Extended Utilization of Constraint-Based Metabolic Model in a Long-Growing Crop
Abstract
:1. Introduction
2. Materials and Methods
2.1. The rMeCBM-KU50 Model
2.2. Model Sensitivity Analysis
3. Results
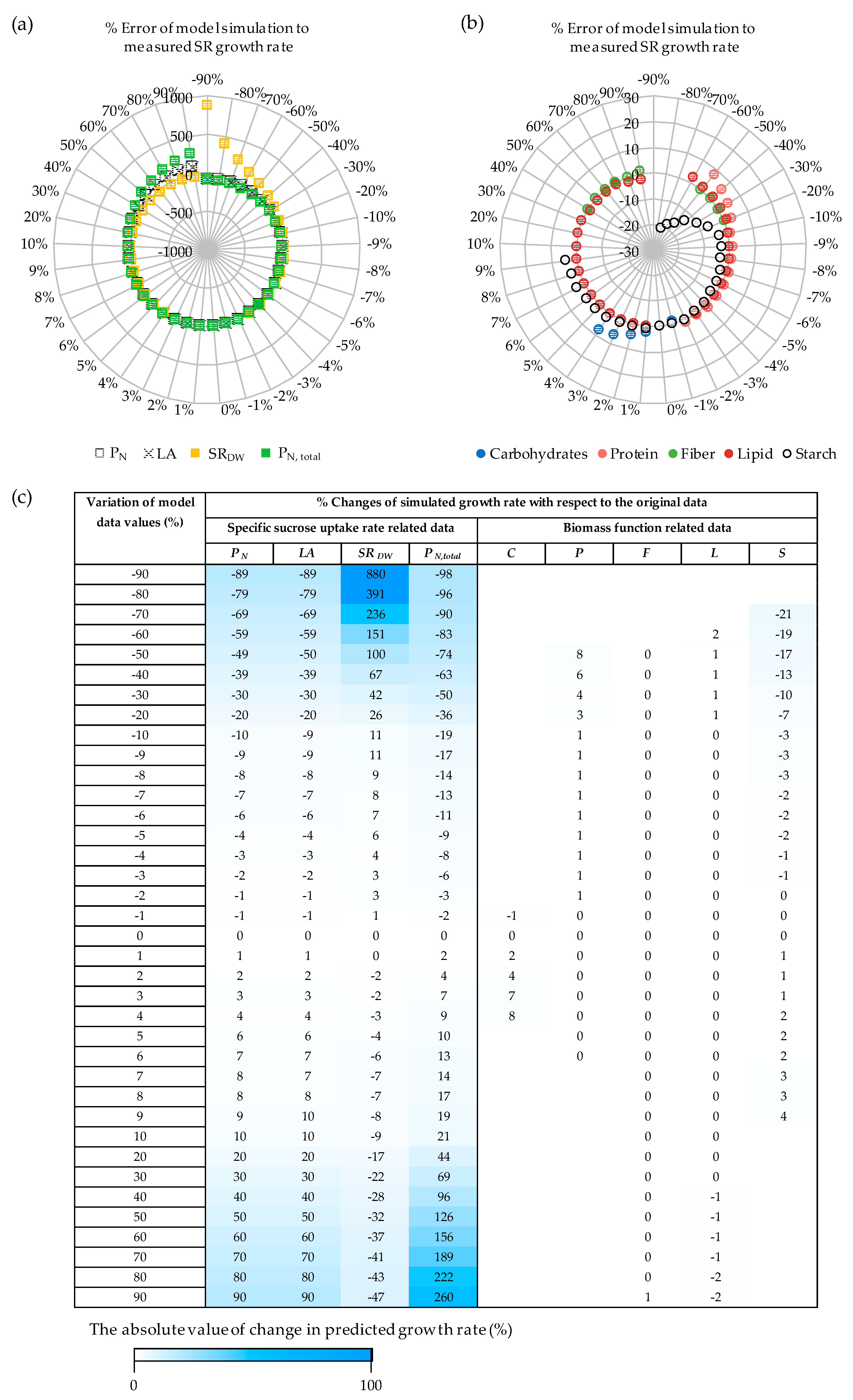
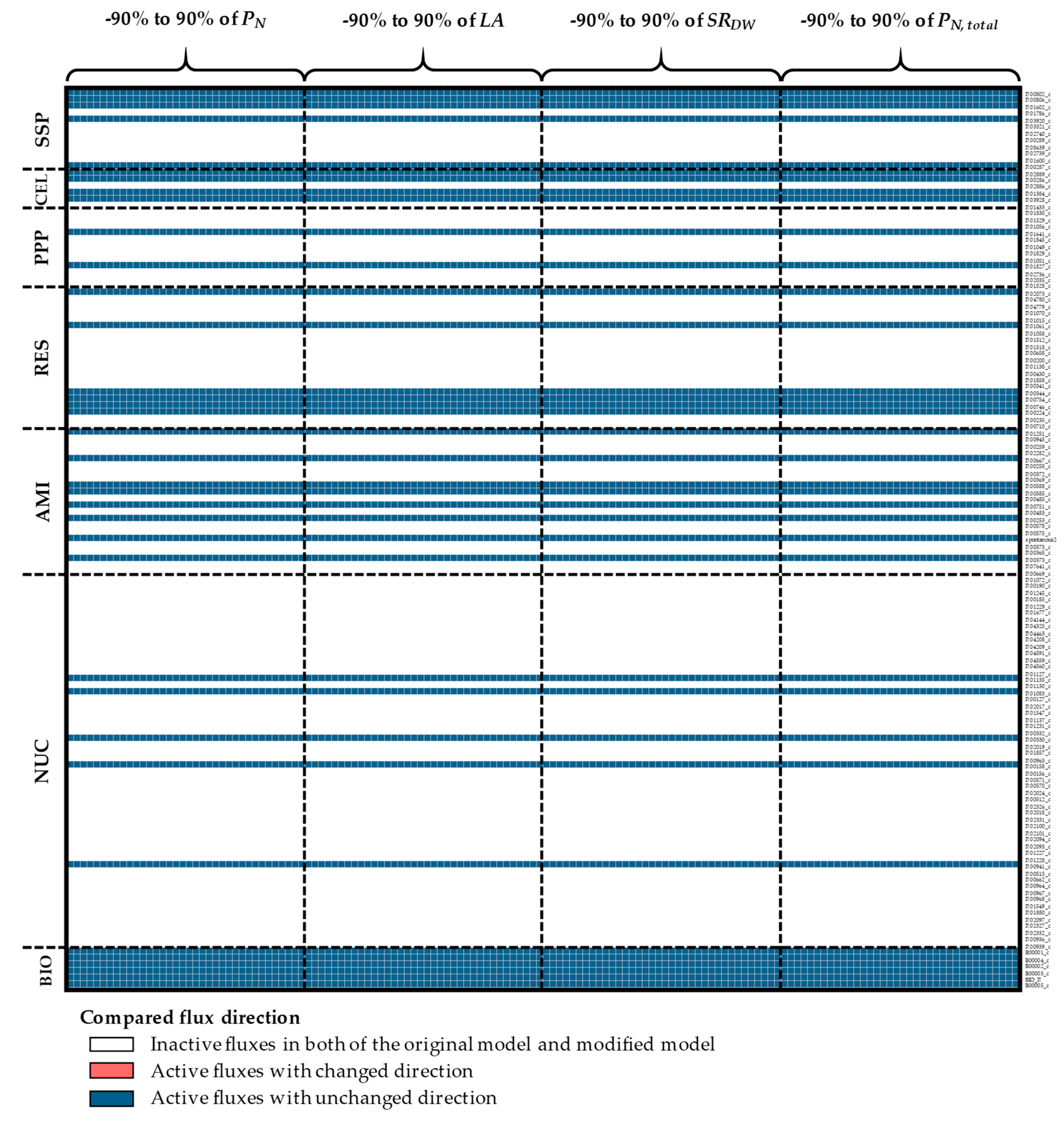
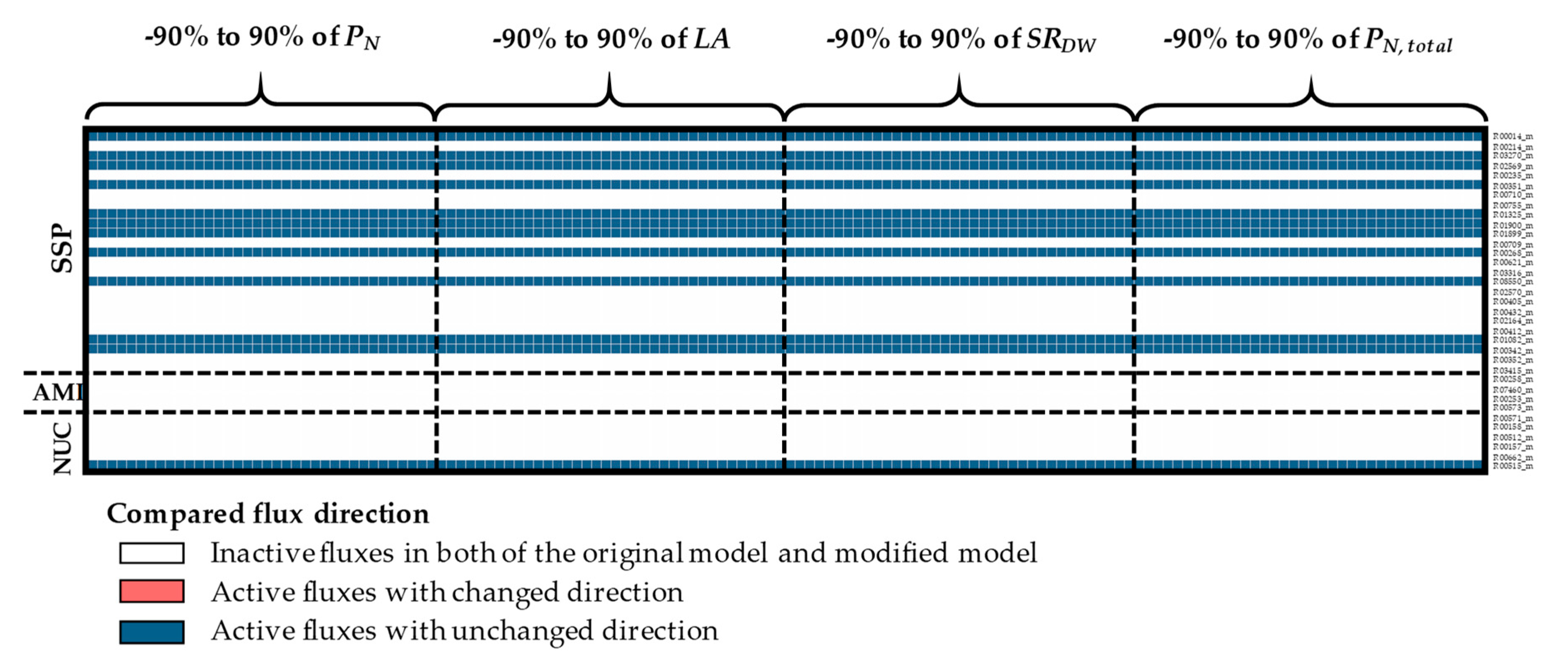
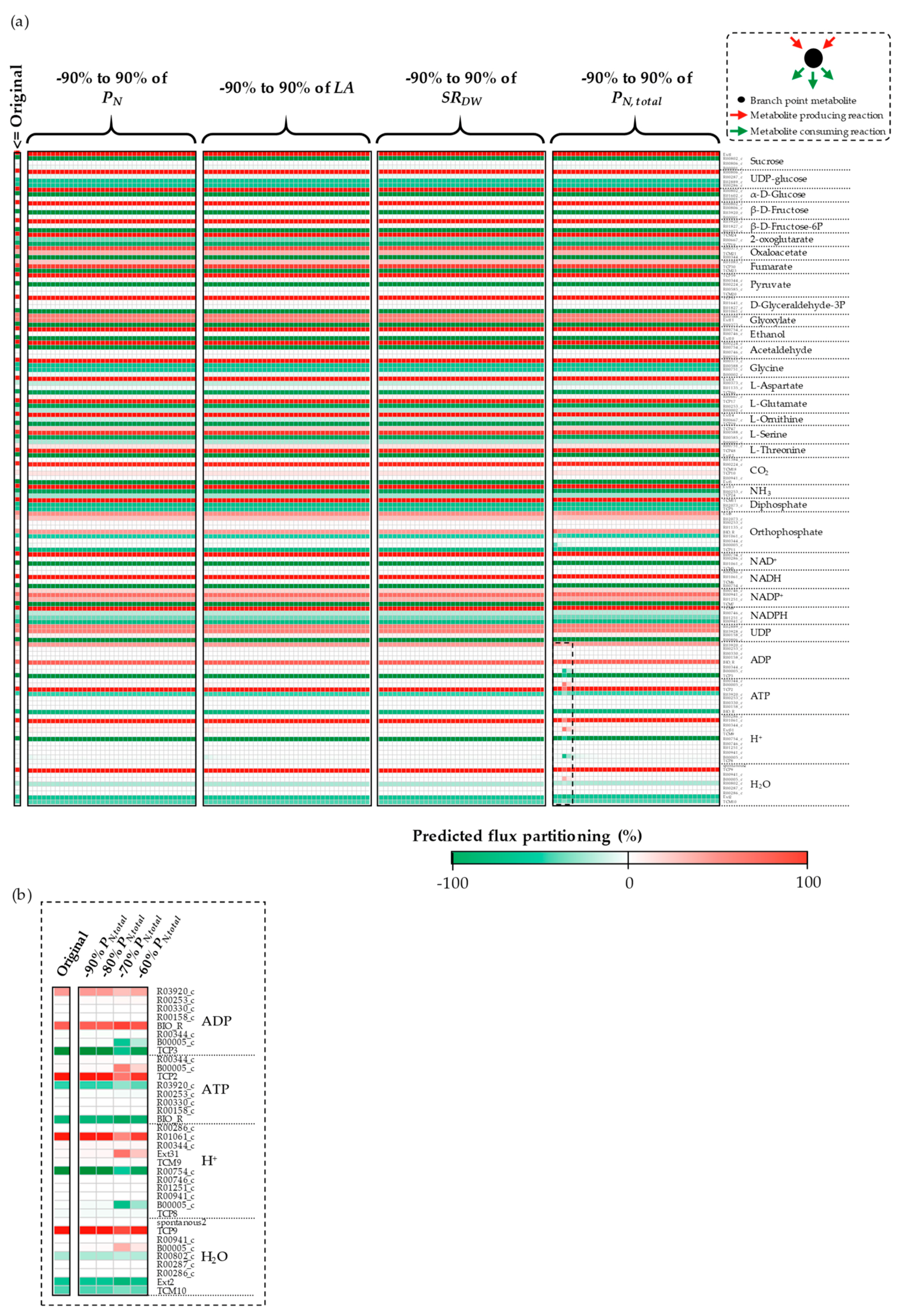
3.1. The Impacts of Substrate–Uptake Related Data on Model Prediction
3.2. The Impacts of Biomass Function-Related Data on Model Prediction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
Appendix A. Supplemental Figures





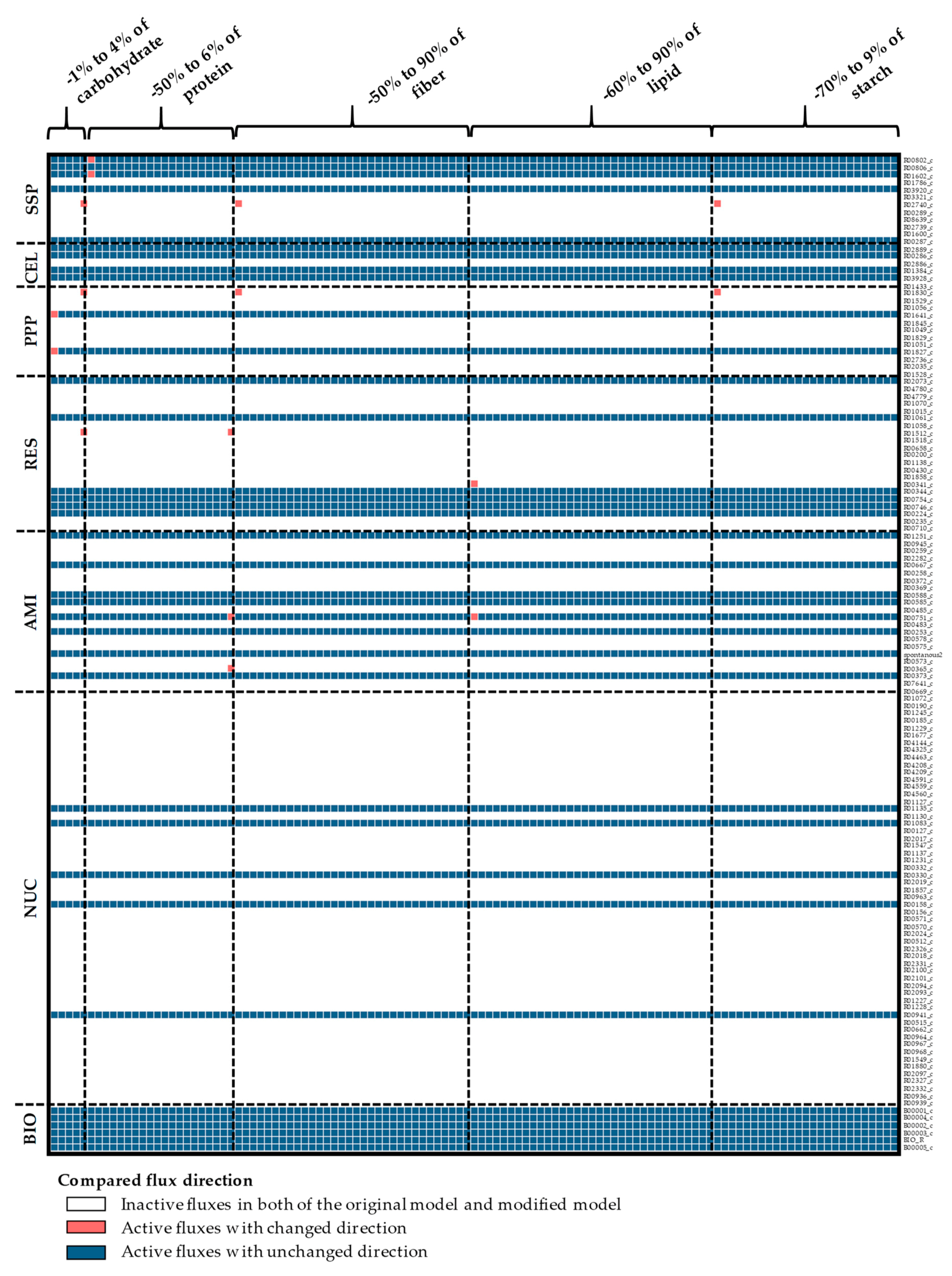
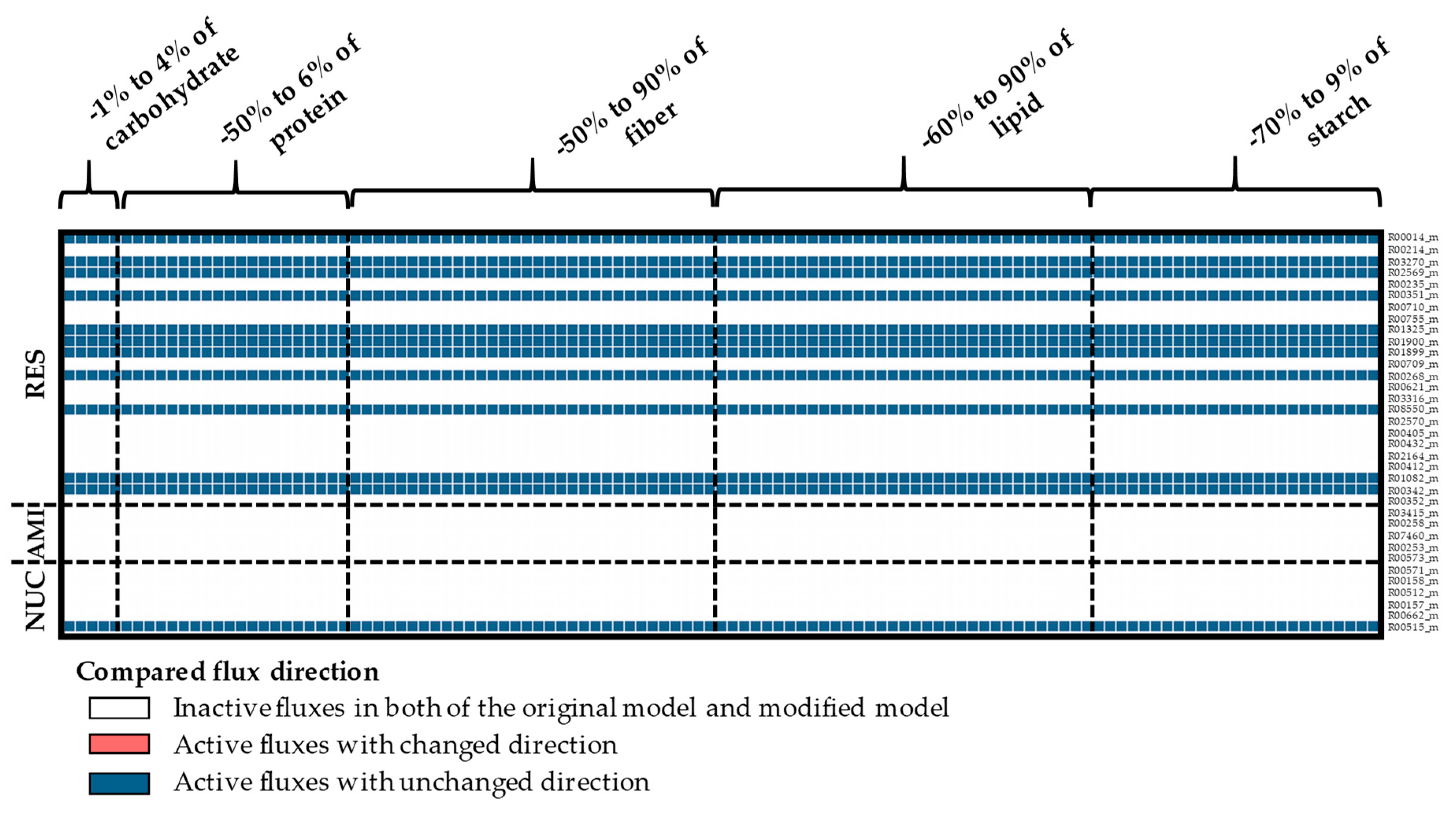
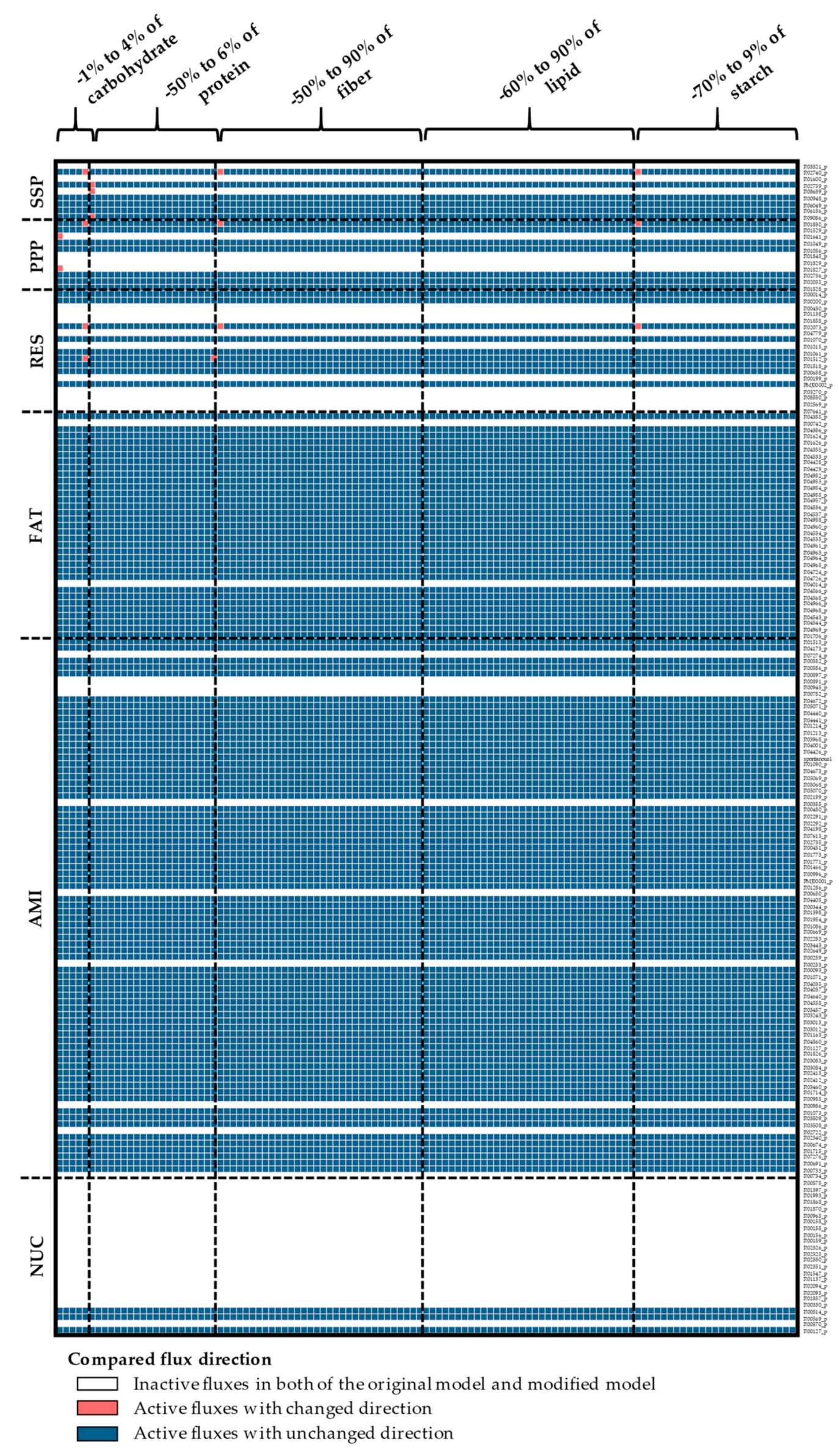


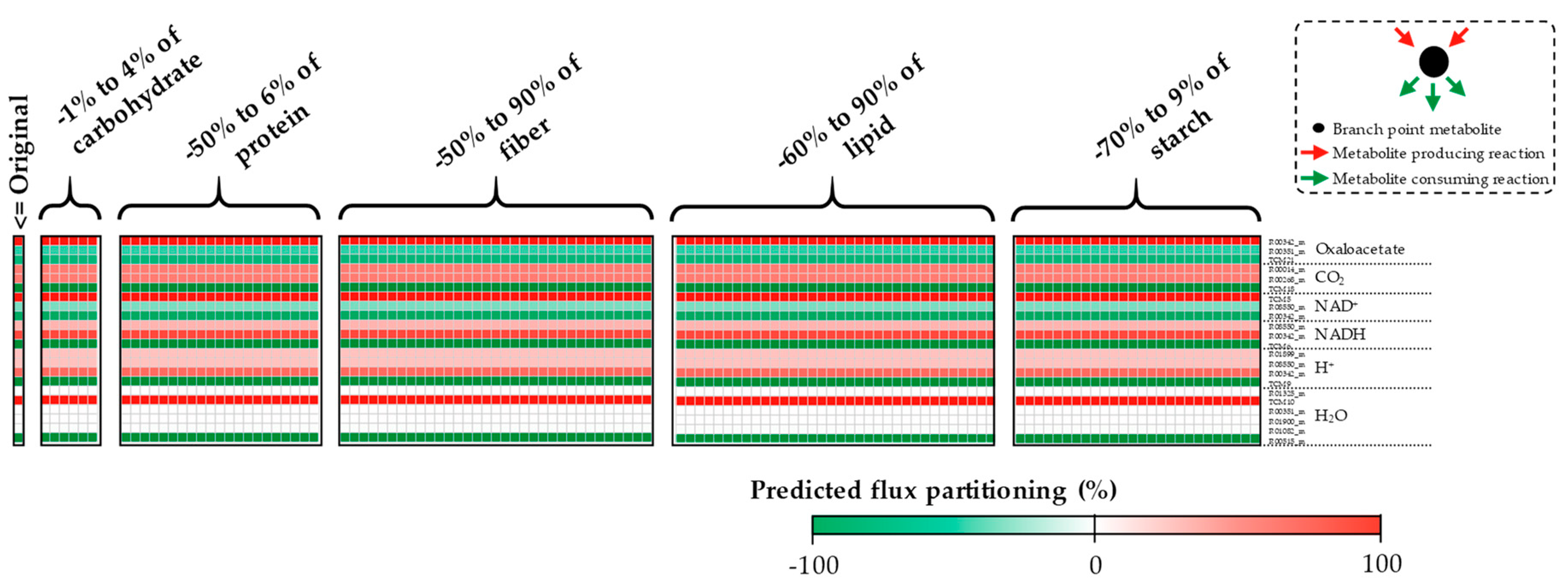


References
- Allen, D.K.; Libourel, I.G.; Shachar-Hill, Y. Metabolic flux analysis in plants: Coping with complexity. Plant Cell Environ. 2009, 32, 1241–1257. [Google Scholar] [CrossRef]
- Orth, J.D.; Thiele, I.; Palsson, B.O. What is flux balance analysis? Nat. Biotechnol. 2010, 28, 245–248. [Google Scholar] [CrossRef]
- Lakshmanan, M.; Zhang, Z.; Mohanty, B.; Kwon, J.Y.; Choi, H.Y.; Nam, H.J.; Kim, D.I.; Lee, D.Y. Elucidating rice cell metabolism under flooding and drought stresses using flux-based modeling and analysis. Plant Physiol. 2013, 162, 2140–2150. [Google Scholar] [CrossRef] [PubMed]
- Chiewchankaset, P.; Siriwat, W.; Suksangpanomrung, M.; Boonseng, O.; Meechai, A.; Tanticharoen, M.; Kalapanulak, S.; Saithong, T. Understanding carbon utilization routes between high and low starch-producing cultivars of cassava through flux balance analysis. Sci. Rep. 2019, 9, 2964. [Google Scholar] [CrossRef]
- Colombié, S.; Nazaret, C.; Bénard, C.; Biais, B.; Mengin, V.; Solé, M.; Fouillen, L.; Dieuaide-Noubhani, M.; Mazat, J.P.; Beauvoit, B.; et al. Modelling central metabolic fluxes by constraint-based optimization reveals metabolic reprogramming of developing Solanum lycopersicum (tomato) fruit. Plant J. 2015, 81, 24–39. [Google Scholar] [CrossRef]
- Hay, J.O.; Schwender, J. Metabolic network reconstruction and flux variability analysis of storage synthesis in developing oilseed rape (Brassica napus L.) embryos. Plant J. 2011, 67, 526–541. [Google Scholar] [CrossRef] [PubMed]
- Rolletschek, H.; Grafahrend-Belau, E.; Munz, E.; Radchuk, V.; Kartausch, R.; Tschiersch, H.; Melkus, G.; Schreiber, F.; Jakob, P.M.; Borisjuk, L. Metabolic architecture of the cereal grain and its relevance to maximize carbon use efficiency. Plant Physiol. 2015, 169, 1698–1713. [Google Scholar] [CrossRef] [PubMed]
- Collakova, E.; Yen, J.Y.; Senger, R.S. Are we ready for genome-scale modeling in plants? Plant Sci. 2012, 191, 53–70. [Google Scholar] [CrossRef]
- Zuniga, C.; Levering, J.; Antoniewicz, M.R.; Guarnieri, M.T.; Betenbaugh, M.J.; Zengler, K. Predicting dynamic metabolic demands in the photosynthetic eukaryote Chlorella vulgaris. Plant Physiol. 2018, 176, 450–462. [Google Scholar] [CrossRef] [PubMed]
- Brandl, J.; Andersen, M.R. Current state of genome-scale modeling in filamentous fungi. Biotechnol. Lett. 2015, 37, 1131–1139. [Google Scholar] [CrossRef]
- Bro, C.; Regenberg, B.; Förster, J.; Nielsen, J. In silico aided metabolic engineering of Saccharomyces cerevisiae for improved bioethanol production. Metab. Eng. 2006, 8, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Edwards, J.S.; Ibarra, R.U.; Palsson, B.O. In silico predictions of Escherichia coli metabolic capabilities are consistent with experimental data. Nat. Biotechnol. 2001, 19, 125–130. [Google Scholar]
- Edwards, J.S.; Palsson, B.O. Systems properties of the Haemophilus influenzaeRd metabolic genotype. J. Biol. Chem. 1999, 274, 17410–17416. [Google Scholar] [CrossRef] [PubMed]
- Edwards, J.S.; Palsson, B.O. The Escherichia coli MG1655 in silico metabolic genotype: Its definition, characteristics, and capabilities. Proc. Natl. Acad. Sci. USA 2000, 97, 5528–5533. [Google Scholar] [CrossRef]
- Förster, J.; Famili, I.; Fu, P.; Palsson, B.O.; Nielsen, J. Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res. 2003, 13, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Rau, M.H.; Zeidan, A.A. Constraint-based modeling in microbial food biotechnology. Biochem. Soc. Trans. 2018, 46, 249–260. [Google Scholar] [CrossRef]
- Teusink, B.; Wiersma, A.; Molenaar, D.; Francke, C.; de Vos, W.M.; Siezen, R.J.; Smid, E.J. Analysis of growth of Lactobacillus plantarum WCFS1 on a complex medium using a genome-scale metabolic model. J. Biol. Chem. 2006, 281, 40041–40048. [Google Scholar] [CrossRef]
- Poolman, M.G.; Miguet, L.; Sweetlove, L.J.; Fell, D.A. A genome-scale metabolic model of Arabidopsis and some of its properties. Plant Physiol. 2009, 151, 1570–1581. [Google Scholar] [CrossRef]
- de Oliveira Dal’Molin, C.G.; Quek, L.E.; Palfreyman, R.W.; Brumbley, S.M.; Nielsen, L.K. AraGEM, a genome-scale reconstruction of the primary metabolic network in Arabidopsis. Plant Physiol. 2010, 152, 579–589. [Google Scholar] [CrossRef]
- Saha, R.; Suthers, P.F.; Maranas, C.D. Zea mays iRS1563: A comprehensive genome-scale metabolic reconstruction of maize metabolism. PLoS ONE 2011, 6, e21784. [Google Scholar]
- Pilalis, E.; Chatziioannou, A.; Thomasset, B.; Kolisis, F. An in silico compartmentalized metabolic model of Brassica napus enables the systemic study of regulatory aspects of plant central metabolism. Biotechnol. Bioeng. 2011, 108, 1673–1682. [Google Scholar] [CrossRef]
- Grafahrend-Belau, E.; Schreiber, F.; Koschützki, D.; Junker, B.H. Flux balance analysis of barley seeds: A computational approach to study systemic properties of central metabolism. Plant Physiol. 2009, 149, 585–598. [Google Scholar] [CrossRef]
- Hamilton, J.J.; Reed, J.L. Software platforms to facilitate reconstructing genome-scale metabolic networks. Environ. Microbiol. 2014, 16, 49–59. [Google Scholar] [CrossRef]
- Thiele, I.; Palsson, B.O. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat. Protoc. 2010, 5, 93–121. [Google Scholar] [CrossRef] [PubMed]
- Zomorrodi, A.R.; Suthers, P.F.; Ranganathan, S.; Maranas, C.D. Mathematical optimization applications in metabolic networks. Metab. Eng. 2012, 14, 672–686. [Google Scholar] [CrossRef] [PubMed]
- Cheung, C.M.; Williams, T.C.; Poolman, M.G.; Fell, D.A.; Ratcliffe, R.G.; Sweetlove, L.J. A method for accounting for maintenance costs in flux balance analysis improves the prediction of plant cell metabolic phenotypes under stress conditions. Plant J. 2013, 75, 1050–1061. [Google Scholar] [CrossRef]
- Feist, A.M.; Palsson, B.O. The biomass objective function. Curr. Opin. Microbiol. 2010, 13, 344–349. [Google Scholar] [CrossRef] [PubMed]
- Schwender, J.; Hay, J.O. Predictive modeling of biomass component tradeoffs in Brassica napus developing oilseeds based on in silico manipulation of storage metabolism. Plant Physiol. 2012, 160, 1218–1236. [Google Scholar] [CrossRef] [PubMed]
- Feist, A.M.; Henry, C.S.; Reed, J.L.; Krummenacker, M.; Joyce, A.R.; Karp, P.D.; Broadbelt, L.J.; Hatzimanikatis, V.; Palsson, B.O. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol. Syst. Biol. 2007, 3, 121. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Cheung, C.Y.M.; Hilbers, P.A.J.; van Riel, N.A.W. Flux balance analysis of plant metabolism: The effect of biomass composition and model structure on model predictions. Front Plant Sci. 2016, 7, 537. [Google Scholar] [CrossRef] [PubMed]
- Nookaew, I.; Jewett, M.C.; Meechai, A.; Thammarongtham, C.; Laoteng, K.; Cheevadhanarak, S.; Nielsen, J.; Bhumiratana, S. The genome-scale metabolic model iIN800 of Saccharomyces cerevisiae and its validation: A scaffold to query lipid metabolism. BMC Syst. Biol. 2008, 2, 71. [Google Scholar] [CrossRef] [PubMed]
- Dikicioglu, D.; Kırdar, B.; Oliver, S.G. Biomass composition: The “elephant in the room” of metabolic modelling. Metabolomics 2015, 11, 1690–1701. [Google Scholar] [CrossRef] [PubMed]
- Howeler, R.H.; Lutaladio, N.; Thomas, G. Cassava, a 21st century crop. In Save and Grow Cassava: A Guide to Sustainable Production Intensification; Howeler, R.H., Lutaladio, N., Thomas, G., Eds.; FAO: Roma, Italy, 2013; pp. 1–18. [Google Scholar]
- Jorge, M.A. Regeneration guidelines: Cassava. In Crop Specific Regeneration Guidelines [CD-ROM]; Dulloo, M.E., Thormann, I., Jorge, M.A., Hanson, J., Eds.; CGIAR Systemwide Genetic Resource Programme (SGRS), Global Crop Diversity Trust: Rome, Italy, 2008; p. 9. [Google Scholar]
- Meléndez-Hevia, E.; de Paz-Lugo, P. Branch-point stoichiometry can generate weak links in metabolism: The case of glycine biosynthesis. J. Biosci. 2008, 33, 771–780. [Google Scholar] [CrossRef]
- Schellenberger, J.; Que, R.; Fleming, R.M.; Thiele, I.; Orth, J.D.; Feist, A.M.; Zielinski, D.C.; Bordbar, A.; Lewis, N.E.; Rahmanian, S.; et al. Quantitative prediction of cellular metabolism with constraint-based models: The COBRA Toolbox v2.0. Nat. Protoc. 2011, 6, 1290–1307. [Google Scholar] [CrossRef]
- El-Sharkawy, M.A.; De Tafur, S.M. Comparative photosynthesis, growth, productivity, and nutrient use efficiency among tall- and short-stemmed rain-fed cassava cultivars. Photosynthetica 2010, 48, 173–188. [Google Scholar] [CrossRef]
- Alves, A.A.C. Cassava botany and physiology. In Cassava: Biology, Production and Utilization; Hillocks, R.J., Thresh, J.M., Bellotti, A.C., Eds.; CAB International Publishing: Wallingford, UK, 2002; pp. 67–89. [Google Scholar]
- Mahon, J.D.; Lowe, S.B.; Hunt, L.A.; Thiagarajah, M. Environmental effects on photosynthesis and transpiration in attached leaves of cassava (Manihot esculenta Crantz). Photosynthetica 1977, 11, 121–130. [Google Scholar]
- Edwards, G.E.; Sheta, E.; Moore, B.D.; Dai, Z.; Franceschi, V.R.; Cheng, S.H.; Lin, C.H.; Ku, M.S. Photosynthetic characteristics of cassava (Manihot esculenta Crantz), a C3 species with chlorenchymatous bundle sheath cells. Plant Cell Physiol. 1990, 31, 1199–1206. [Google Scholar]
- El-Sharkawy, M.A.; Cock, J.H. Photosynthesis of cassava (Manihot esculenta). Exp Agr. 1990, 26, 325–340. [Google Scholar] [CrossRef]
- Ramanujam, T.; Indira, P. Canopy structure on growth and development of cassava (Manihot esculenta Crantz). Turrialba 1983, 33, 321–327. [Google Scholar]
- El-Sharkawy, M.A.; Cock, J.H.; Lynam, J.K.; del Pilar Hernàndez, A.; Cadavid, L.F.L. Relationships between biomass, root-yield and single-leaf photosynthesis in field-grown cassava. Field Crops Res. 1990, 25, 183–201. [Google Scholar] [CrossRef]
- Boonseng, O.; Tungsakol, S.; Chuthangkha, S.; Hansethasuk, J. Evaluation of cassava germplasm for database in breeding and utilization. In Cassava Research Report in 1999-2001 (4215500022-4415500060); Department of Agriculture, Planning and Technical Division, Department of Agriculture (DOA): Bangkok, Thailand, 1999; pp. 73–106. [Google Scholar]
- Nuwamanya, E.; Baguma, Y.; Kawuki, R.S.; Rubaihayo, P.R. Quantification of starch physicochemical characteristics in a cassava segregating population. Afr. Crop Sci. J. 2008, 16, 191–202. [Google Scholar] [CrossRef]
- Stincone, A.; Prigione, A.; Cramer, T.; Wamelink, M.M.; Campbell, K.; Cheung, E.; Olin-Sandoval, V.; Grüning, N.M.; Krüger, A.; Tauqeer Alam, M.; et al. The return of metabolism: Biochemistry and physiology of the pentose phosphate pathway. Biol. Rev. Camb. Philos. Soc. 2015, 90, 927–963. [Google Scholar] [PubMed]
- De Souza, A.P.; Massenburg, L.N.; Jaiswal, D.; Cheng, S.; Shekar, R.; Long, S.P. Rooting for cassava: Insights into photosynthesis and associated physiology as a route to improve yield potential. New Phytol. 2017, 213, 50–65. [Google Scholar] [CrossRef] [PubMed]

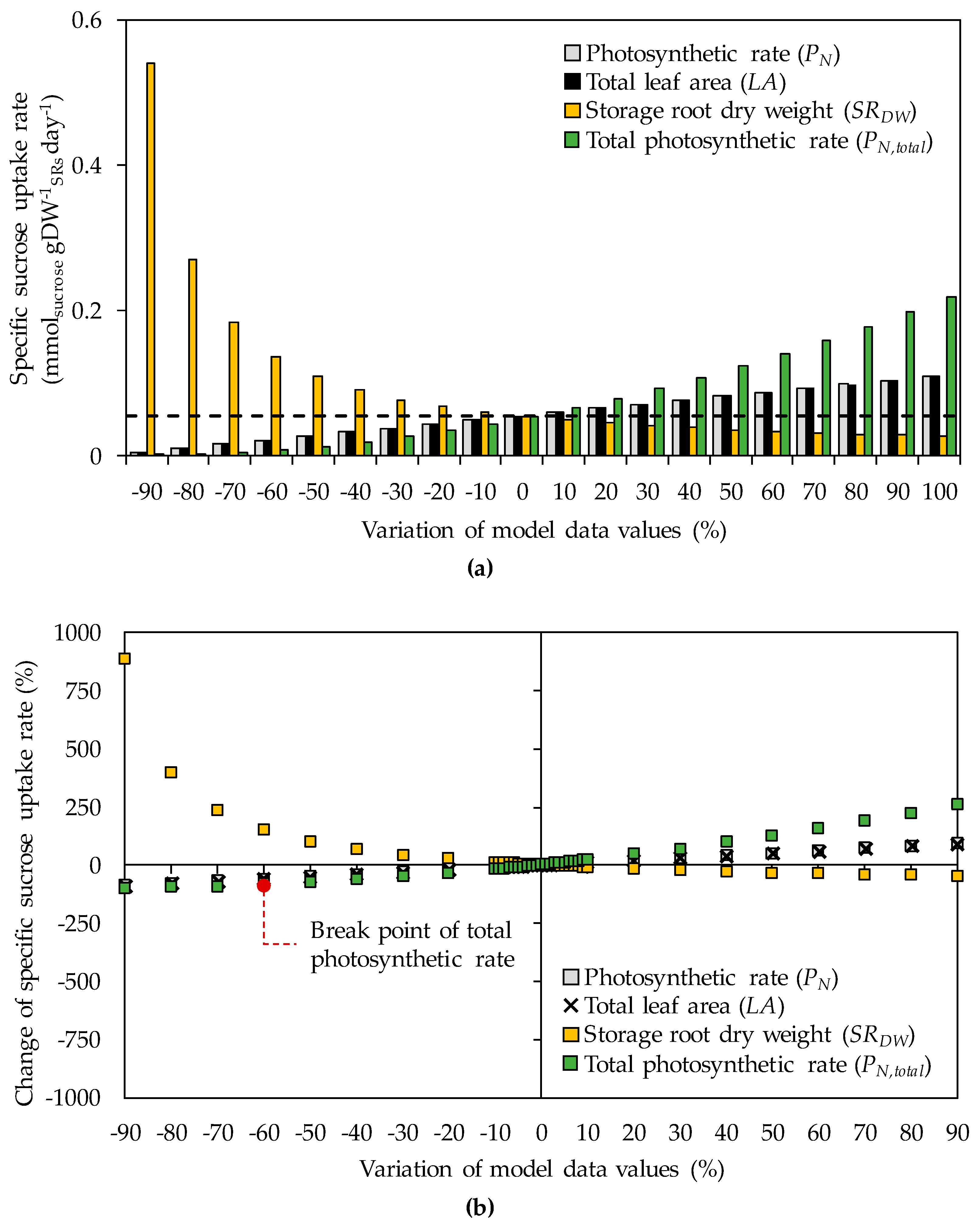
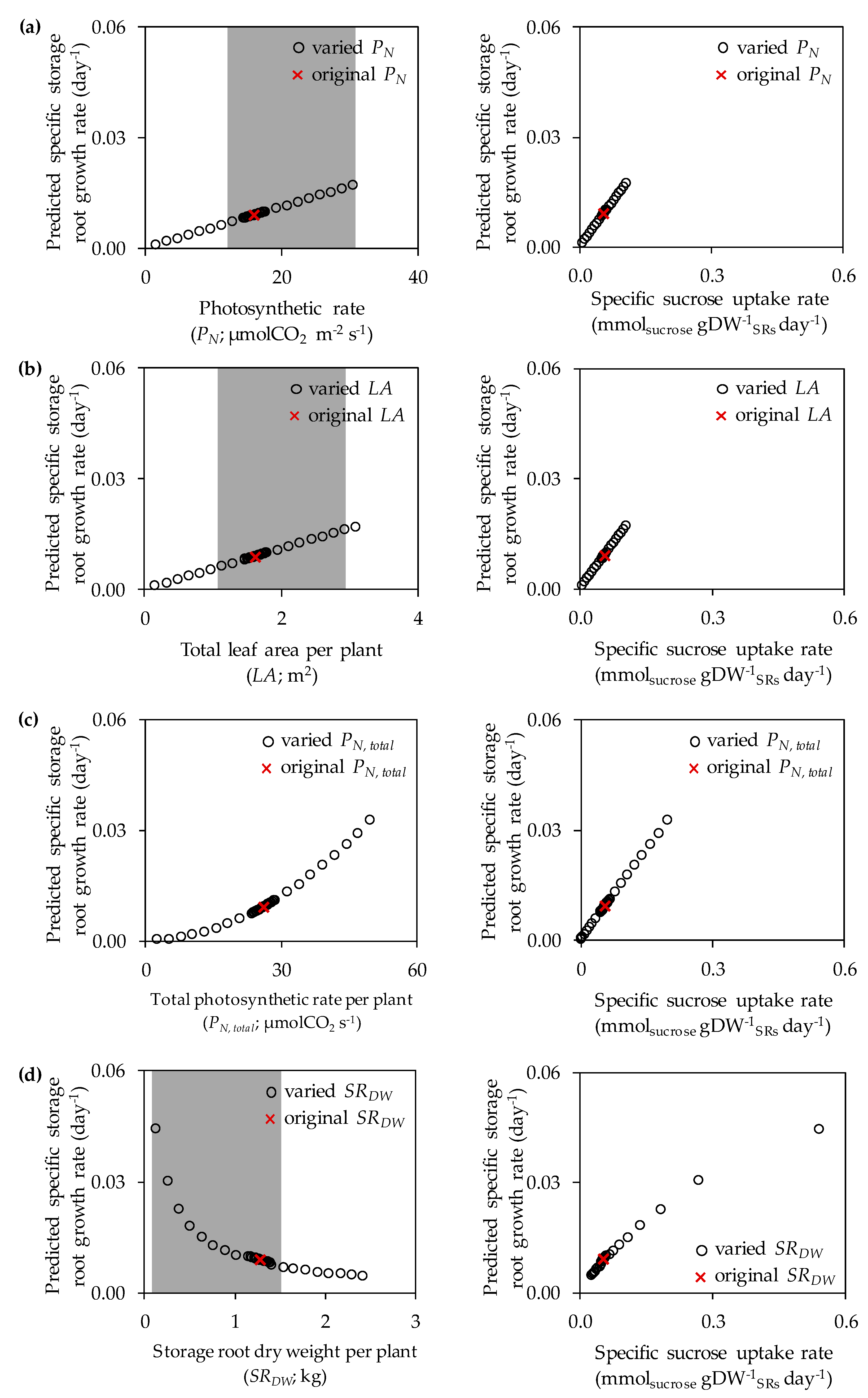
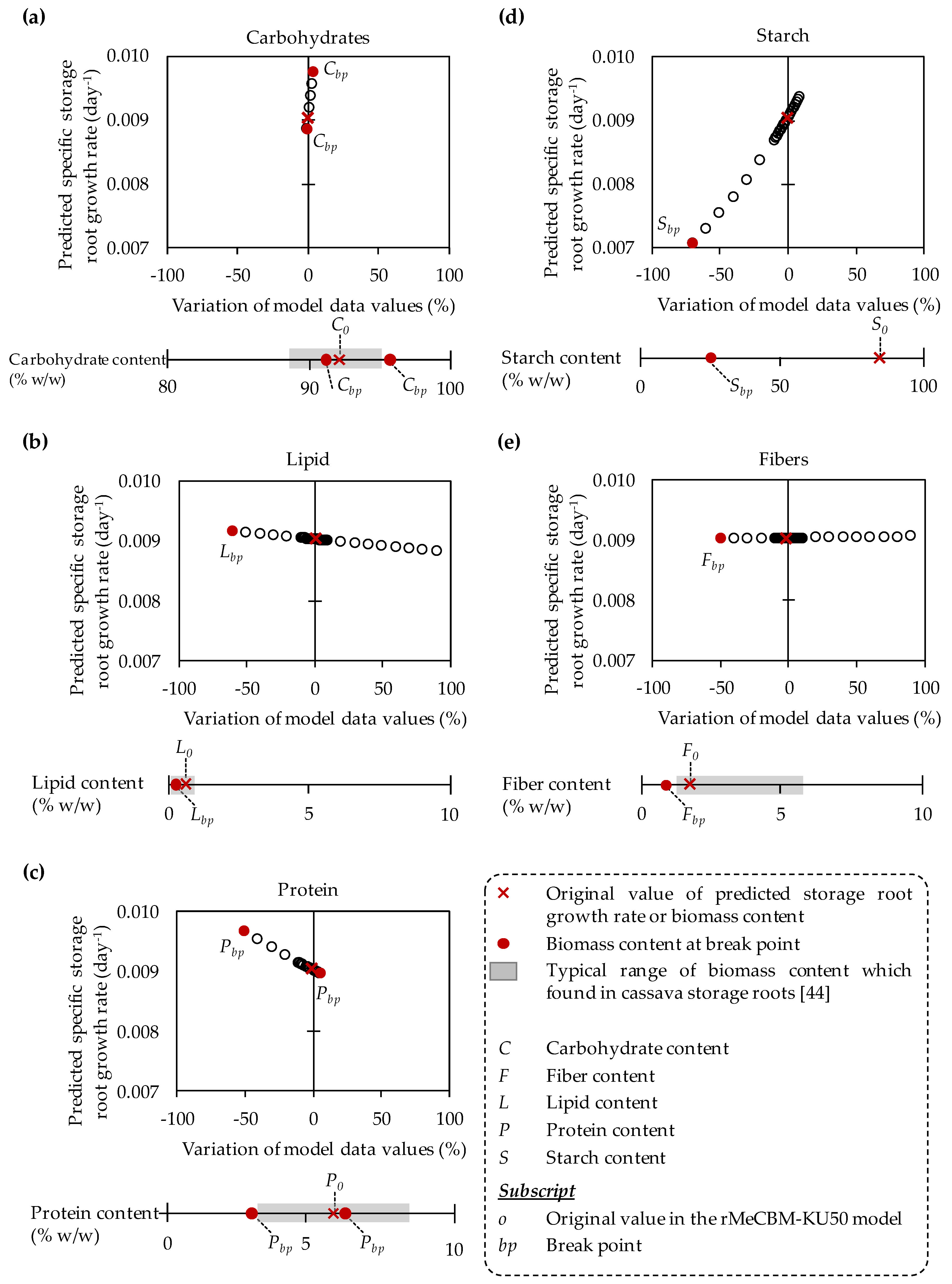


| Model Input Data | Original Values a | Range of Values in Sensitivity Analysis b |
|---|---|---|
| Specific sucrose uptake rate (mmolsucrose gDW−1SRs day−1) | 0.0548 | 0.0005–0.5400 d |
| Photosynthetic rate (µmolCO2 m−2 s−1) | 16.08 | 1.61–30.55 |
| Total leaf area (m2) | 1.63 | 0.16–3.10 |
| Storage root dry weight (kgDW) | 1.28 | 0.13–2.43 |
| Total photosynthetic rate (µmolCO2 s−1) | 26.21 | 0.26–94.91 c |
| Model Input Data | Original Values a | Range of Values in Sensitivity Analysis b |
|---|---|---|
| Carbohydrates | 92.07 | 9.21–99.44 |
| Starch | 84.06 | 8.41–91.62 |
| Proteins | 5.78 | 0.58–10.99 |
| Fibers | 1.69 | 0.17–3.22 |
| Lipid | 0.45 | 0.05–0.86 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chiewchankaset, P.; Kalapanulak, S.; Saithong, T. Extended Utilization of Constraint-Based Metabolic Model in a Long-Growing Crop. Processes 2019, 7, 259. https://doi.org/10.3390/pr7050259
Chiewchankaset P, Kalapanulak S, Saithong T. Extended Utilization of Constraint-Based Metabolic Model in a Long-Growing Crop. Processes. 2019; 7(5):259. https://doi.org/10.3390/pr7050259
Chicago/Turabian StyleChiewchankaset, Porntip, Saowalak Kalapanulak, and Treenut Saithong. 2019. "Extended Utilization of Constraint-Based Metabolic Model in a Long-Growing Crop" Processes 7, no. 5: 259. https://doi.org/10.3390/pr7050259





