061170 | rs1410996 | Frequency | OR * (95% CI) | p-Value |
|---|---|---|---|---|---|---|---|
| Haplotype associations with early AMD | |||||||
| 1 | G | T | - | - | 0.3692 | 1.00 | - |
| 2 | A | T | - | - | 0.3295 | 1.24 (0.91–1.69) | 0.17 |
| 3 | A | A | - | - | 0.1904 | 1.05 (0.76–1.46) | 0.77 |
| 4 | G | A | - | - | 0.1109 | 1.28 (0.80–2.03) | 0.3 |
| 5 | - | - | C | G | 0.3737 | 1.00 | - |
| 6 | - | - | T | A | 0.3357 | 0.76 (0.58–1.00) | 0.049 |
| 7 | - | - | T | G | 0.2620 | 0.60 (0.45–0.82) | 0.0011 |
| Haplotype associations with exudative AMD | |||||||
| 1 | G | T | - | - | 0.3673 | 1.00 | - |
| 2 | A | T | - | - | 0.3298 | 1.28 (0.93–1.77) | 0.13 |
| 3 | A | A | - | - | 0.1926 | 1.10 (0.78–1.53) | 0.59 |
| 4 | G | A | - | - | 0.1102 | 1.28 (0.81–2.03) | 0.3 |
| 5 | - | - | C | G | 0.4402 | 1.00 | - |
| 6 | - | - | T | A | 0.2738 | 0.29 (0.20–0.40) | <0.001 |
| 7 | - | - | T | G | 0.257 | 0.42 (0.30–0.58) | <0.001 |
| 8 | - | - | C | A | 0.0287 | 0.03 (0.00–0.21) | <0.001 |
| Characteristic | Non-Responders n = 22 | Responders n = 99 | p-Value |
|---|---|---|---|
| Gender | 7 (31.8) 15 (68.2) | 32 (32.3) 67 (67.7) | 0.963 |
| Males, n (%) | |||
| Females, n (%) | |||
| Age years; median (IQR) | 77 (10) | 78 (10) | 0.184 |
| Response parameter | |||
| VA, median (IQR) | |||
| Baseline | 0.42 (0.45) | 0.26 (0.26) | 0.020 * |
| Treated(final) | 0.31 (0.31) | 0.35 (0.32) | <0.001 ** |
| CRT (μm), median (IQR) | |||
| Baseline | 272.5 (86.5) | 321 (114) | 0.386 * |
| Treated (final) | 314.5 (87.75) | 274 (94) | <0.001 ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cebatoriene, D.; Vilkeviciute, A.; Gedvilaite, G.; Bruzaite, A.; Kriauciuniene, L.; Zaliuniene, D.; Liutkeviciene, R. CFH (rs1061170, rs1410996), KDR (rs2071559, rs1870377) and KDR and CFH Serum Levels in AMD Development and Treatment Efficacy. Biomedicines 2024, 12, 948. https://doi.org/10.3390/biomedicines12050948
Cebatoriene D, Vilkeviciute A, Gedvilaite G, Bruzaite A, Kriauciuniene L, Zaliuniene D, Liutkeviciene R. CFH (rs1061170, rs1410996), KDR (rs2071559, rs1870377) and KDR and CFH Serum Levels in AMD Development and Treatment Efficacy. Biomedicines. 2024; 12(5):948. https://doi.org/10.3390/biomedicines12050948
Chicago/Turabian StyleCebatoriene, Dzastina, Alvita Vilkeviciute, Greta Gedvilaite, Akvile Bruzaite, Loresa Kriauciuniene, Dalia Zaliuniene, and Rasa Liutkeviciene. 2024. "CFH (rs1061170, rs1410996), KDR (rs2071559, rs1870377) and KDR and CFH Serum Levels in AMD Development and Treatment Efficacy" Biomedicines 12, no. 5: 948. https://doi.org/10.3390/biomedicines12050948






