Probing the Reactivity of [4Fe-4S] Fumarate and Nitrate Reduction (FNR) Regulator with O2 and NO: Increased O2 Resistance and Relative Specificity for NO of the [4Fe-4S] L28H FNR Cluster
Abstract
:1. Introduction
2. Experimental Section
2.1. Purification of EcFNR Proteins
2.2. Purification of AfFNR Proteins
2.3. Crystallization and X-ray Structure Solution of L28H-AfFNR
2.4. Spectroscopy
2.5. Rapid Reaction Kinetics of FNR Proteins
2.6. Mass Spectrometry
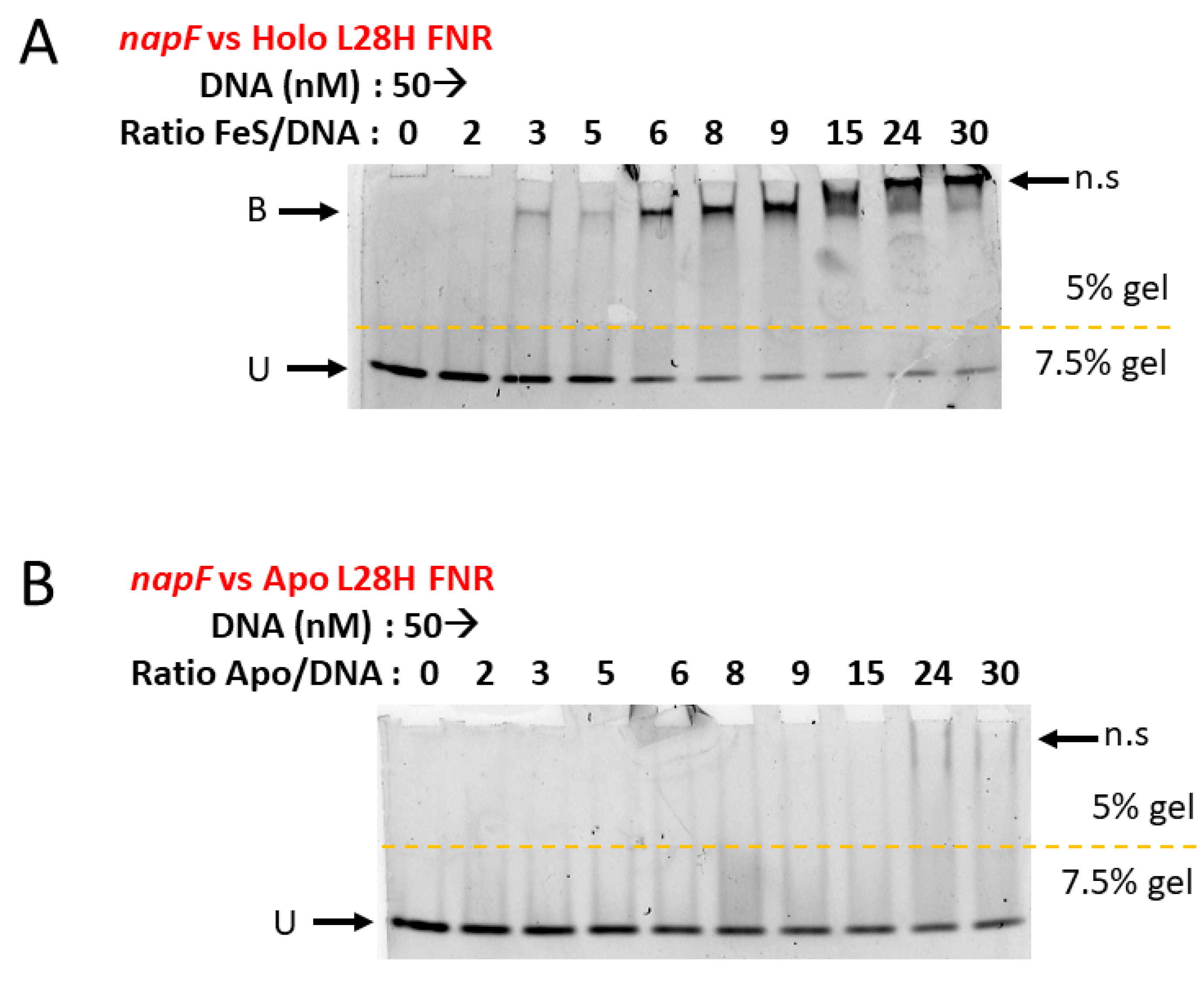
2.7. Electrophoretic Mobility Shift Assays (EMSAs)
2.8. Other Analytical Methods
2.9. Molecular Dynamics (MD) Simulations of wt and L28H-AfFNR
3. Results
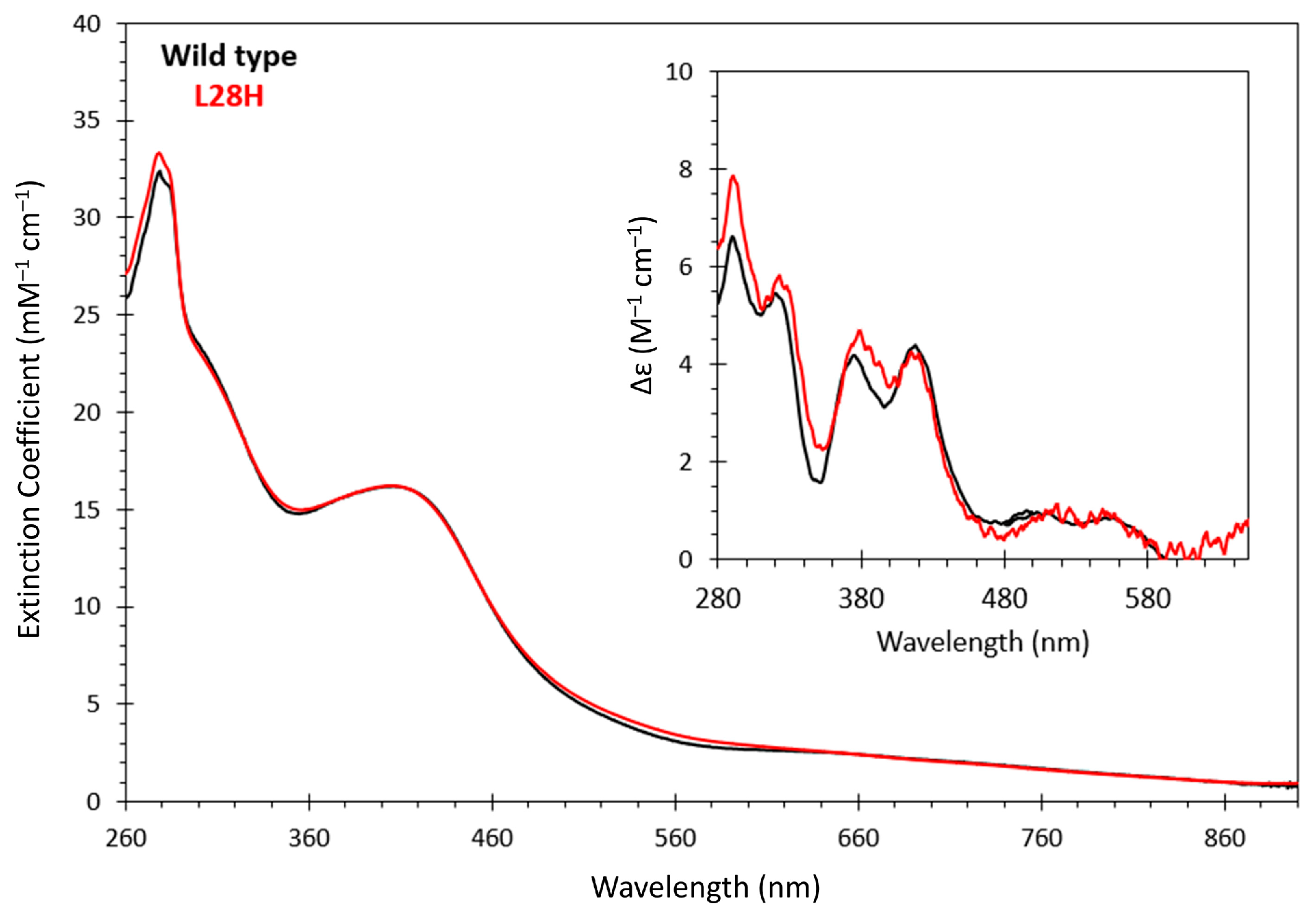
3.1. Spectroscopic, O2 Reactivity and DNA-Binding Properties of [4Fe-4S] L28H-EcFNR
3.2. [4Fe-4S] L28H-EcFNR Reacts with Multiple NO Molecules
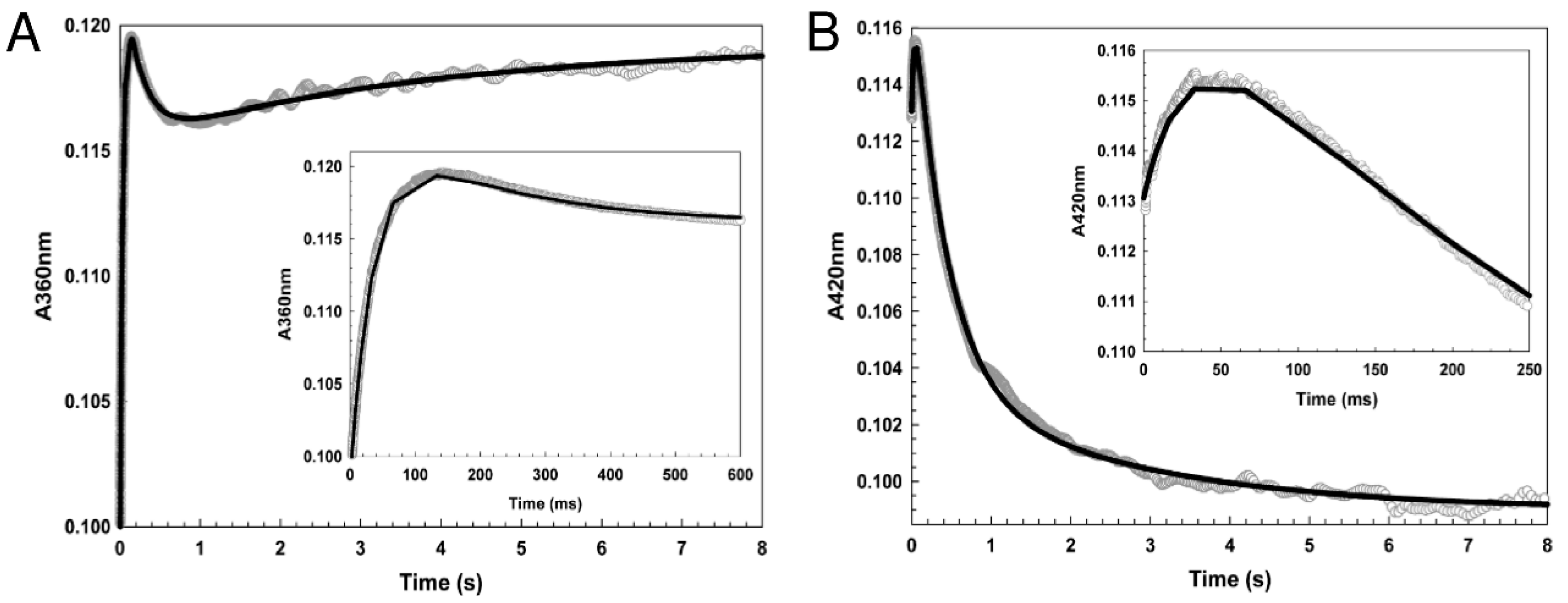
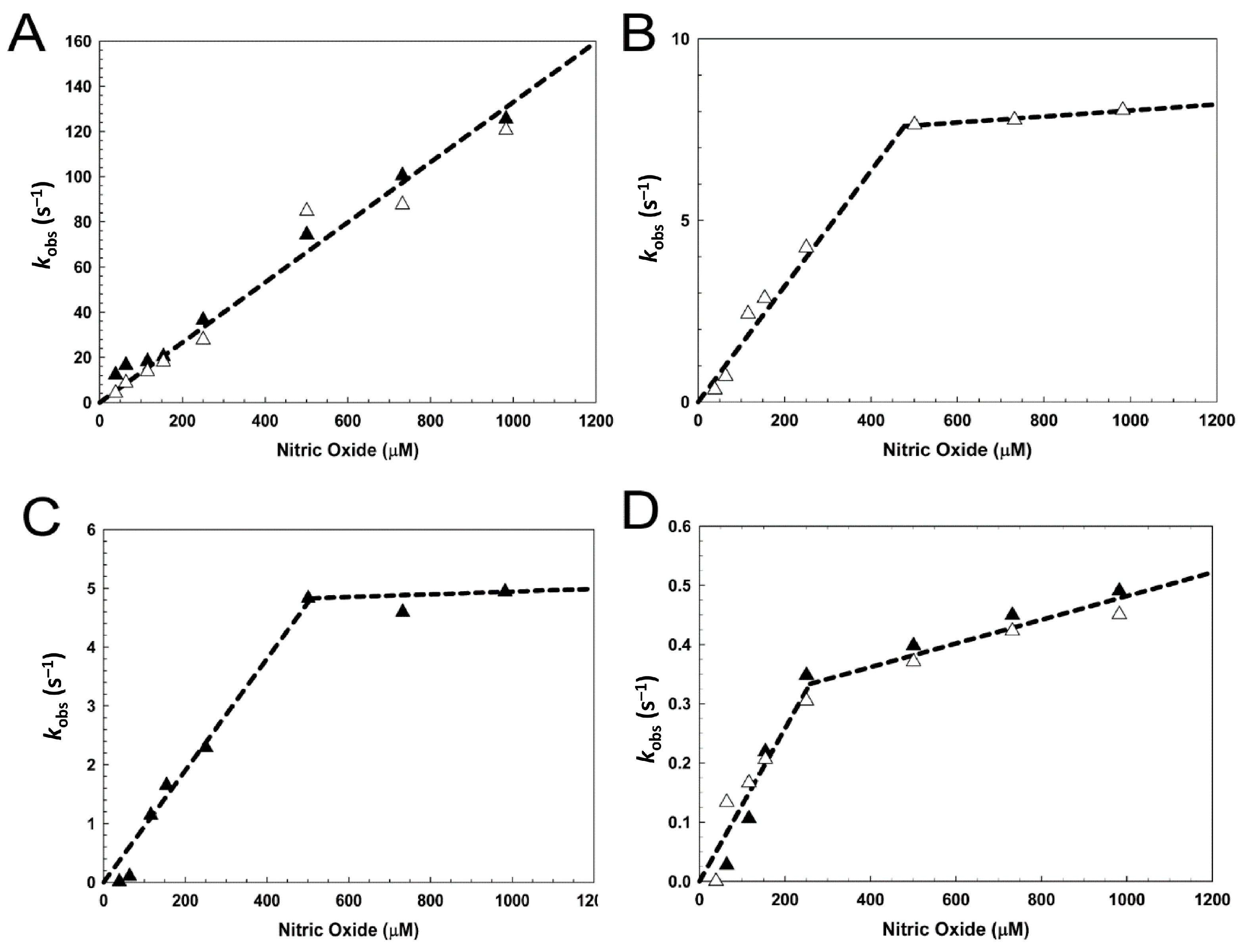
3.3. [4Fe-4S] L28H-EcFNR Reacts Rapidly with NO
3.4. Nitrosylation of L28H-EcFNR Results in Cluster Sulfide Oxidation and Formation of Protein-Associated Persulfides
3.5. Crystal Structure and O2 Reactivity of [4Fe-4S] L28H-AfFNR
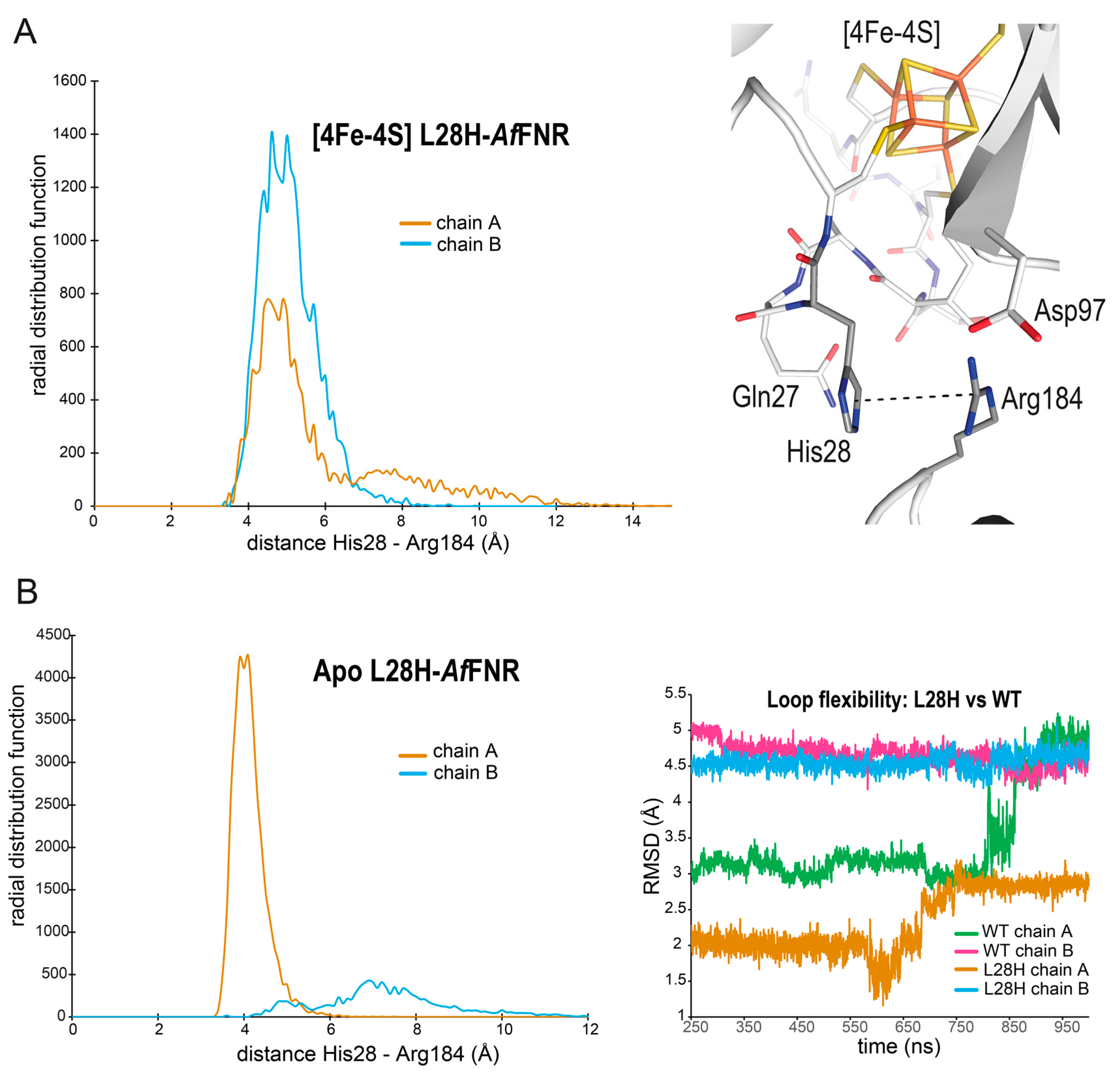
3.6. MD Simulations of wt and L28H-AfFNR Provide a Possible Explanation for the O2 Resistance Phenotype of the Variant
4. Discussion
5. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fleischhacker, A.S.; Kiley, P.J. Iron-containing transcription factors and their roles as sensors. Curr. Opin. Chem. Biol. 2011, 15, 335–341. [Google Scholar] [CrossRef]
- Crack, J.C.; Green, J.; Thomson, A.J.; Le Brun, N.E. Iron-sulfur clusters as biological sensors: The chemistry of reactions with molecular oxygen and nitric oxide. Acc. Chem. Res. 2014, 47, 3196–3205. [Google Scholar] [CrossRef]
- Guest, J.R.; Russell, G.C. Complexes and complexities of the citric acid cycle in Escherichia coli. Curr. Top. Cell Regul. 1992, 33, 231–247. [Google Scholar]
- Khoroshilova, N.; Beinert, H.; Kiley, P.J. Association of a polynuclear iron-sulfur center with a mutant FNR protein enhances DNA-binding. Proc. Natl. Acad. Sci. USA 1995, 92, 2499–2503. [Google Scholar] [CrossRef]
- Guest, J.R. The Leeuwenhoek Lecture, 1995. Adaptation to life without oxygen. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1995, 350, 189–202. [Google Scholar]
- Khoroshilova, N.; Popescu, C.; Munck, E.; Beinert, H.; Kiley, P.J. Iron-sulfur cluster disassembly in the FNR protein of Escherichia coli by O2: [4Fe-4S] to [2Fe-2S] conversion with loss of biological activity. Proc. Natl. Acad. Sci. USA 1997, 94, 6087–6092. [Google Scholar] [CrossRef]
- Beinert, H.; Kiley, P.J. Fe-S proteins in sensing and regulatory functions. Curr. Opin. Chem. Biol. 1999, 3, 152–157. [Google Scholar] [CrossRef]
- Guest, J.R. Oxygen-regulated gene expression in Escherichia coli. The 1992 Marjory Stephenson Prize Lecture. J. Gen. Microbiol. 1992, 138, 2253–2263. [Google Scholar] [CrossRef]
- Dunn, A.K. Vibrio fischeri metabolism: Symbiosis and beyond. Adv. Microb. Physiol. 2012, 61, 37–68. [Google Scholar]
- Septer, A.N.; Bose, J.L.; Dunn, A.K.; Stabb, E.V. FNR-mediated regulation of bioluminescence and anaerobic respiration in the light-organ symbiont Vibrio fischeri. FEMS Microbiol. Lett. 2010, 306, 72–81. [Google Scholar] [CrossRef]
- Volbeda, A.; Darnault, C.; Renoux, O.; Nicolet, Y.; Fontecilla-Camps, J.C. The crystal structure of the global anaerobic transcriptional regulator FNR explains its extremely fine-tuned monomer-dimer equilibrium. Sci. Adv. 2015, 1, e1501086. [Google Scholar] [CrossRef]
- Green, J.; Guest, J.R. A role for iron in transcriptional activation by FNR. FEBS Lett. 1993, 329, 55–58. [Google Scholar] [CrossRef]
- Green, J.; Irvine, A.S.; Meng, W.; Guest, J.R. FNR-DNA interactions at natural and semi-synthetic promoters. Mol. Microbiol. 1996, 19, 125–137. [Google Scholar] [CrossRef]
- Kiley, P.J.; Beinert, H. Oxygen sensing by the global regulator, FNR: The role of the iron-sulfur cluster. FEMS Microbiol. Revs. 1999, 22, 341–352. [Google Scholar] [CrossRef]
- Lazazzera, B.A.; Bates, D.M.; Kiley, P.J. The activity of the Escherichia coli transcription factor FNR is regulated by a change in oligomeric state. Genes Dev. 1993, 7, 1993–2005. [Google Scholar] [CrossRef]
- Crack, J.C.; Gaskell, A.A.; Green, J.; Cheesman, M.R.; Le Brun, N.E.; Thomson, A.J. Influence of the environment on the [4Fe-4S]2+ to [2Fe-2S]2+ cluster switch in the transcriptional regulator FNR. J. Am. Chem. Soc. 2008, 130, 1749–1758. [Google Scholar] [CrossRef]
- Crack, J.C.; Green, J.; Cheesman, M.R.; Le Brun, N.E.; Thomson, A.J. Superoxide-mediated amplification of the oxygen-induced switch from [4Fe-4S] to [2Fe-2S] clusters in the transcriptional regulator FNR. Proc. Natl. Acad. Sci. USA 2007, 104, 2092–2097. [Google Scholar] [CrossRef]
- Crack, J.C.; Thomson, A.J.; Le Brun, N.E. Mass spectrometric identification of intermediates in the O2-driven [4Fe-4S] to [2Fe-2S] cluster conversion in FNR. Proc. Natl. Acad. Sci. USA 2017, 114, E3215–E3223. [Google Scholar] [CrossRef]
- Popescu, C.V.; Bates, D.M.; Beinert, H.; Munck, E.; Kiley, P.J. Mössbauer spectroscopy as a tool for the study of activation/inactivation of the transcription regulator FNR in whole cells of Escherichia coli. Proc. Natl. Acad. Sci. USA 1998, 95, 13431–13435. [Google Scholar] [CrossRef]
- Vine, C.E.; Cole, J.A. Unresolved sources, sinks, and pathways for the recovery of enteric bacteria from nitrosative stress. FEMS Microbiol. Lett. 2011, 325, 99–107. [Google Scholar] [CrossRef]
- Nunoshiba, T.; deRojas-Walker, T.; Wishnok, J.S.; Tannenbaum, S.R.; Demple, B. Activation by nitric oxide of an oxidative-stress response that defends Escherichia coli against activated macrophages. Proc. Natl. Acad. Sci. USA 1993, 90, 9993–9997. [Google Scholar] [CrossRef]
- Cruz-Ramos, H.; Crack, J.; Wu, G.G.; Hughes, M.N.; Scott, C.; Thomson, A.J.; Green, J.; Poole, R.K. NO sensing by FNR: Regulation of the Escherichia coli NO-detoxifying flavohaemoglobin, Hmp. EMBO J. 2002, 21, 3235–3244. [Google Scholar] [CrossRef]
- Pullan, S.T.; Gidley, M.D.; Jones, R.A.; Barrett, J.; Stevanin, T.A.; Read, R.C.; Green, J.; Poole, R.K. Nitric oxide in chemostat-cultured Escherichia coli is sensed by Fnr and other global regulators: Unaltered methionine biosynthesis indicates lack of S nitrosation. J. Bacteriol. 2007, 189, 1845–1855. [Google Scholar] [CrossRef]
- Crack, J.C.; Stapleton, M.R.; Green, J.; Thomson, A.J.; Le Brun, N.E. Mechanism of [4Fe-4S](Cys)4 cluster nitrosylation is conserved among NO-responsive regulators. J. Biol. Chem. 2013, 288, 11492–11502. [Google Scholar] [CrossRef]
- Crack, J.C.; Munnoch, J.; Dodd, E.L.; Knowles, F.; Al Bassam, M.M.; Kamali, S.; Holland, A.A.; Cramer, S.P.; Hamilton, C.J.; Johnson, M.K.; et al. NsrR from Streptomyces coelicolor is a nitric oxide-sensing [4Fe-4S] cluster protein with a specialized regulatory function. J. Biol. Chem. 2015, 290, 12689–12704. [Google Scholar] [CrossRef]
- Partridge, J.D.; Bodenmiller, D.M.; Humphrys, M.S.; Spiro, S. NsrR targets in the Escherichia coli genome: New insights into DNA sequence requirements for binding and a role for NsrR in the regulation of motility. Mol. Microbiol. 2009, 73, 680–694. [Google Scholar] [CrossRef]
- Mukhopadhyay, P.; Zheng, M.; Bedzyk, L.A.; LaRossa, R.A.; Storz, G. Prominent roles of the NorR and Fur regulators in the Escherichia coli transcriptional response to reactive nitrogen species. Proc. Natl. Acad. Sci. USA 2004, 101, 745–750. [Google Scholar] [CrossRef]
- Crack, J.C.; Smith, L.J.; Stapleton, M.R.; Peck, J.; Watmough, N.J.; Buttner, M.J.; Buxton, R.S.; Green, J.; Oganesyan, V.S.; Thomson, A.J.; et al. Mechanistic insight into the nitrosylation of the [4Fe-4S] cluster of WhiB-like proteins. J. Am. Chem. Soc. 2011, 133, 1112–1121. [Google Scholar] [CrossRef]
- Jervis, A.J.; Crack, J.C.; White, G.; Artymiuk, P.J.; Cheesman, M.R.; Thomson, A.J.; Le Brun, N.E.; Green, J. The O2 sensitivity of the transcription factor FNR is controlled by Ser24 modulating the kinetics of [4Fe-4S] to [2Fe-2S] conversion. Proc. Natl. Acad. Sci. USA 2009, 106, 4659–4664. [Google Scholar] [CrossRef]
- Bates, D.M.; Popescu, C.V.; Khoroshilova, N.; Vogt, K.; Beinert, H.; Munck, E.; Kiley, P.J. Substitution of leucine 28 with histidine in the Escherichia coli transcription factor FNR results in increased stability of the [4Fe-4S]2+ cluster to oxygen. J. Biol. Chem. 2000, 275, 6234–6240. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Beinert, H. Semi-micro methods for analysis of labile sulfide and of labile sulfide plus sulfane sulfur in unusually stable iron-sulfur proteins. Anal. Biochem. 1983, 131, 373–378. [Google Scholar] [CrossRef]
- Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef]
- Evans, P.R.; Murshudov, G.N. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 2013, 69, 1204–1214. [Google Scholar] [CrossRef]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef]
- Murshudov, G.N.; Skubak, P.; Lebedev, A.A.; Pannu, N.S.; Steiner, R.A.; Nicholls, R.A.; Winn, M.D.; Long, F.; Vagin, A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 355–367. [Google Scholar] [CrossRef]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Kuzmic, P. Program DYNAFIT for the analysis of enzyme kinetic data: Application to HIV proteinase. Anal. Biochem. 1996, 237, 260–273. [Google Scholar] [CrossRef]
- Lewis, R.S.; Deen, W.M. Kinetics of the reaction of nitric oxide with oxygen in aqueous solutions. Chem. Res. Toxicol. 1994, 7, 568–574. [Google Scholar] [CrossRef]
- Saavedra, J.E.; Southan, G.J.; Davies, K.M.; Lundell, A.; Markou, C.; Hanson, S.R.; Adrie, C.; Hurford, W.E.; Zapol, W.M.; Keefer, L.K. Localizing antithrombotic and vasodilatory activity with a novel, ultrafast nitric oxide donor. J. Med. Chem. 1996, 39, 4361–4365. [Google Scholar] [CrossRef]
- Keefer, L.K.; Nims, R.W.; Davies, K.M.; Wink, D.A. “NONOates” (1-substituted diazen-1-ium-1,2-diolates) as nitric oxide donors: Convenient nitric oxide dosage forms. Meth. Enzymol. 1996, 268, 281–293. [Google Scholar]
- Schrödinger Release 2023-1; Schrödinger, LLC: New York, NY, USA, 2023.
- Mouesca, J.M.; Chen, J.L.; Noodleman, L.; Bashford, D.; Case, D.A. Density-functional Poisson-Boltzmann calculations of redox potentials for iron-sulfur clusters. J. Am. Chem. Soc. 1994, 116, 11898–11914. [Google Scholar] [CrossRef]
- Lu, C.; Wu, C.J.; Ghoreishi, D.; Chen, W.; Wang, L.L.; Damm, W.; Ross, G.A.; Dahlgren, M.K.; Russell, E.; Von Bargen, C.D.; et al. OPLS4: Improving force field accuracy on challenging regimes of chemical space. J. Chem. Theory Comput. 2021, 17, 4291–4300. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Schrödinger. Maestro, Desmond-Interoperability Tools; Schrödinger: New York, NY, USA, 2019. [Google Scholar]
- Kiley, P.J.; Reznikoff, W.S. FNR mutants that activate gene expression in the presence of oxygen. J. Bacteriol. 1991, 173, 16–22. [Google Scholar] [CrossRef]
- Vasilieva, S.V.; Streltsova, D.A.; Vlaskina, A.V.; Mikoian, V.D.; Vanin, A.F. Sources of divalent sulfur allow recovery of the Fnr [4Fe-4S]2+ center in Escherichia coli incubated with nitric oxide donors. Biophysics 2012, 57, 166–169. [Google Scholar] [CrossRef]
- Frolov, E.N.; Vanin, A.F. New type of paramagnetic nitrosyl complexes of non-heme iron. Biofizika 1973, 18, 605–610. [Google Scholar]
- Lazazzera, B.A.; Beinert, H.; Khoroshilova, N.; Kennedy, M.C.; Kiley, P.J. DNA binding and dimerization of the Fe-S-containing FNR protein from Escherichia coli are regulated by oxygen. J. Biol. Chem. 1996, 271, 2762–2768. [Google Scholar] [CrossRef]
- Smith, A.D.; Agar, J.N.; Johnson, K.A.; Frazzon, J.; Amster, I.J.; Dean, D.R.; Johnson, M.K. Sulfur transfer from IscS to IscU: The first step in iron-sulfur cluster biosynthesis. J. Am. Chem. Soc. 2001, 123, 11103–11104. [Google Scholar] [CrossRef]
- Zhang, B.; Crack, J.C.; Subramanian, S.; Green, J.; Thomson, A.J.; Le Brun, N.E.; Johnson, M.K. Reversible cycling between cysteine persulfide-ligated [2Fe-2S] and cysteine-ligated [4Fe-4S] clusters in the FNR regulatory protein. Proc. Natl. Acad. Sci. USA 2012, 109, 15734–15739. [Google Scholar] [CrossRef]
- Volbeda, A.; Nicolet, Y.; Fontecilla-Camps, J.C. Fumarate and nitrate reduction regulator (FNR). In Encyclopedia of Inorganic and Bioinorganic Chemistry; Wiley: Hoboken, NJ, USA, 2017. [Google Scholar]
- Heyda, J.; Mason, P.E.; Jungwirth, P. Attractive interactions between side chains of histidine-histidine and histidine-arginine-based cationic dipeptides in water. J. Phys. Chem. B 2010, 114, 8744–8749. [Google Scholar] [CrossRef]
- Mettert, E.L.; Kiley, P.J. Reassessing the structure and function relationship of the O2 sensing transcription factor FNR. Antiox. Redox Signal. 2018, 29, 1830–1840. [Google Scholar] [CrossRef]
- Pellicer Martinez, M.T.; Crack, J.C.; Stewart, M.Y.; Bradley, J.M.; Svistunenko, D.A.; Johnston, A.W.; Cheesman, M.R.; Todd, J.D.; Le Brun, N.E. Mechanisms of iron- and O2-sensing by the [4Fe-4S] cluster of the global iron regulator RirA. Elife 2019, 8, e47804. [Google Scholar] [CrossRef]
- Butler, A.R.; Glidewell, C.; Li, M. Nitrosyl complexes of iron-sulfur clusters. Adv. Inorg. Chem. 1988, 32, 335–393. [Google Scholar]
- Bruckdorfer, R. The basics about nitric oxide. Mol. Aspects Med. 2005, 26, 3–31. [Google Scholar] [CrossRef]
- Bryar, T.R.; Eaton, D.R. Electronic configuration and structure of paramagnetic iron dinitrosyl complexes. Can. J. Chem. 1992, 70, 1917–1926. [Google Scholar] [CrossRef]
- Costanzo, S.; Menage, S.; Purrello, R.; Bonomo, R.P.; Fontecave, M. Re-examination of the formation of dinitrosyl-iron complexes during reaction of S-nitrosothiols with Fe(II). Inorg. Chim. Acta 2001, 318, 1–7. [Google Scholar] [CrossRef]
- Kim, S.O.; Orii, Y.; Lloyd, D.; Hughes, M.N.; Poole, R.K. Anoxic function for the Escherichia coli flavohaemoglobin (Hmp): Reversible binding of nitric oxide and reduction to nitrous oxide. FEBS Lett. 1999, 445, 389–394. [Google Scholar] [CrossRef]
- Pieper, G.M.; Halligan, N.L.; Hilton, G.; Konorev, E.A.; Felix, C.C.; Roza, A.M.; Adams, M.B.; Griffith, O.W. Non-heme iron protein: A potential target of nitric oxide in acute cardiac allograft rejection. Proc. Natl. Acad. Sci. USA 2003, 100, 3125–3130. [Google Scholar] [CrossRef]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and better reference data for improved all-atom structure validation. Prot. Sci. 2018, 27, 293–315. [Google Scholar] [CrossRef]
- Morin, A.; Eisenbraun, B.; Key, J.; Sanschagrin, P.C.; Timony, M.A.; Ottaviano, M.; Sliz, P. Collaboration gets the most out of software. Elife 2013, 2, e01456. [Google Scholar] [CrossRef]



| Before Treatment with NO | After Treatment with NO (~530 μM) a | |||||||
|---|---|---|---|---|---|---|---|---|
| Sample | Conc/ μM | [4Fe-4S]/ μM | S2−/ μM a | S2−/ [4Fe-4S] | S2−/ μM | S2−/ [4Fe-4S] | S0 /μM | S0/ [4Fe-4S] |
| 1 | 44.2 | 25.3 | 101.12 (±3.00) | 4.00 (±0.12) | 23.79 (±0.73) | 0.94 (±0.03) | 96.94 (±11.46) | 3.84 (±0.45) |
| 2 | 62.9 | 36.0 | 143.80 (±4.26) | 4.00 (±0.12) | n.d. | n.d. | 108.89 (±10.14) | 3.03 (±0.28) |
| 3 b | 40.3 | 23.0 | 92.20 (±2.73) | 4.00 (±0.12) | 2.76 (±0.00) | 0.12 (±0.00) | 53.33 (±5.01) | 2.31 (±0.22) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Crack, J.C.; Amara, P.; de Rosny, E.; Darnault, C.; Stapleton, M.R.; Green, J.; Volbeda, A.; Fontecilla-Camps, J.C.; Le Brun, N.E. Probing the Reactivity of [4Fe-4S] Fumarate and Nitrate Reduction (FNR) Regulator with O2 and NO: Increased O2 Resistance and Relative Specificity for NO of the [4Fe-4S] L28H FNR Cluster. Inorganics 2023, 11, 450. https://doi.org/10.3390/inorganics11120450
Crack JC, Amara P, de Rosny E, Darnault C, Stapleton MR, Green J, Volbeda A, Fontecilla-Camps JC, Le Brun NE. Probing the Reactivity of [4Fe-4S] Fumarate and Nitrate Reduction (FNR) Regulator with O2 and NO: Increased O2 Resistance and Relative Specificity for NO of the [4Fe-4S] L28H FNR Cluster. Inorganics. 2023; 11(12):450. https://doi.org/10.3390/inorganics11120450
Chicago/Turabian StyleCrack, Jason C., Patricia Amara, Eve de Rosny, Claudine Darnault, Melanie R. Stapleton, Jeffrey Green, Anne Volbeda, Juan C. Fontecilla-Camps, and Nick E. Le Brun. 2023. "Probing the Reactivity of [4Fe-4S] Fumarate and Nitrate Reduction (FNR) Regulator with O2 and NO: Increased O2 Resistance and Relative Specificity for NO of the [4Fe-4S] L28H FNR Cluster" Inorganics 11, no. 12: 450. https://doi.org/10.3390/inorganics11120450






