Copper(I)/Triphenylphosphine Complexes Containing Naphthoquinone Ligands as Potential Anticancer Agents
Abstract
:1. Introduction
2. Results and Discussion
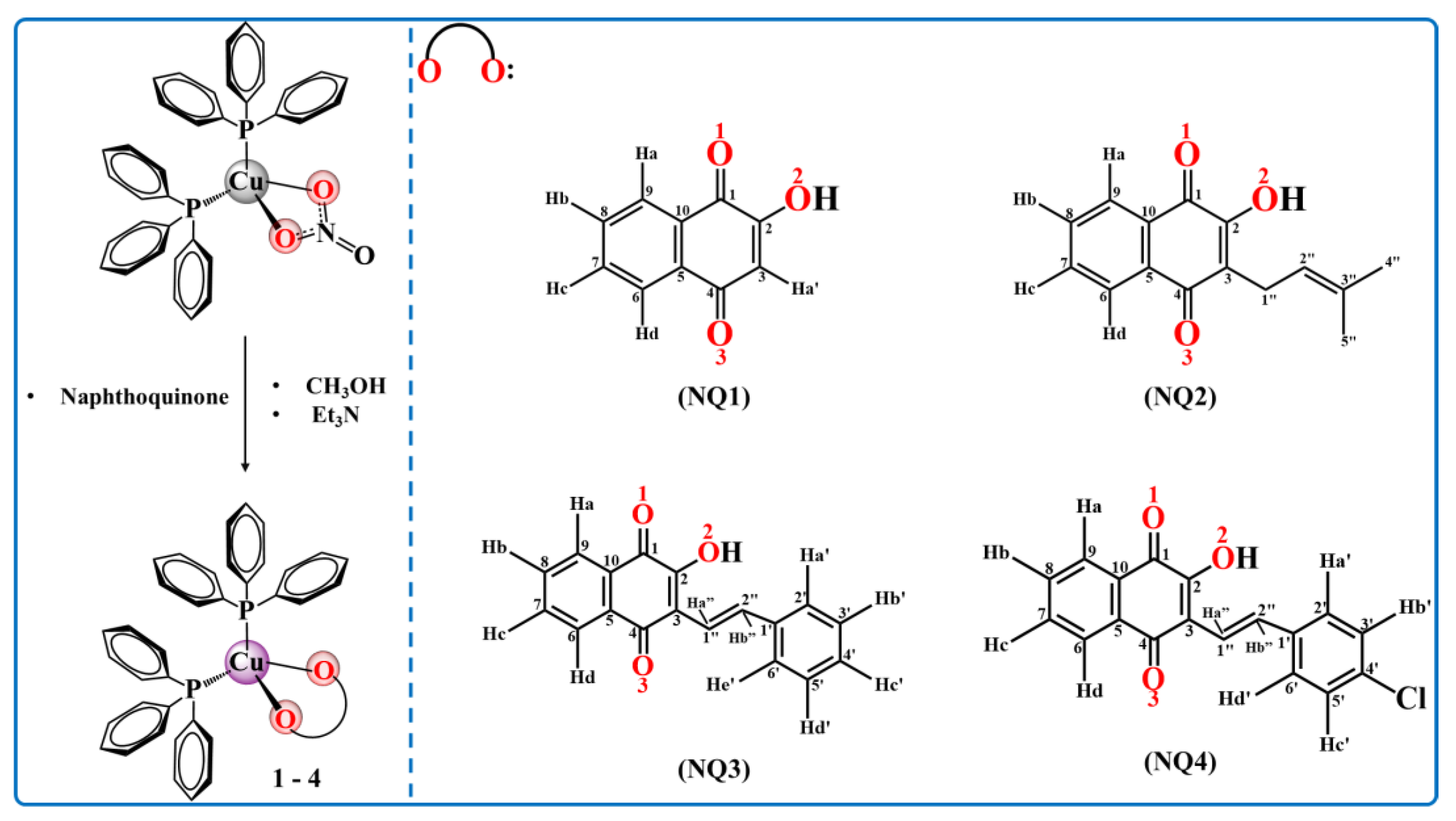
2.1. Syntheses of the Compounds
2.2. Structural Studies
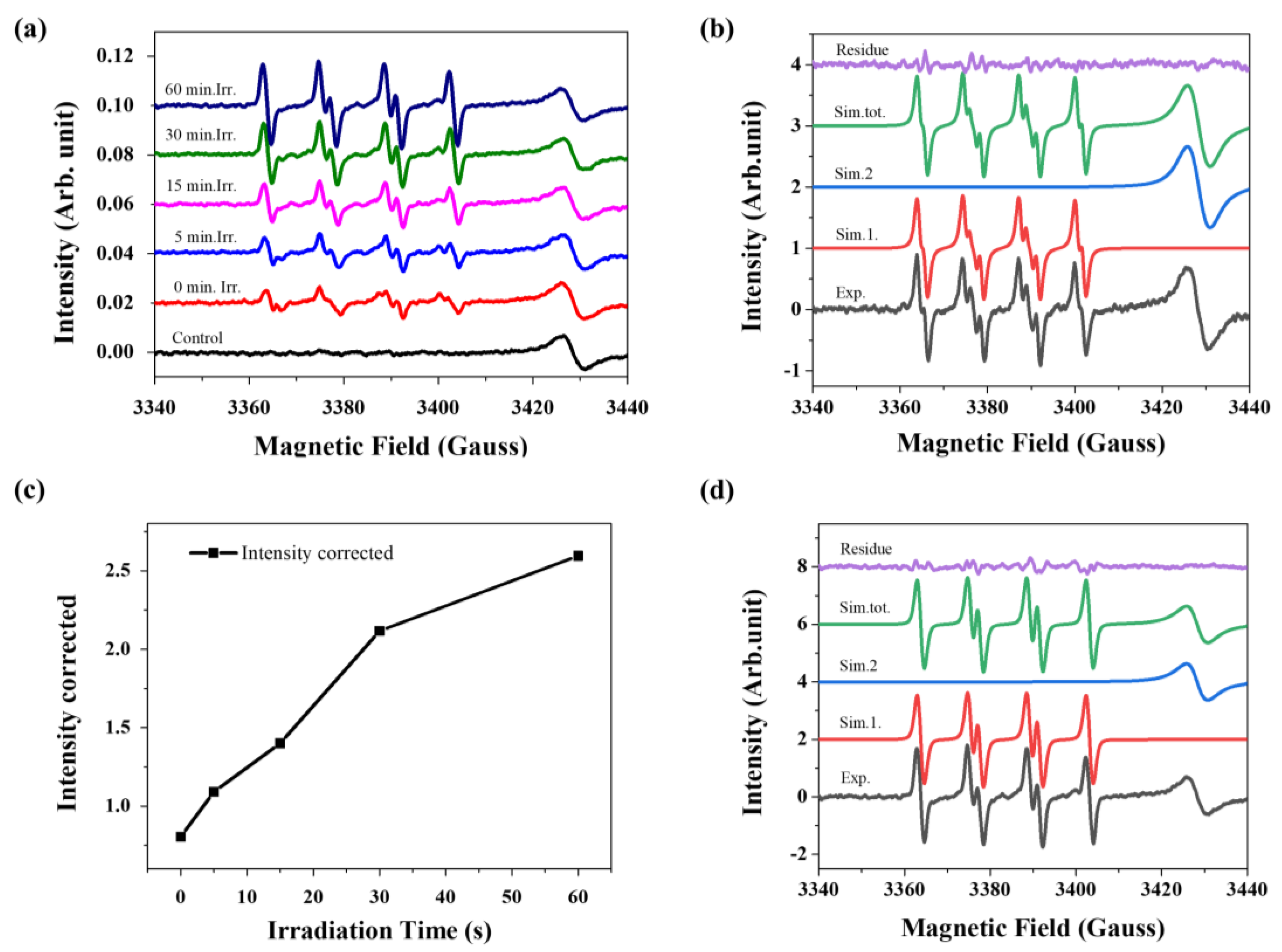
3. Biological Studies
4. Experimental Section
4.1. Materials
4.2. Physical Measurements
4.3. Syntheses of the Copper Complexes
4.4. Stability of Complexes in Culture Medium
4.5. X-ray Crystallography
4.6. EPR Measurements
4.7. Cell Culture
4.8. Cell Viability Assay
4.9. Cell Morphology
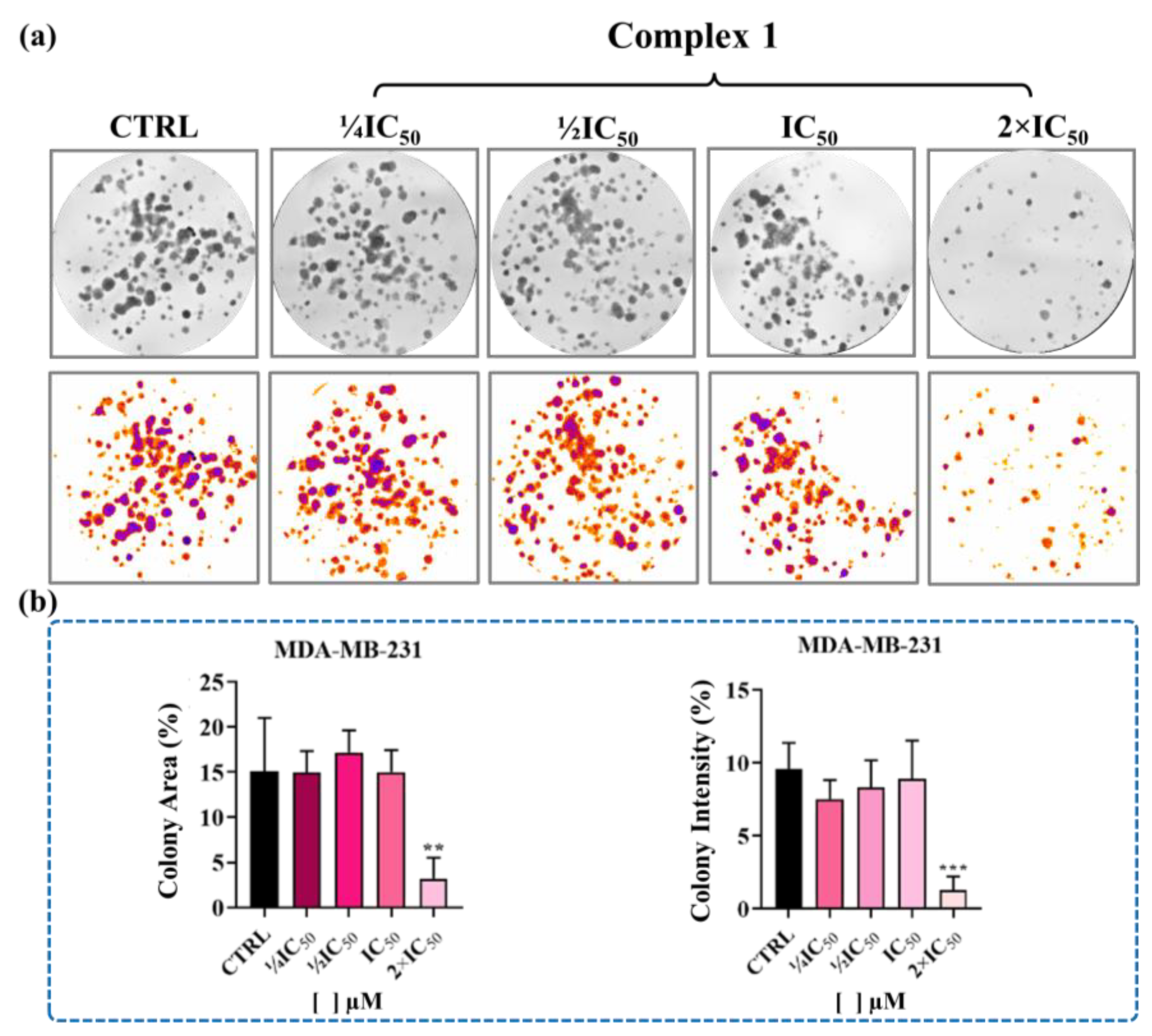
4.10. Colony Formation
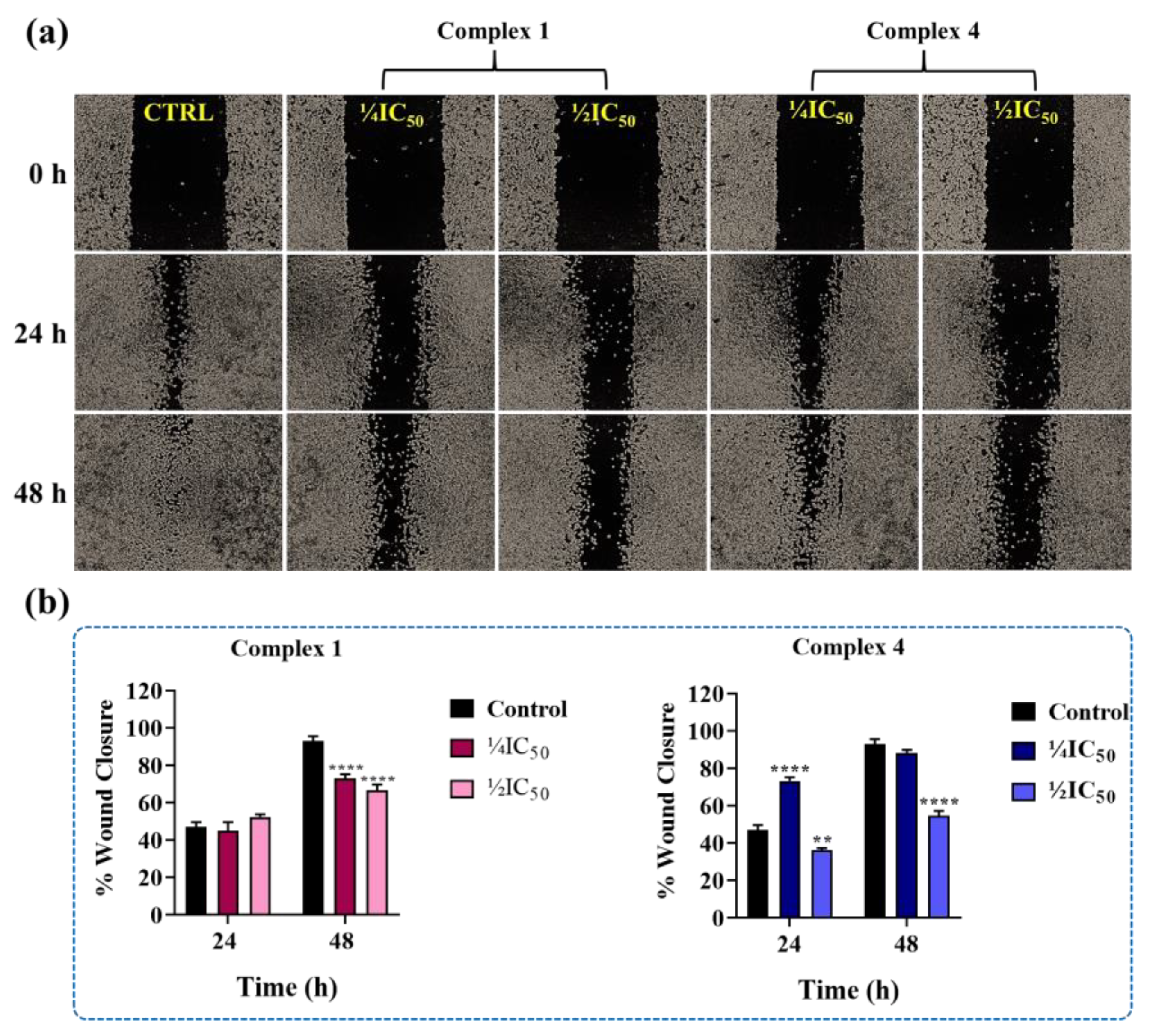
4.11. Scratch Assay (Wound Healing)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Weiderpass, E.; Soerjomataram, I. The Ever-Increasing Importance of Cancer as a Leading Cause of Premature Death Worldwide. Cancer 2021, 127, 3029–3030. [Google Scholar] [CrossRef]
- Crosby, D.; Bhatia, S.; Brindle, K.M.; Coussens, L.M.; Dive, C.; Emberton, M.; Esener, S.; Fitzgerald, R.C.; Gambhir, S.S.; Kuhn, P.; et al. Early Detection of Cancer. Science 2022, 375, eaay9040. [Google Scholar] [CrossRef] [PubMed]
- Somarelli, J.A.; DeGregori, J.; Gerlinger, M.; Heng, H.H.; Marusyk, A.; Welch, D.R.; Laukien, F.H. Questions to Guide Cancer Evolution as a Framework for Furthering Progress in Cancer Research and Sustainable Patient Outcomes. Med. Oncol. 2022, 39, 137. [Google Scholar] [CrossRef] [PubMed]
- Garreffa, E.; Arora, D. Breast Cancer in the Elderly, in Men and during Pregnancy. Surgery 2022, 40, 139–146. [Google Scholar] [CrossRef]
- Smolarz, B.; Nowak, A.Z.; Romanowicz, H. Breast Cancer—Epidemiology, Classification, Pathogenesis and Treatment (Review of Literature). Cancers 2022, 14, 2569. [Google Scholar] [CrossRef]
- Rossi, C.; Cicalini, I.; Cufaro, M.C.; Consalvo, A.; Upadhyaya, P.; Sala, G.; Antonucci, I.; Del Boccio, P.; Stuppia, L.; De Laurenzi, V. Breast Cancer in the Era of Integrating “Omics” Approaches. Oncogenesis 2022, 11, 17. [Google Scholar] [CrossRef]
- Kavan, S.; Kruse, T.A.; Vogsen, M.; Hildebrandt, M.G.; Thomassen, M. Heterogeneity and Tumor Evolution Reflected in Liquid Biopsy in Metastatic Breast Cancer Patients: A Review. Cancer Metastasis Rev. 2022, 41, 433–446. [Google Scholar] [CrossRef]
- Mehraj, U.; Mushtaq, U.; Mir, M.A.; Saleem, A.; Macha, M.A.; Lone, M.N.; Hamid, A.; Zargar, M.A.; Ahmad, S.M.; Wani, N.A. Chemokines in Triple-Negative Breast Cancer Heterogeneity: New Challenges for Clinical Implications. Semin. Cancer Biol. 2022, 86, 769–783. [Google Scholar] [CrossRef]
- Fusco, N.; Sajjadi, E.; Venetis, K.; Ivanova, M.; Andaloro, S.; Guerini-Rocco, E.; Montagna, E.; Caldarella, P.; Veronesi, P.; Colleoni, M.; et al. Low-Risk Triple-Negative Breast Cancers: Clinico-Pathological and Molecular Features. Crit. Rev. Oncol. Hematol. 2022, 172, 103643. [Google Scholar] [CrossRef]
- Herbst, R.S.; Morgensztern, D.; Boshoff, C. The Biology and Management of Non-Small Cell Lung Cancer. Nature 2018, 553, 446–454. [Google Scholar] [CrossRef]
- Marino, F.Z.; Bianco, R.; Accardo, M.; Ronchi, A.; Cozzolino, I.; Morgillo, F.; Rossi, G.; Franco, R. Molecular Heterogeneity in Lung Cancer: From Mechanisms of Origin to Clinical Implications. Int. J. Med. Sci. 2019, 16, 981–989. [Google Scholar] [CrossRef] [PubMed]
- De Sousa, V.M.L.; Carvalho, L. Heterogeneity in Lung Cancer. Pathobiology 2018, 85, 96–107. [Google Scholar] [CrossRef] [PubMed]
- Testa, U.; Castelli, G.; Pelosi, E. Lung Cancers: Molecular Characterization, Clonal Heterogeneity and Evolution, and Cancer Stem Cells. Cancers 2018, 10, 248. [Google Scholar] [CrossRef] [PubMed]
- Gamberi, T.; Hanif, M. Metal-Based Complexes in Cancer Treatment. Biomedicines 2022, 10, 2573. [Google Scholar] [CrossRef] [PubMed]
- Ndagi, U.; Mhlongo, N.; Soliman, M.E. Metal Complexes in Cancer Therapy—An Update from Drug Design Perspective. Drug Des. Devel. Ther. 2017, 11, 599–616. [Google Scholar] [CrossRef]
- Paprocka, R.; Wiese-Szadkowska, M.; Janciauskiene, S.; Kosmalski, T.; Kulik, M.; Helmin-Basa, A. Latest Developments in Metal Complexes as Anticancer Agents. Coord. Chem. Rev. 2022, 452, 85–94. [Google Scholar] [CrossRef]
- Li, X.; Wang, Y.; Li, M.; Wang, H.; Dong, X. Metal Complexes or Chelators with ROS Regulation Capacity: Promising Candidates for Cancer Treatment. Molecules 2022, 27, 148. [Google Scholar] [CrossRef]
- Neethu, K.S.; Sivaselvam, S.; Theetharappan, M.; Ranjitha, J.; Bhuvanesh, N.S.P.; Ponpandian, N.; Neelakantan, M.A.; Kaveri, M.V. In Vitro Evaluations of Biomolecular Interactions, Antioxidant and Anticancer Activities of Nickel(II) and Copper(II) Complexes with 1:2 Coordination of Anthracenyl Hydrazone Ligands. Inorganica Chim. Acta 2021, 524, 120419. [Google Scholar] [CrossRef]
- Guan, D.; Zhao, L.; Shi, X.; Ma, X.; Chen, Z. Copper in Cancer: From Pathogenesis to Therapy. Biomed. Pharmacother. 2023, 163, 114791. [Google Scholar] [CrossRef]
- Wang, C.; Yang, X.; Dong, C.; Chai, K.; Ruan, J.; Shi, S. Cu-Related Agents for Cancer Therapies. Coord. Chem. Rev. 2023, 487, 215156. [Google Scholar] [CrossRef]
- Ji, P.; Wang, P.; Chen, H.; Xu, Y.; Ge, J.; Tian, Z.; Yan, Z. Potential of Copper and Copper Compounds for Anticancer Applications. Pharmaceuticals 2023, 16, 234. [Google Scholar] [CrossRef]
- Singh, N.K.; Kumbhar, A.A.; Pokharel, Y.R.; Yadav, P.N. Anticancer Potency of Copper(II) Complexes of Thiosemicarbazones. J. Inorg. Biochem. 2020, 210, 111134. [Google Scholar] [CrossRef] [PubMed]
- Castillo-Rodríguez, R.A.; Palencia, G.; Anaya-Rubio, I.; Gallardo-Pérez, J.C.; Jiménez-Farfán, D.; Escamilla-Ramírez, Á.; Zavala-Vega, S.; Cruz-Salgado, A.; Cervantes-Rebolledo, C.; Gracia-Mora, I.; et al. Anti-Proliferative, pro-Apoptotic and Anti-Invasive Effect of the Copper Coordination Compound Cas III-La through the Induction of Reactive Oxygen Species and Regulation of Wnt/β-Catenin Pathway in Glioma. J. Cancer 2021, 12, 5693–5711. [Google Scholar] [CrossRef] [PubMed]
- da Silva, D.A.; De Luca, A.; Squitti, R.; Rongioletti, M.; Rossi, L.; Machado, C.M.L.; Cerchiaro, G. Copper in Tumors and the Use of Copper-Based Compounds in Cancer Treatment. J. Inorg. Biochem. 2022, 226, 111634. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Chen, X.; Huang, G.; Jiang, Y.; Gou, Y.; Deng, J. Synthesis, Anti-Tumour Activity, and Mechanism of Benzoyl Hydrazine Schiff Base-Copper Complexes. J. Mol. Struct. 2022, 1268, 133730. [Google Scholar] [CrossRef]
- Jiang, Y.; Huo, Z.; Qi, X.; Zuo, T.; Wu, Z. Copper-Induced Tumor Cell Death Mechanisms and Antitumor Theragnostic Applications of Copper Complexes. Nanomedicine 2022, 17, 303–324. [Google Scholar] [CrossRef]
- Jung, H.S.; Koo, S.; Won, M.; An, S.; Park, H.; Sessler, J.L.; Han, J.; Kim, J.S. Cu(Ii)-BODIPY Photosensitizer for CAIX Overexpressed Cancer Stem Cell Therapy. Chem. Sci. 2023, 14, 1808–1819. [Google Scholar] [CrossRef]
- Wellington, K.W. Understanding Cancer and the Anticancer Activities of Naphthoquinones-a Review. RSC Adv. 2015, 5, 20309–20338. [Google Scholar] [CrossRef]
- Hong, S.; Huber, S.M.; Gagliardi, L.; Cramer, C.C.; Tolman, W.B. Copper(I)-α-Ketocarboxylate Complexes: Characterization and O 2 Reactions That Yield Copper-Oxygen Intermediates Capable of Hydroxylating Arenes. J. Am. Chem. Soc. 2007, 129, 14190–14192. [Google Scholar] [CrossRef]
- Mohd Zubir, M.Z.; Jamaludin, N.S.; Abdul Halim, S.N. Hirshfeld Surface Analysis of Some New Heteroleptic Copper(I) Complexes. J. Mol. Struct. 2019, 1193, 141–150. [Google Scholar] [CrossRef]
- Cabrera, A.R.; Gonzalez, I.A.; Cortés-Arriagada, D.; Natali, M.; Berke, H.; Daniliuc, C.G.; Camarada, M.B.; Toro-Labbé, A.; Rojas, R.S.; Salas, C.O. Synthesis of New Phosphorescent Imidoyl-Indazol and Phosphine Mixed Ligand Cu(i) Complexes-Structural Characterization and Photophysical Properties. RSC Adv. 2016, 6, 5141–5153. [Google Scholar] [CrossRef]
- Villarreal, W.; Colina-Vegas, L.; Visbal, G.; Corona, O.; Corrêa, R.S.; Ellena, J.; Cominetti, M.R.; Batista, A.A.; Navarro, M. Copper(I)-Phosphine Polypyridyl Complexes: Synthesis, Characterization, DNA/HSA Binding Study, and Antiproliferative Activity. Inorg. Chem. 2017, 56, 3781–3793. [Google Scholar] [CrossRef] [PubMed]
- Meza-Morales, W.; Machado-Rodriguez, J.C.; Alvarez-Ricardo, Y.; Obregón-Mendoza, M.A.; Nieto-Camacho, A.; Toscano, R.A.; Soriano-García, M.; Cassani, J.; Enríquez, R.G. A New Family of Homoleptic Copper Complexes of Curcuminoids: Synthesis, Characterization and Biological Properties. Molecules 2019, 24, 910. [Google Scholar] [CrossRef] [PubMed]
- Małecki, J.G.; Maroń, A.; Palion, J.; Nycz, J.E.; Szala, M. A Copper(I) Phosphine Complex with 5,7-Dinitro-2-Methylquinolin-8-Ol as Co-Ligand. Transit. Met. Chem. 2014, 39, 755–762. [Google Scholar] [CrossRef]
- Aliaga-Alcalde, N.; Marqués-Gallego, P.; Kraaijkamp, M.; Herranz-Lancho, C.; Den Dulk, H.; Görner, H.; Roubeau, O.; Teat, S.J.; Weyhermüller, T.; Reedijk, J. Copper Curcuminoids Containing Anthracene Groups: Fluorescent Molecules with Cytotoxic Activity. Inorg. Chem. 2010, 49, 9655–9663. [Google Scholar] [CrossRef] [PubMed]
- Aguirrechu-Comerón, A.; Oramas-Royo, S.; Pérez-Acosta, R.; Hernández-Molina, R.; Gonzalez-Platas, J.; Estévez-Braun, A. Preparation of New Metallic Complexes from 2-Hydroxy-3-((5-Methylfuran-2-Yl)Methyl)-1,4-Naphthoquinone. Polyhedron 2020, 177, 114280. [Google Scholar] [CrossRef]
- Oliveira, K.M.; Peterson, E.J.; Carroccia, M.C.; Cominetti, M.R.; Deflon, V.M.; Farrell, N.P.; Batista, A.A.; Correa, R.S. Ru(Ii)-Naphthoquinone Complexes with High Selectivity for Triple-Negative Breast Cancer. Dalt. Trans. 2020, 49, 16193–16203. [Google Scholar] [CrossRef]
- Oliveira, K.M.; Liany, L.-D.; Corrêa, R.S.; Deflon, V.M.; Cominetti, M.R.; Batista, A.A. Selective Ru(II)/Lawsone Complexes Inhibiting Tumor Cell Growth by Apoptosis. J. Inorg. Biochem. 2017, 176, 66–76. [Google Scholar] [CrossRef]
- Chaquin, P.; Canac, Y.; Lepetit, C.; Zargarian, D.; Chauvin, R. Estimating Local Bonding/Antibonding Character of Canonical Molecular Orbitals from Their Energy Derivatives. The Case of Coordinating Lone Pair Orbitals. Int. J. Quantum Chem. 2016, 116, 1285–1295. [Google Scholar] [CrossRef]
- Salunke-Gawali, S.; Pereira, E.; Dar, U.A.; Bhand, S. Metal Complexes of Hydroxynaphthoquinones: Lawsone, Bis-Lawsone, Lapachol, Plumbagin and Juglone. J. Mol. Struct. 2017, 1148, 435–458. [Google Scholar] [CrossRef]
- Majdi, C.; Duvauchelle, V.; Meffre, P.; Benfodda, Z. An Overview on the Antibacterial Properties of Juglone, Naphthazarin, Plumbagin and Lawsone Derivatives and Their Metal Complexes. Biomed. Pharmacother. 2023, 162, 114690. [Google Scholar] [CrossRef]
- Kosiha, A.; Parthiban, C.; Ciattini, S.; Chelazzi, L.; Elango, K.P. Metal Complexes of Naphthoquinone Based Ligand: Synthesis, Characterization, Protein Binding, DNA Binding/Cleavage and Cytotoxicity Studies. J. Biomol. Struct. Dyn. 2018, 36, 4170–4181. [Google Scholar] [CrossRef] [PubMed]
- Selvaraj, F.S.S.; Samuel, M.; Karuppiah, A.K.; Raman, N. Transition Metal Complexes Incorporating Lawsone: A Review. J. Coord. Chem. 2022, 75, 2509–2532. [Google Scholar] [CrossRef]
- Oladipo, S.D.; Mocktar, C.; Omondi, B. In Vitro Biological Studies of Heteroleptic Ag(I) and Cu(I) Unsymmetrical N,N′-Diarylformamidine Dithiocarbamate Phosphine Complexes; the Effect of the Metal Center. Arab. J. Chem. 2020, 13, 6379–6394. [Google Scholar] [CrossRef]
- Fan, W.W.; Li, Z.F.; Li, J.B.; Yang, Y.P.; Yuan, Y.; Tang, H.Q.; Gao, L.X.; Jin, Q.H.; Zhang, Z.W.; Zhang, C.L. Synthesis, Structure, Terahertz Spectroscopy and Luminescent Properties of Copper (I) Complexes with Bis(Diphenylphosphino)Methane and N-Donor Ligands. J. Mol. Struct. 2015, 1099, 351–358. [Google Scholar] [CrossRef]
- Wu, Y.; Han, X.; Qu, Y.; Zhao, K.; Wang, C.; Huang, G.; Wu, H. Two Cu(I) Complexes Constructed by Different N-Heterocyclic Benzoxazole Ligands: Syntheses, Structures and Fluorescent Properties. J. Mol. Struct. 2019, 1191, 95–100. [Google Scholar] [CrossRef]
- Leite, C.M.; Honorato, J.; Martin, A.C.B.M.; Silveira, R.G.; Colombari, F.M.; Amaral, J.C.; Costa, A.R.; Cominetti, M.R.; Plutin, A.M.; Aguiar, D.; et al. Experimental and Theoretical DFT Study of Cu(I)/N,N-Disubstituted-N′-acylthioureato Anticancer Complexes: Actin Cytoskeleton and Induction of Death by Apoptosis in Triple-Negative Breast Tumor Cells. Inorg. Chem. 2022, 61, 664–677. [Google Scholar] [CrossRef]
- Gunasekaran, N.; Bhuvanesh, N.S.P.; Karvembu, R. Synthesis, Characterization and Catalytic Oxidation Property of Copper(I) Complexes Containing Monodentate Acylthiourea Ligands and Triphenylphosphine. Polyhedron 2017, 122, 39–45. [Google Scholar] [CrossRef]
- Idriss, K.A.; Sedaira, H.; Hashem, E.Y.; Saleh, M.S.; Soliman, S.A. The Visible Absorbance Maximum of 2-Hydroxy-1,4-Naphthoquinone as a Novel Probe for the Hydrogen Bond Donor Abilities of Solvents and Solvent Mixtures. Monatshefte Fur Chem. 1996, 127, 29–42. [Google Scholar] [CrossRef]
- McCormick, T.; Jia, W.L.; Wang, S. Phosphorescent Cu(I) Complexes of 2-(2′-Pyridylbenzimidazolyl) Benzene: Impact of Phosphine Ancillary Ligands on Electronic and Photophysical Properties of the Cu(I) Complexes. Inorg. Chem. 2006, 45, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Ibis, C.; Sahinler Ayla, S.; Babayeva, E. Reactions of Quinones with Some Amino Alcohols, Thiols and a UV-Vis Study. Phosphorus Sulfur Silicon Relat. Elem. 2020, 195, 474–480. [Google Scholar] [CrossRef]
- Sørensen, T.; Nielsen, M. Synthesis, UV/Vis Spectra and Electrochemical Characterisation of Arylthio and Styryl Substituted Ferrocenes. Open Chem. 2011, 9, 610–618. [Google Scholar] [CrossRef]
- Verma, S.K.; Singh, V.K. Synthesis, Electrochemical, Fluorescence and Antimicrobial Studies of 2-Chloro-3-Amino-1,4-Naphthoquinone Bearing Mononuclear Transition Metal Dithiocarbamate Complexes [M{κ2S,S-S2C–Piperazine–C2H4N(H)ClNQ}n]. RSC Adv. 2015, 5, 53036–53046. [Google Scholar] [CrossRef]
- Win, T.; Bittner, S. Novel 2-Amino-3-(2,4-Dinitrophenylamino) Derivatives of 1,4-Naphthoquinone. Tetrahedron Lett. 2005, 46, 3229–3231. [Google Scholar] [CrossRef]
- Záliš, S.; Fiedler, J.; Pospíšil, L.; Fanelli, N.; Lanza, C.; Lampugnani, L. Electron Transfer in Donor–Acceptor Molecules of Substituted Naphtoquinones: Spectral and Redox Properties of Internal Charge Transfer Complexes. Microchem. J. 1996, 54, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Sutovsky, Y.; Likhtenshtein, G.I.; Bittner, S. Synthesis and Photochemical Behavior of Donor-Acceptor Systems Obtained from Chloro-1,4-Naphthoquinone Attached to Trans-Aminostilbenes. Tetrahedron 2003, 59, 2939–2945. [Google Scholar] [CrossRef]
- Nowicka, B.; Walczak, J.; Kapsiak, M.; Barnaś, K.; Dziuba, J.; Suchoń, A. Impact of Cytotoxic Plant Naphthoquinones, Juglone, Plumbagin, Lawsone and 2-Methoxy-1,4-Naphthoquinone, on Chlamydomonas Reinhardtii Reveals the Biochemical Mechanism of Juglone Toxicity by Rapid Depletion of Plastoquinol. Plant Physiol. Biochem. 2023, 197, 107660. [Google Scholar] [CrossRef]
- Pieta, P.; Petr, A.; Kutner, W.; Dunsch, L. In Situ ESR Spectroscopic Evidence of the Spin-Trapped Superoxide Radical, O2{radical Dot}-, Electrochemically Generated in DMSO at Room Temperature. Electrochim. Acta 2008, 53, 3412–3415. [Google Scholar] [CrossRef]
- Clément, J.L.; Ferré, N.; Siri, D.; Karoui, H.; Rockenbauer, A.; Tordo, P. Assignment of the EPR Spectrum of 5,5-Dimethyl-1-Pyrroline N-Oxide (DMPO) Superoxide Spin Adduct. J. Org. Chem. 2005, 70, 1198–1203. [Google Scholar] [CrossRef] [PubMed]
- Shoji, T.; Li, L.; Abe, Y.; Ogata, M.; Ishimoto, Y.; Gonda, R.; Mashino, T.; Mochizuki, M.; Uemoto, M.; Miyata, N. DMPO-OH Radical Formation from 5,5-Dimethyl-1-Pyrroline N-Oxide (DMPO) in Hot Water. Anal. Sci. 2007, 23, 219–221. [Google Scholar] [CrossRef]
- Zalibera, M.; Rapta, P.; Staško, A.; Brindzová, L.; Brezová, V. Thermal Generation of Stable Spin Trap Adducts with Super-Hyperfine Structure in Their EPR Spectra: An Alternative EPR Spin Trapping Assay for Radical Scavenging Capacity Determination in Dimethylsulphoxide. Free Radic. Res. 2009, 43, 457–469. [Google Scholar] [CrossRef] [PubMed]
- Massagué, J.; Obenauf, A.C. Metastatic Colonization by Circulating Tumour Cells. Nature 2016, 529, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Fares, J.; Fares, M.Y.; Khachfe, H.H.; Salhab, H.A.; Fares, Y. Molecular Principles of Metastasis: A Hallmark of Cancer Revisited. Signal Transduct. Target. Ther. 2020, 5, 28. [Google Scholar] [CrossRef] [PubMed]
- Jonkman, J.E.N.; Cathcart, J.A.; Xu, F.; Bartolini, M.E.; Amon, J.E.; Stevens, K.M.; Colarusso, P. An Introduction to the Wound Healing Assay Using Live-Cell Microscopy. Cell Adhes. Migr. 2014, 8, 440–451. [Google Scholar] [CrossRef]
- Stamm, A.; Reimers, K.; Strauß, S.; Vogt, P.; Scheper, T.; Pepelanova, I. In Vitro Wound Healing Assays—State of the Art. BioNanoMaterials 2016, 17, 79–87. [Google Scholar] [CrossRef]
- Al Habash, A.; Aljasim, L.A.; Owaidhah, O.; Edward, D.P. A Review of the Efficacy of Mitomycin C in Glaucoma Filtration Surgery. Clin. Ophthalmol. 2015, 9, 1945–1951. [Google Scholar] [CrossRef]
- Jardine, F.H.; Vohra, A.G.; Young, F.J. Copper(I) Nitrato and Nitrate Complexes. J. Inorg. Nucl. Chem. 1971, 33, 2941–2945. [Google Scholar] [CrossRef]
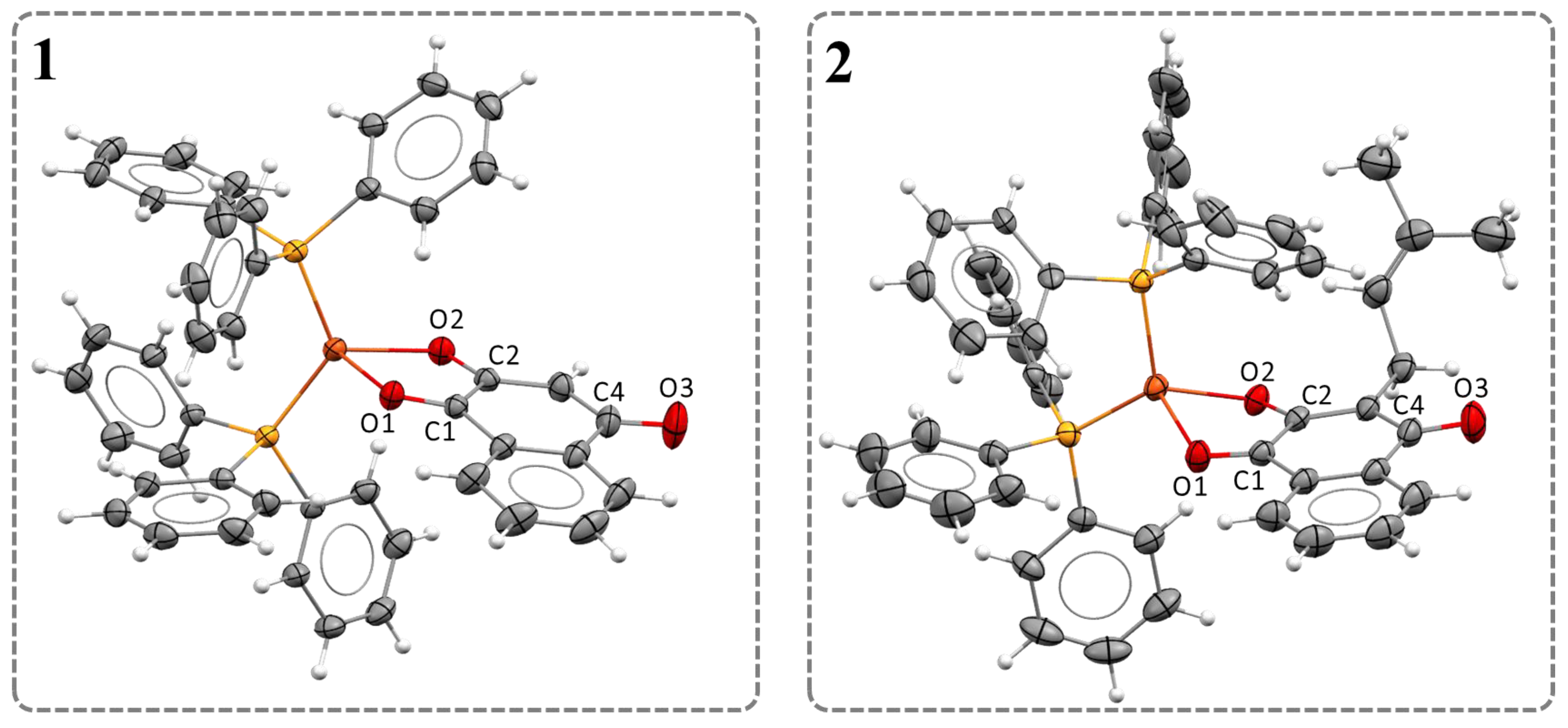



| Bond Lengths (Å) | Ligands | Complexes | Bond Angles (°) | Complexes | |||
|---|---|---|---|---|---|---|---|
| NQ1 * | NQ2 ** | 1 | 2 | 1 | 2 | ||
| Cu1–P1 | - | - | 2.243(4) | 2.208(5) | P1–Cu1–P2 | 127.67(2) | 118.84(2) |
| Cu1–P2 | - | - | 2.244(4) | 2.259(5) | P1–Cu1–O1 | 102.92(4) | 114.28(4) |
| Cu1–O1 | - | - | 2.227(1) | 2.228(2) | O2–Cu1–P2 | 116.03(4) | 109.92(5) |
| Cu1–O2 | - | - | 2.034(1) | 2.015(1) | O2–Cu1–P1 | 114.65(4) | 123.69(5) |
| C1–O1 | 1.217 | 1.226 | 1.224(2) | 1.227(2) | O2–Cu1–O1 | 77.14(4) | 76.81(6) |
| C2–O2 | 1.335 | 1.346 | 1.273(2) | 1.278(2) | O1–Cu1–P2 | 100.04(4) | 103.99(5) |
| C4–O3 | 1.226 | 1.225 | 1.230(2) | 1.237(3) | - | - | - |
| Inhibitory Concentration of 50% of Cell Viability, IC50 (μM) | ||||||||
|---|---|---|---|---|---|---|---|---|
| MCF7 | MDA-MB-231 | MCF-10A | A549 | MRC-5 | SI1 * | SI2 * | SI3 * | |
| 1 | 15 ± 3 | 5.5 ± 0.3 | 5.9 ± 0.2 | 3.6 ± 0.3 | 2.7 ± 0.2 | 0.4 | 1.1 | 0.8 |
| 2 | 9 ± 1 | 6.5 ± 0.3 | 4.4 ± 0.8 | 6.1 ± 0.4 | 3.3 ± 0.2 | 0.5 | 0.7 | 0.5 |
| 3 | 11 ± 3 | 7.2 ± 0.3 | 5.4 ± 0.3 | 4.4 ± 0.1 | 2.8 ± 0.1 | 0.5 | 0.8 | 0.6 |
| 4 | 7 ± 2 | 7.9 ± 0.1 | 4.0 ± 0.2 | 4.5 ± 0.1 | 3.4 ± 0.2 | 0.6 | 0.5 | 0.8 |
| Cu(NO3)2·3H2O | >25 | >25 | >25 | >25 | >25 | - | - | - |
| PPh3 | >25 | >25 | >25 | >25 | >25 | - | - | - |
| NQ | >25 | >25 | >25 | >25 | >25 | - | - | - |
| Cisplatin | 13.9 ± 2.0 | 10.2 ± 0.2 | 23.9 ± 0.7 | 14.4 ± 1.4 | 29.9 ± 0.8 | 1.7 | 2.3 | 2.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leite, C.M.; Araujo-Neto, J.H.; Guedes, A.P.M.; Costa, A.R.; Demidoff, F.C.; Netto, C.D.; Castellano, E.E.; Nascimento, O.R.; Batista, A.A. Copper(I)/Triphenylphosphine Complexes Containing Naphthoquinone Ligands as Potential Anticancer Agents. Inorganics 2023, 11, 367. https://doi.org/10.3390/inorganics11090367
Leite CM, Araujo-Neto JH, Guedes APM, Costa AR, Demidoff FC, Netto CD, Castellano EE, Nascimento OR, Batista AA. Copper(I)/Triphenylphosphine Complexes Containing Naphthoquinone Ligands as Potential Anticancer Agents. Inorganics. 2023; 11(9):367. https://doi.org/10.3390/inorganics11090367
Chicago/Turabian StyleLeite, Celisnolia M., João H. Araujo-Neto, Adriana P. M. Guedes, Analu R. Costa, Felipe C. Demidoff, Chaquip D. Netto, Eduardo E. Castellano, Otaciro R. Nascimento, and Alzir A. Batista. 2023. "Copper(I)/Triphenylphosphine Complexes Containing Naphthoquinone Ligands as Potential Anticancer Agents" Inorganics 11, no. 9: 367. https://doi.org/10.3390/inorganics11090367