Identification and Characterization of Malolactic Bacteria Isolated from the Eastern Foothills of Helan Mountain in China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains, Medium, and Growth Condition
2.2. Isolation of MLB from Wine
2.3. Identification of MLB by 16S rRNA
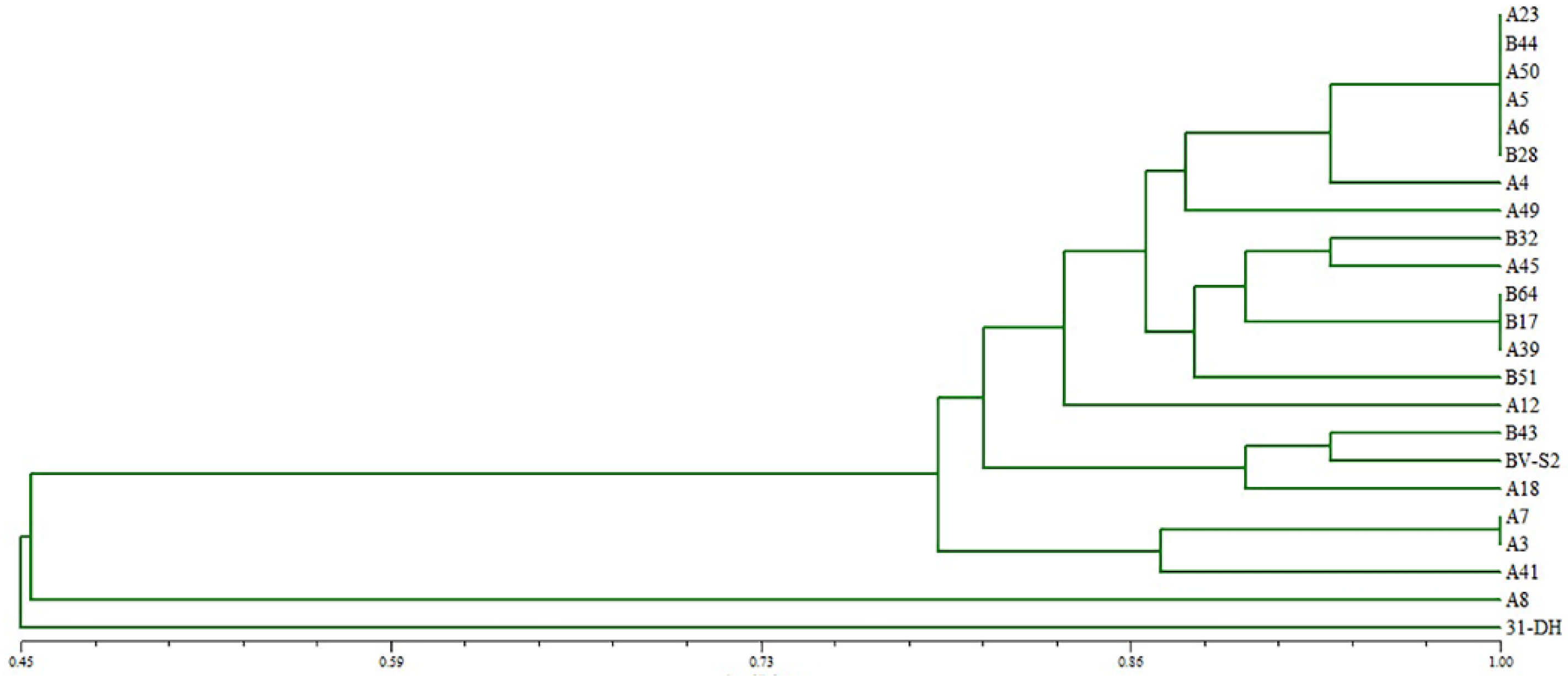
2.4. RAPD Analysis
2.5. Stress Tolerance of L. plantarum
2.6. Determination of the β-D-Glucosidase Activity of L. plantarum
2.7. MLF Assay and Basic Chemical Parameters
2.8. Biogenic Amine
2.9. Analysis of Volatile Compounds
2.10. Statistical Analysis
3. Results
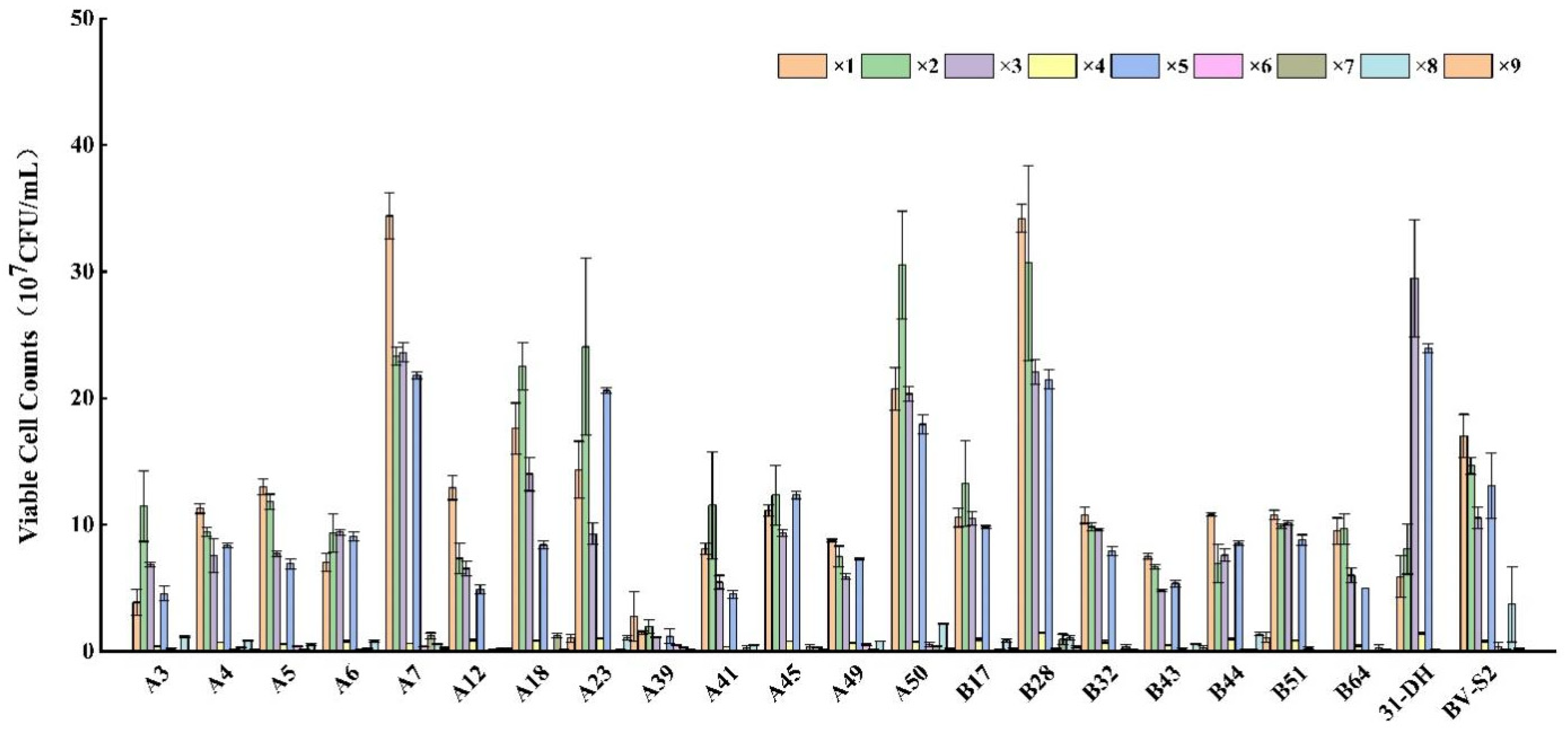
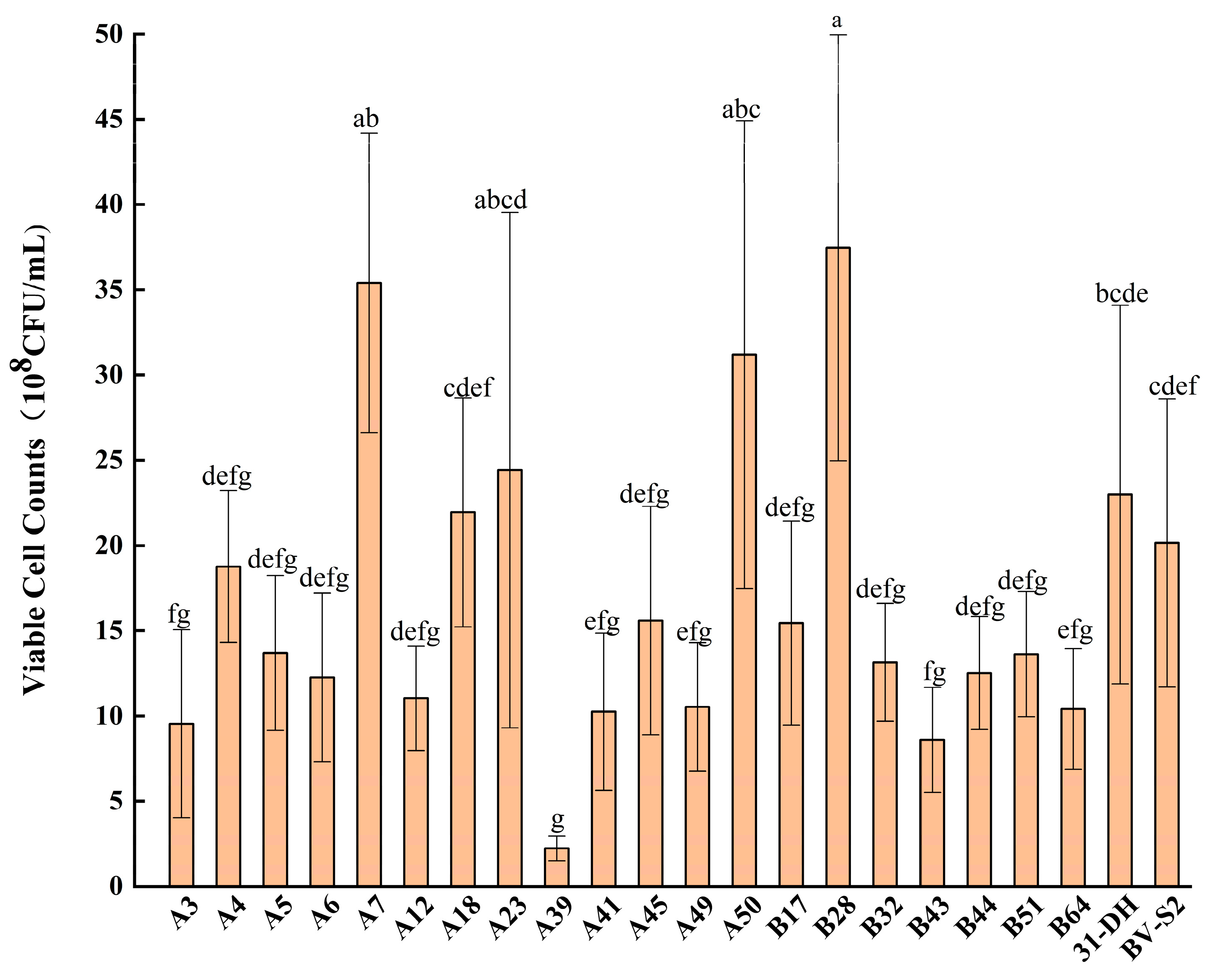
3.1. Isolation and Identification of Strains
3.2. Stress Tolerance of L. plantarum
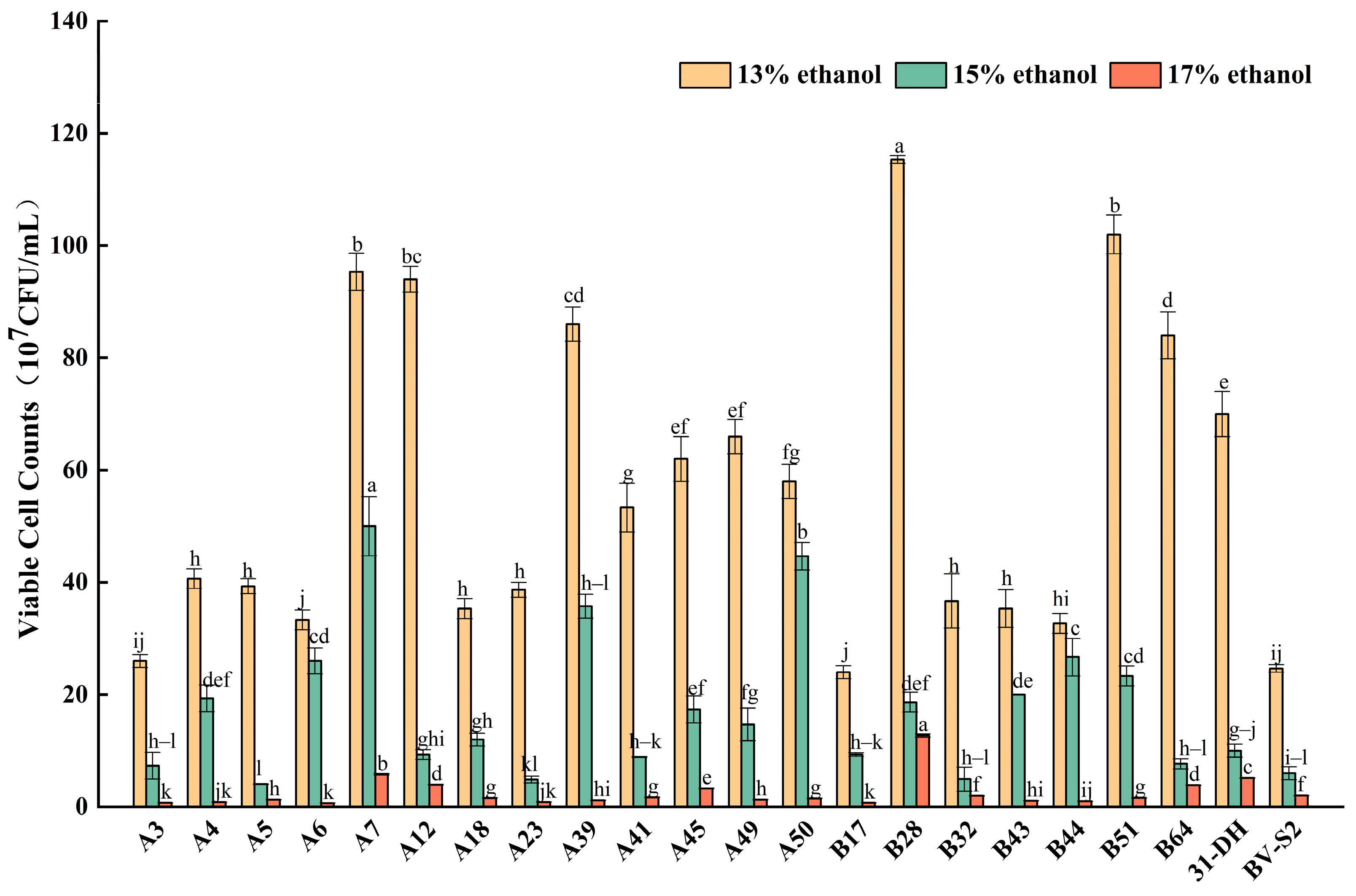
3.2.1. Tolerance of L. plantarum to Ethanol
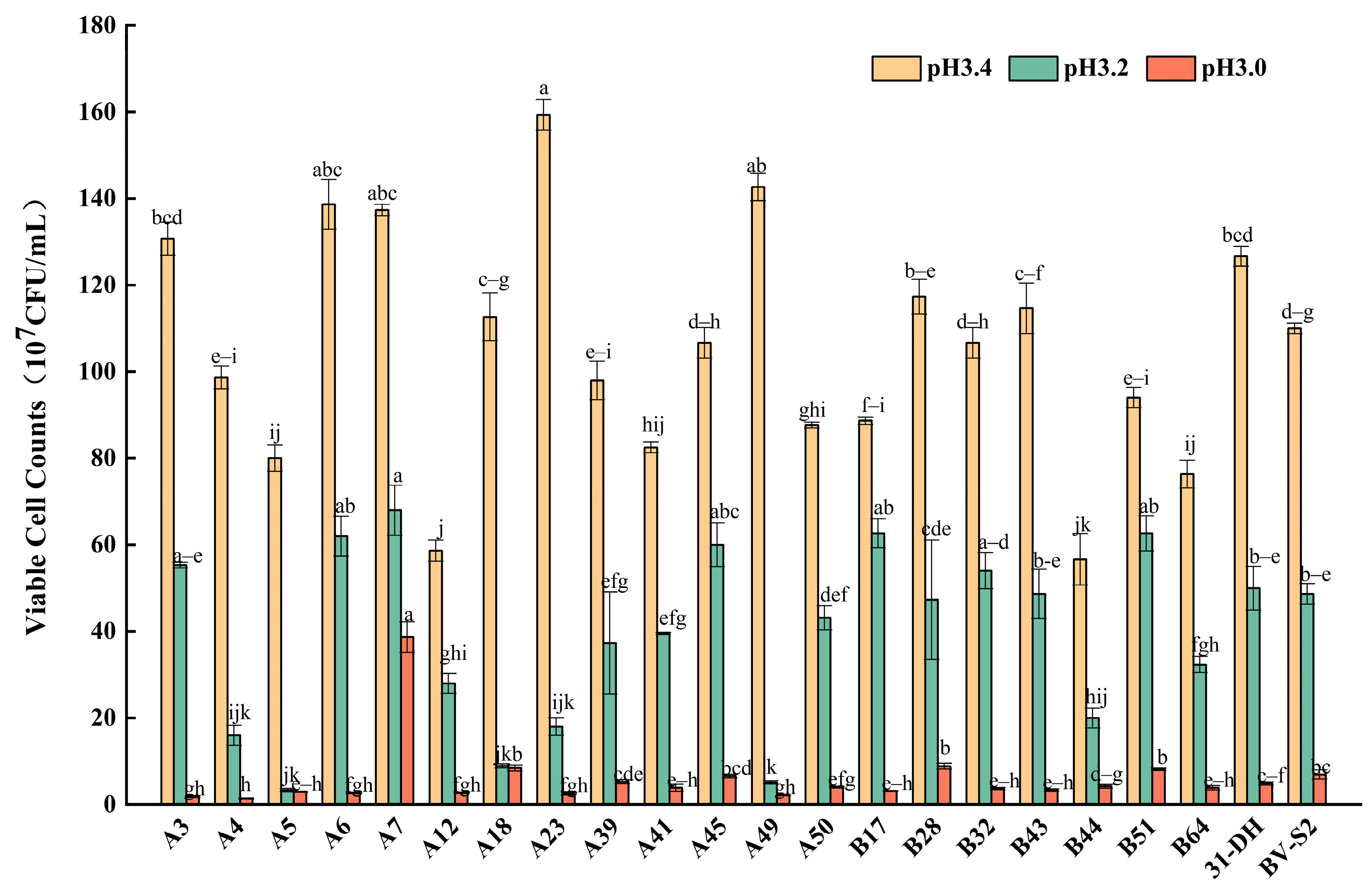
3.2.2. Tolerance of L. plantarum to pH
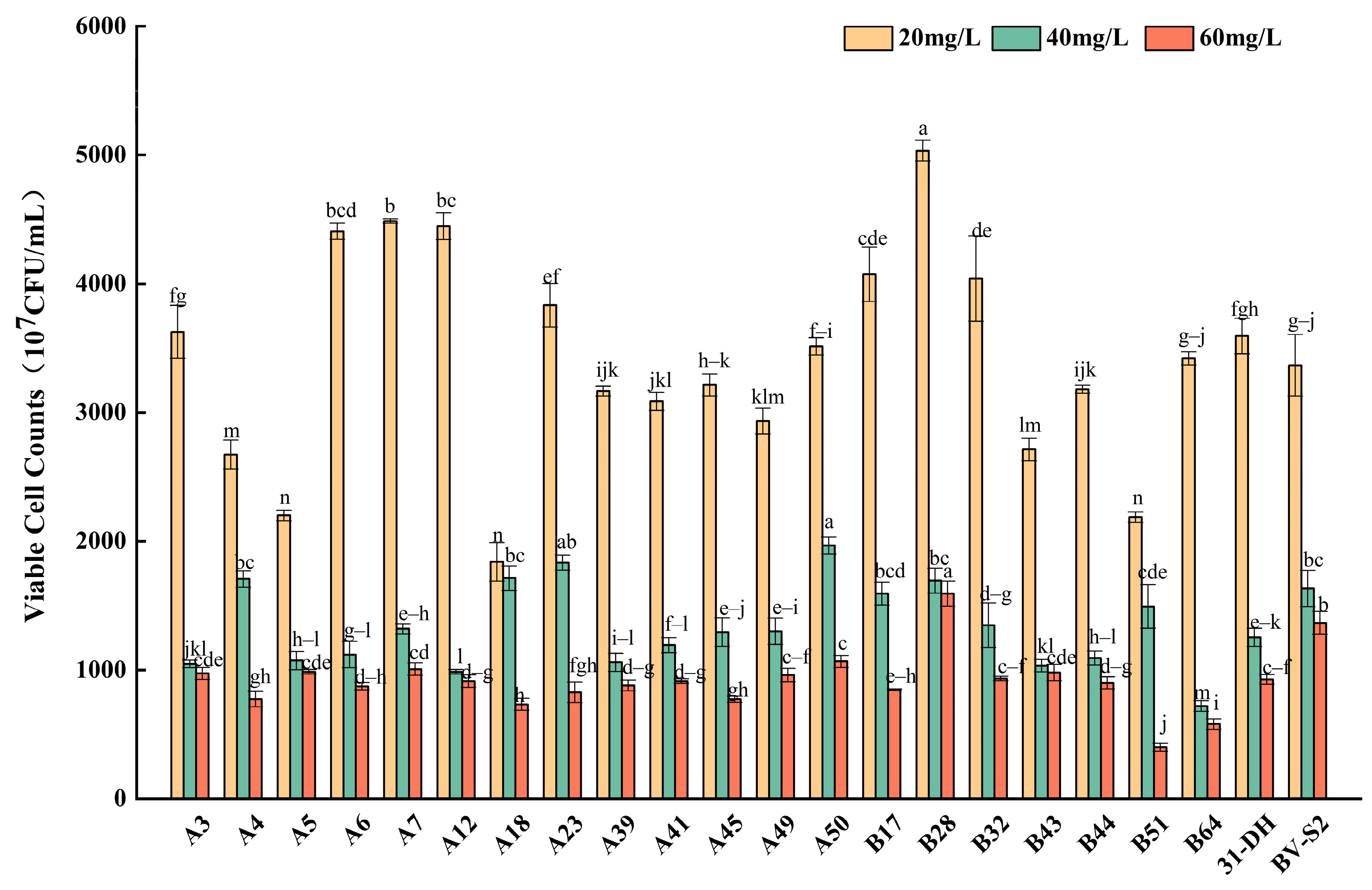
3.2.3. Tolerance of L. plantarum to SO2
3.2.4. Tolerance of L. plantarum to Recombinational Factor
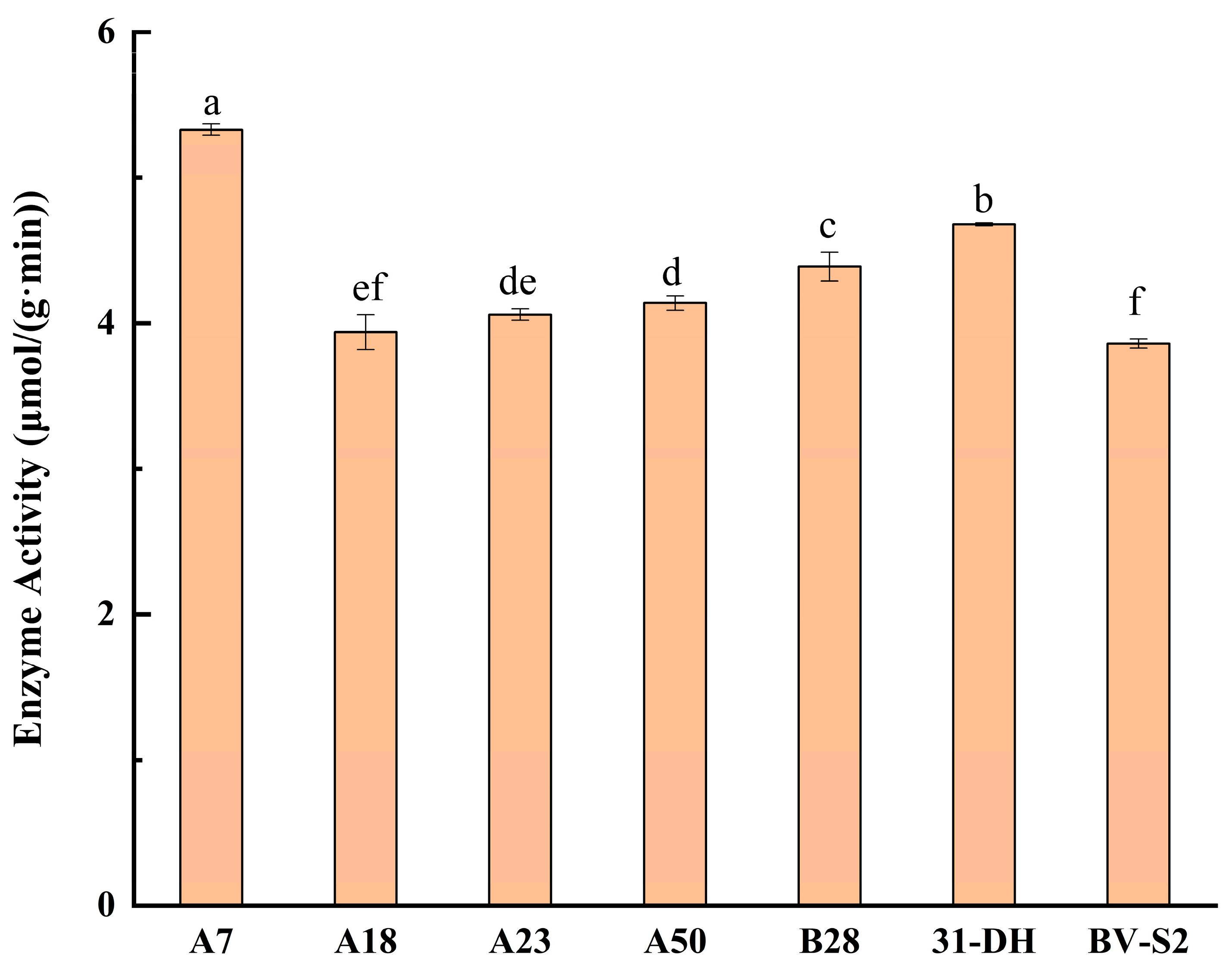
3.3. Determination of β-D-Glucosidase Activity
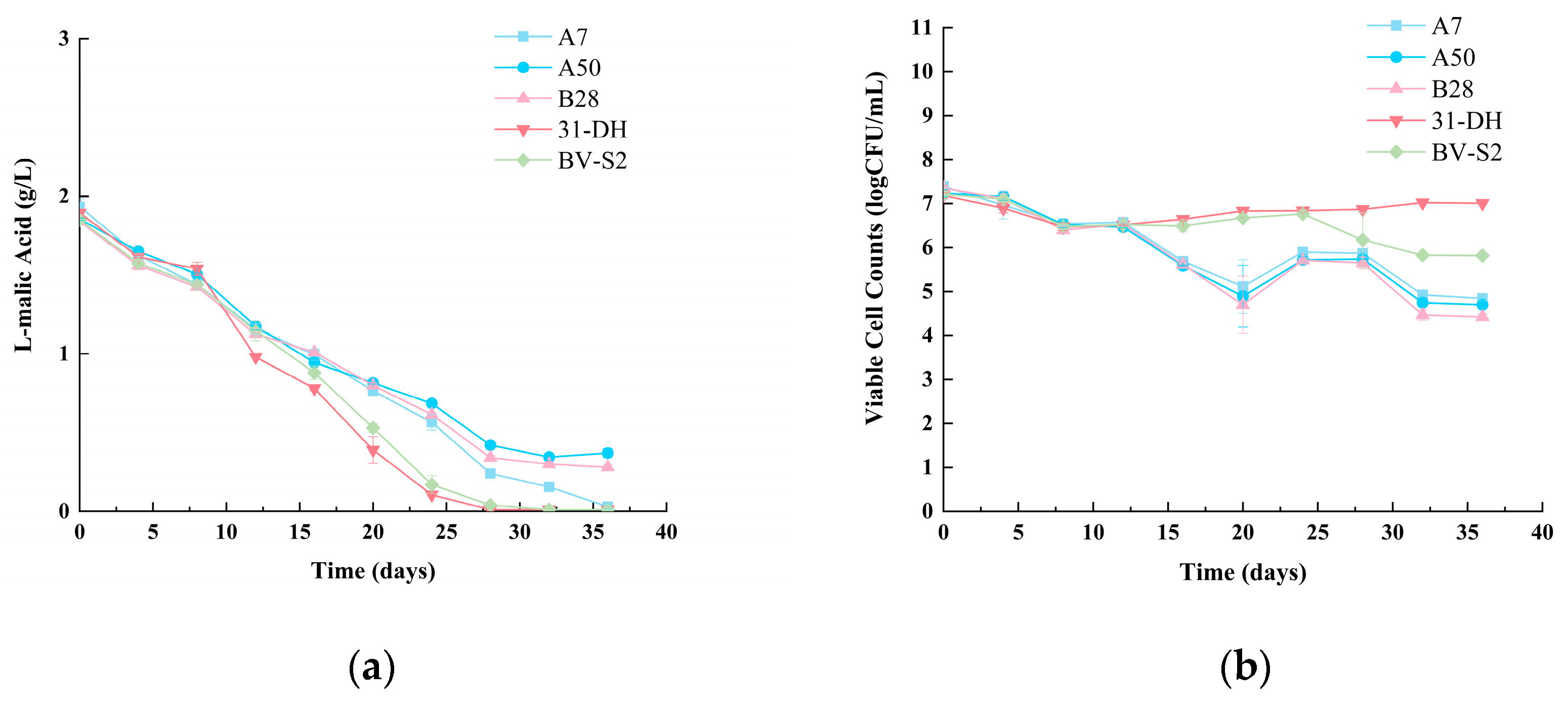
3.4. MLF Assay and Basic Chemical Parameters
3.4.1. MLF Assay of Cabernet Sauvignon Wines
3.4.2. Analysis of Basic Chemical Parameters of Wine
3.4.3. Biogenic Amine Analysis
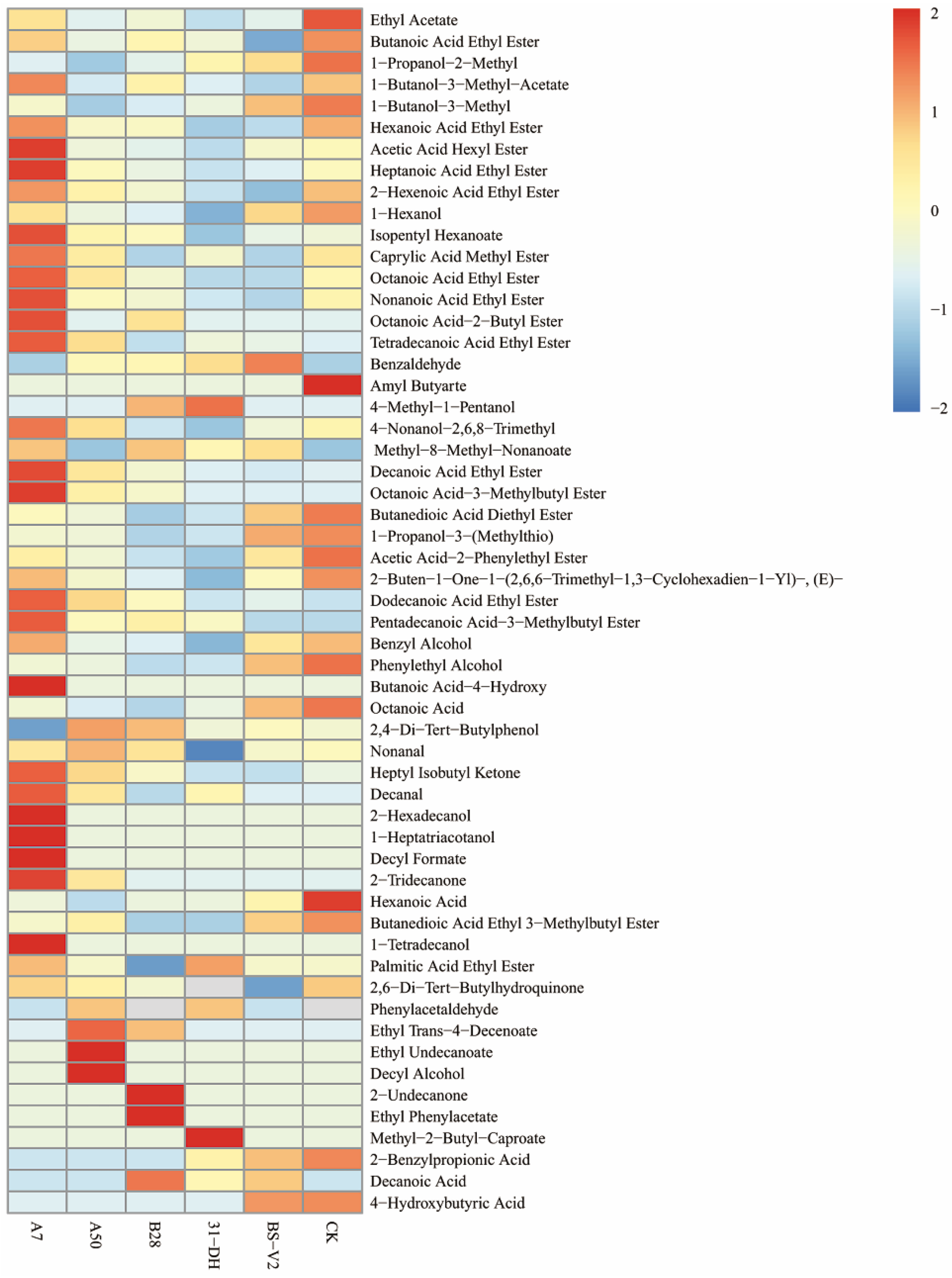
3.4.4. Aroma Components Analysis before and after MLF
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brizuela, N.; Bravo-Ferrada, B.R.; Pozo-Bayón, A.; Maria; Semorile, L.; Tymczyszyn, E. Changes in the volatile profile of Pinot noir wines caused by Patagonian Lactobacillus plantarum and Oenococcus oeni strains. Food Res. Int. 2018, 106, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Brizuela, N.; Bravo-Ferrada, B.R.; Hens, D.; Hollmann, A.; Delfederico, L.; Caballero, A.; Tymczyszyn, E.; Semorile, L. Comparative vinification assays with selected Patagonian strains of Oenococcus oeni and Lactobacillus plantarum. LWT 2016, 77, 348–355. [Google Scholar] [CrossRef]
- Cappello, M.S.; Zapparoli, G.; Logrieco, A.; Bartowsky, E.J. Linking wine lactic acid bacteria diversity with wine aroma and flavour. Int. J. Food Microbiol. 2017, 243, 16–27. [Google Scholar] [CrossRef]
- Genisheva, Z.; Mussatto, S.I.; Oliveira, J.M.; Teixeira, J.A. Malolactic fermentation of wines with immobilised lactic acid bacteria—Influence of concentration, type of support material and storage conditions. Food Chem. 2013, 138, 1510–1514. [Google Scholar] [CrossRef]
- Ribéreau-Gayon, P.; Dubourdieu, D.; Doneche, B.; Lonvaud-Funel, A. Handbook of Enology: The Microbiology of Wine and Vinifications, 2nd ed.; Wiley and Sons: Chichester, UK; New York, NY, USA, 2006; p. 325. [Google Scholar]
- Bravo-Ferrada, B.R.; Hollmann, A.; Delfederico, L.; Valdés La Hens, D.; Caballero, A.; Semorile, L. Patagonian red wines: Selection of Lactobacillus plantarum isolates as potential starter cultures for malolactic fermentation. World J. Microbiol. Biotechnol. 2013, 29, 1537–1549. [Google Scholar] [CrossRef]
- Liu, S.Q. Malolactic fermentation in wine—Beyond deacidification. J. Appl. Microbiol. 2010, 92, 589–601. [Google Scholar] [CrossRef]
- Iorizzo, M.; Testa, B.; Lombardi, S.J.; García-Ruiz, A.; Muñoz-González, C.; Bartolomé, B.; Moreno-Arribas, M.V.; Silvia Jane Lombardi, S.G. Selection and technological potential of Lactobacillus plantarum bacteria suitable for wine malolactic fermentation and grape aroma release. LWT 2016, 73, 557–566. [Google Scholar] [CrossRef]
- Behera, S.S.; Ray, R.C.; Nevijo, Z. Lactobacillus plantarum with Functional Properties: An Approach to Increase Safety and Shelf-Life of Fermented Foods. Biomed Res. Int. 2018, 2018, 9361614. [Google Scholar] [CrossRef]
- Felis, G.E.; Dellaglio, F. Taxonomy of Lactobacilli and Bifidobacteria. Curr. Issues Intest. Microbiol. 2007, 8, 44–61. [Google Scholar]
- Dicks, L.; Endo, A. Taxonomic Status of Lactic Acid Bacteria in Wine and Key Characteristics to Differentiate Species. S. Afr. J. Enol. Vitic. 2008, 30, 72–90. [Google Scholar] [CrossRef]
- Kumara, M.; Dhillonb, S.; Singhala, A.; Sooda, A.; Ghosh, D.M.; Ganguli, A. Cell surface and stress tolerance properties of a newly isolated Lactobacillus plantarum Ch1. Acta Mater. 2011, 40, 38–44. [Google Scholar] [CrossRef]
- Toit, M.; Engelbrecht, L.; Lerm, E.; Krieger-Weber, S. Lactobacillus: The Next Generation of Malolactic Fermentation Starter Cultures-an Overview. Food Bioproc. Tech. 2011, 4, 876–906. [Google Scholar] [CrossRef]
- Grimaldi, A.; Bartowsky, E.J.; Jiranek, V. Screening of Lactobacillus spp. and Pediococcus spp. for glycosidase activities that are important in oenology. J. Appl. Microbiol. 2010, 99, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- Grimaldi, A.; Bartowsky, E.; Jiranek, V. A survey of glycosidase activities of commercial wine strains of Oenococcus oeni. Int. J. Food Microbiol. 2005, 105, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, S.; Zhao, H.; Gu, P.; Chen, Y.; Zhang, B.; Zhu, B. Acetaldehyde released by Lactobacillus plantarum enhances accumulation of pyranoanthocyanins in wine during malolactic fermentation. Food Res. Int. 2018, 108, 254–263. [Google Scholar] [CrossRef]
- Curiel, J.A.; Muñoz, R.; de Felipe, F.L. Delaying Effect of a Wine Lactobacillus plantarum Strain on the Coloration and Xanthylium Pigment Formation Occurring in (+)-Catechin and (−)-Epicatechin Wine Model Solutions. J. Agric. Food Chem. 2010, 58, 11318–11324. [Google Scholar] [CrossRef] [PubMed]
- Mtshali, P.S.; Divol, B.; Van Rensburg, P.; Du Toit, M. Genetic screening of wine-related enzymes in Lactobacillus species isolated from South African wines. J. Appl. Microbiol. 2010, 108, 1389–1397. [Google Scholar] [CrossRef] [PubMed]
- Costello, P.J.; Siebert, T.E.; Solomon, M.R.; Bartowsky, E.J. Synthesis of fruity ethyl esters by acyl coenzyme A: Alcohol acyltransferase and reverse esterase activities in Oenococcus oeni and Lactobacillus plantarum. J. Appl. Microbiol. 2013, 114, 797–806. [Google Scholar] [CrossRef] [PubMed]
- Lerm, E.; Engelbrecht, L.; Du Toit, M. Selection and Characterisation of Oenococcus oeni and Lactobacillus plantarum South African Wine Isolates for Use as Malolactic Fermentation Starter Cultures. S. Afr. J. Enol. Vitic. 2011, 32, 280–295. [Google Scholar] [CrossRef]
- Vaquero, I.; Marcobal, Á.; Muñoz, R. Tannase activity by lactic acid bacteria isolated from grape must and wine. Int. J. Food Microbiol. 2004, 96, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Vataščinová, T.; Pipová, M.; Fraqueza, M.J.R.; Mala, P.; Dudriková, E.; Drážovská, M.; Lauková, A. Short communication: Antimicrobial potential of Lactobacillus plantarum strains isolated from Slovak raw sheep milk cheeses. J. Dairy Sci. 2020, 103, 6900–6903. [Google Scholar] [CrossRef]
- Seddik, H.A.; Bendali, F.; Gancel, F.; Fliss, I.; Spano, G.; Drider, D. Lactobacillus plantarum and Its Probiotic and Food Potentialities. Probiotics Antimicrob. Proteins 2017, 9, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Chen, O.; Hong, Y.; Ma, J.; Deng, L.; Yi, L.; Zeng, K. Screening lactic acid bacteria from pickle and cured meat as biocontrol agents of Penicillium digitatum on citrus fruit. Biol. Control 2021, 158, 104606. [Google Scholar] [CrossRef]
- Fayol-Messaoudi, D.; Coconnier-Polter, M.-H.; Moal, V.L.-L.; Atassi, F.; Berger, C.N.; Servin, A.L. The Lactobacillus plantarum strain ACA-DC287 isolated from a Greek cheese demonstrates antagonistic activity in vitro and in vivo against Salmonella enterica serovar Typhimurium. J. Appl. Microbiol. 2007, 103, 657–665. [Google Scholar] [CrossRef]
- Jin, G.; Jiranek, V.; Hayes, A.M.; Grbin, P.R. Isolation and Characterization of High-Ethanol-Tolerance Lactic Acid Bacteria from Australian Wine. Foods 2022, 11, 1231. [Google Scholar] [CrossRef]
- Xia, S.; Lin, R.; Cui, X.; Shan, J. The application of orthogonal test method in the parameters optimization of PEMFC under steady working condition. Int. J. Hydrog. Energy 2016, 41, 11380–11390. [Google Scholar] [CrossRef]
- Chen, Q.; Ren, X.; Wang, L.; Tian, Y.; Zhao, M.; Song, Z.; Liu, S. Correlation between β-Glycosidase Activity and Acid Stress Tolerance in Oenococcus oeni. Food Sci. 2017, 38, 115–120. (In Chinese) [Google Scholar]
- Bai, X.; Jin, G.; Liu, S.; Ma, W.; Zhang, Z.; Wang, H.; Zang, J. Malolactic Fermentation Characteristics of Lactobacillus hilgardii Q19 at Low Temperature and Its Effect on Aroma Components in Wine. Food Sci. 2020, 41, 146–152. (In Chinese) [Google Scholar]
- Berbegal, C.; Peña, N.; Russo, P.; Grieco, F.; Pardo, I.; Ferrer, S.; Spano, G.; Capozzi, V. Technological properties of Lactobacillus plantarum strains isolated from grape must fermentation. Food Microbiol. 2016, 57, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Succi, M.; Pannella, G.; Tremonte, P.; Tipaldi, L.; Coppola, R.; Iorizzo, M.; Lombardi, S.J.; Sorrentino, E. Sub-optimal pH Preadaptation Improves the Survival of Lactobacillus plantarum Strains and the Malic Acid Consumption in Wine-Like Medium. Food Microbiol. 2017, 8, 470. [Google Scholar] [CrossRef] [PubMed]
- Matthews, A.; Grimaldi, A.; Walker, M.; Bartowsky, E.; Grbin, P.; Jiranek, V. Lactic Acid Bacteria as a Potential Source of Enzymes for Use in Vinification. Appl. Environ. Microbiol. 2004, 70, 5715–5731. [Google Scholar] [CrossRef] [PubMed]
- Bartowsky, E.J.; Costello, P.J.; Chambers, P.J. Emerging trends in the application of malolactic fermentation. Aust. J. Grape Wine Res. 2015, 21, 663–669. [Google Scholar] [CrossRef]
- Spano, G.; Rinaldi, A.; Ugliano, M.; Moio, L.; Beneduce, L.; Massa, S. A β-glucosidase gene isolated from wine Lactobacillus plantarum is regulated by abiotic stresses. J. Appl. Microbiol. 2005, 98, 855–861. [Google Scholar] [CrossRef]
- Renchinkhand, G.; Park, Y.W.; Cho, S.-H.; Song, G.-Y.; Bae, H.C.; Choi, S.-J.; Nam, M.S. Identification of β-Glucosidase Activity of Lactobacillus plantarum CRNB22 in Kimchi and Its Potential to Convert Ginsenoside Rb1 from Panax Ginseng. J. Food Biochem. 2015, 39, 155–163. [Google Scholar] [CrossRef]
- Bravo-Ferrada, B.M.; Hollmann, A.; Brizuela, N.; La Hens, D.V.; Tymczyszyn, E.; Semorile, L. Growth and consumption of l-malic acid in wine-like medium by acclimated and non-acclimated cultures of Patagonian Oenococcus oeni strains. Folia Microbiol. 2016, 61, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Miller, B.J.; Franz, C.M.A.P.; Cho, G.S.; Du Toit, M. Expression of the Malolactic Enzyme Gene (mle) from Lactobacillus plantarum Under Winemaking Conditions. Curr. Microbiol. 2011, 62, 1682–1688. [Google Scholar] [CrossRef]
- Diez-Ozaeta, I.; Lavilla, M.; Amárita, F. Wine aroma profile modification by Oenococcus oeni strains from Rioja Alavesa region: Selection of potential malolactic starters—ScienceDirect. Int. J. Food Microbiol. 2021, 356, 109324. [Google Scholar] [CrossRef]
- López, I.; Tenorio, C.; Zarazaga, M.; Dizy, M.; Torres, C.; Ruiz-Larrea, F. Evidence of mixed wild populations of Oenococcus oeni strains during wine spontaneous malolactic fermentations. Eur. Food Res. Technol. 2007, 226, 215–223. [Google Scholar] [CrossRef]
- Ruiz, P.; Izquierdo, P.M.; Seseña, S.; Palop, M.L. Intraspecific genetic diversity of lactic acid bacteria from malolactic fermentation of Cencibel wines as derived from combined analysis of RAPD-PCR and PFGE patterns. Food Microbiol. 2008, 25, 942–948. [Google Scholar] [CrossRef]
- Ruiz, P.; Izquierdo, P.M.; Seseña, S.; Palop, M.L. Analysis of lactic acid bacteria populations during spontaneous malolactic fermentation of Tempranillo wines at five wineries during two consecutive vintages. Food Control 2010, 21, 70–75. [Google Scholar] [CrossRef]
- Beneduce, L.; Spano, G.; Vernile, A.; Tarantino, D.; Massa, S. Molecular characterization of lactic acid populations associated with wine spoilage. J. Basic Microbiol. 2004, 44, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Fang, Z.; Pai, A.; Luo, J.; Gan, R.; Gao, Y.; Lu, J.; Zhang, P. Glycosidically bound aroma precursors in fruits: A comprehensive review. Crit. Rev. Food Sci. Nutr. 2020, 62, 215–243. [Google Scholar] [CrossRef] [PubMed]
- Todaro, A.; Palmeri, R.; Barbagallo, R.N.; Pifferi, P.G.; Spagna, G. Increase of trans-resveratrol in typical Sicilian wine using β-Glucosidase from various sources. Food Chem. 2008, 107, 1570–1575. [Google Scholar] [CrossRef]
- Pozo-Bayón, M.A.; G-Alegría, E.; Polo, M.C.; Tenorio, C.; Martín-Alvarez, P.J.; Calvo de la Banda, M.T.; Ruiz-Larrea, F.; Moreno-Arribas, M.V. Wine Volatile and Amino Acid Composition after Malolactic Fermentation: Effect of Oenococcus oeni and Lactobacillus plantarum Starter Cultures. J. Agric. Food Chem. 2005, 53, 8729–8735. [Google Scholar] [CrossRef]
| Levels | Stress | ||
|---|---|---|---|
| Ethanol (%(v/v)) | pH | SO2 (mg/L) | |
| 1 | 11 | 3.2 | 20 |
| 2 | 13 | 3.4 | 30 |
| 3 | 15 | 3.6 | 40 |
| The Isolated Strains | Identification | 16S rRNA Sequencing Similarity (%) |
|---|---|---|
| A3, A4, A6, A7, A12, A18, A23, A39, A41, A45, A49, A50, A52, B17, B28, B32, B43, B44, B51, B64 | Lactiplantibacillus plantarum | 99.93–100 |
| A8 | Lentilactobacillus hilgardii | 99.93 |
| Experimental Number | Ethanol (% (v/v)) | pH | SO2 (mg/L) | The Viable Cell Counts (107 CFU/mL) | ||||
|---|---|---|---|---|---|---|---|---|
| A7 | A18 | A23 | A50 | B28 | ||||
| 1 | 1 | 1 | 1 | 24.07 | 17.60 | 14.33 | 16.00 | 28.53 |
| 2 | 1 | 2 | 2 | 13.33 | 22.53 | 24.07 | 19.13 | 24.00 |
| 3 | 1 | 3 | 3 | 13.60 | 14.00 | 9.27 | 9.13 | 15.73 |
| 4 | 2 | 2 | 1 | 0.51 | 0.82 | 0.99 | 0.68 | 0.86 |
| 5 | 2 | 3 | 2 | 11.47 | 8.40 | 20.60 | 7.60 | 15.47 |
| 6 | 2 | 1 | 3 | 0.23 | 0.08 | 0.09 | 0.40 | 0.17 |
| 7 | 3 | 3 | 1 | 0.93 | 1.22 | 0.11 | 0.11 | 0.69 |
| 8 | 3 | 1 | 2 | 0.49 | 0.14 | 1.05 | 1.18 | 0.61 |
| 9 | 3 | 2 | 3 | 0.19 | 1.04 | 2.77 | 0.03 | 0.26 |
| Strains | Stresses | Levels | R | ||
|---|---|---|---|---|---|
| 1 | 2 | 3 | |||
| A7 | Ethanol | δA1 = 9.80 | δA2 = −3.13 | δA3 = −6.67 | 16.47 |
| pH | δB1 = 1.06 | δB2 = −2.52 | δB3 = 1.47 | 3.99 | |
| SO2 | δC1 = 1.30 | δC2 = 1.23 | δC3 = −2.53 | 3.83 | |
| A18 | Ethanol | δA1 = 10.73 | δA2 = −4.21 | δA3 = −6.51 | 17.24 |
| pH | δB1 = −1.37 | δB2 = 0.82 | δB3 = 0.56 | 2.19 | |
| SO2 | δC1 = −0.76 | δC2 = 3.06 | δC3 = −2.27 | 5.33 | |
| A23 | Ethanol | δA1 = 7.75 | δA2 = −0.91 | δA3 = −6.83 | 14.58 |
| pH | δB1 = −2.98 | δB2 = 1.14 | δB3 = 1.85 | 4.83 | |
| SO2 | δC1 = −3.00 | δC2 = 7.10 | δC3 = −4.10 | 11.20 | |
| A50 | Ethanol | δA1 = 8.72 | δA2 = −3.14 | δA3 = −5.59 | 14.31 |
| pH | δB1 = −0.17 | δB2 = 0.58 | δB3 = −0.42 | 1.00 | |
| SO2 | δC1 = −0.43 | δC2 = 3.27 | δC3 = −2.84 | 6.11 | |
| B28 | Ethanol | δA1 = 13.16 | δA2 = −4.09 | δA3 = −9.07 | 22.23 |
| pH | δB1 = 0.18 | δB2 = −1.22 | δB3 = 1.04 | 2.26 | |
| SO2 | δC1 = 0.44 | δC2 = 3.77 | δC3 = −4.20 | 7.97 | |
| Strains | Ethanol (%) | pH | Total Sugar (g/L) | Titratable Acid (g/L) | Volatile Acid (g/L) | L-Lactic Acid (g/L) | Chromaticity | Hue | Glycerol (g/L) |
|---|---|---|---|---|---|---|---|---|---|
| Before MLF | 14.75 ± 0.03 abc | 3.52 ± 0.01 d | 3.63 ± 0.05 a | 8.19 ± 0.25 a | 0.28 ± 0.08 b | 0.08 ± 0.01 f | 17.89 ± 2.07 a | 0.81 ± 0.54 a | 1.03 ± 0.03 a |
| A7 | 14.81 ± 0.01 c | 3.68 ± 0.01 a | 3.53 ± 0.18 ab | 7.13 ± 0.15 ab | 0.58 ± 0.05 a | 1.38 ± 0.01 b | 20.50 ± 0.87 a | 0.68 ± 0.68 bc | 1.10 ± 0.05 a |
| A50 | 14.76 ± 0.01 b | 3.66 ± 0.01 ab | 3.45 ± 0.03 ab | 7.13 ± 0.01 ab | 0.59 ± 0.11 a | 1.14 ± 0.02 e | 20.92 ± 2.25 a | 0.67 ± 0.02 bc | 1.31 ± 0.02 a |
| B28 | 14.72 ± 0.01 bc | 3.67 ± 0.01 ab | 3.22 ± 0.07 ab | 7.31 ± 0.05 ab | 0.54 ± 0.03 a | 1.20 ± 0.01 d | 22.14 ± 2.14 a | 0.69 ± 0.03 b | 1.06 ± 0.24 a |
| 31-DH | 14.81 ± 0.01 a | 3.65 ± 0.01 bc | 3.25 ± 0.29 ab | 6.19 ± 0.05 b | 0.36 ± 0.01 b | 1.45 ± 0.01 a | 21.09 ± 1.83 a | 0.65 ± 0.07 bc | 1.21 ± 0.03 a |
| BV-S2 | 14.71 ± 0.00 c | 3.62 ± 0.01 c | 3.13 ± 0.05 b | 7.31 ± 0.01 ab | 0.34 ± 0.02 b | 1.34 ± 0.02 c | 21.62 ± 0.81 a | 0.59 ± 0.02 c | 1.08 ± 0.01 a |
| Strains | Tryptamine (mg/L) | β-Phenylethylamine (mg/L) | 1,4-Butanediamine (mg/L) | Cadaverine (mg/L) | Tyramine (mg/L) | Histamine (mg/L) |
|---|---|---|---|---|---|---|
| Before MLF | 0.64 ± 0.01 ab | 1.04 ± 0.08 a | 1.46 ± 0.22 a | 0.36 ± 0.02 a | 0.30 ± 0.02 ab | 0.35 ± 0.02 a |
| A7 | 0.58 ± 0.04 ab | 0.46 ± 0.00 bcd | 0.12 ± 0.00 c | 1.11 ± 0.09 a | 0.58 ± 0.08 a | 0.79 ± 0.055 c |
| A50 | 0.42 ± 0.01 b | 0.18 ± 0.18 d | 0.76 ± 0.05 b | 0.39 ± 0.17 b | 0.14 ± 0.02 b | 0.68 ± 0.004 b |
| B28 | 0.48 ± 0.04 ab | 0.34 ± 0.07 cd | 0.86 ± 0.04 b | 0.49 ± 0.08 ab | 0.11 ± 0.01 b | 0.38 ± 0.053 b |
| 31-DH | 0.72 ± 0.18 a | 0.61 ± 0.04 abc | 1.22 ± 0.28 ab | 0.62 ± 0.14 ab | 0.11 ± 0.00 b | 0.38 ± 0.08 ab |
| BV-S2 | 0.56 ± 0.02 ab | 0.86 ± 0.10 ab | 1.26 ± 0.02 ab | 0.31 ± 0.00 b | 0.11 ± 0.00 b | 0.27 ± 0.00 ab |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, J.; Ge, Y.; Gu, X.; Li, R.; Ma, W.; Jin, G. Identification and Characterization of Malolactic Bacteria Isolated from the Eastern Foothills of Helan Mountain in China. Foods 2022, 11, 2455. https://doi.org/10.3390/foods11162455
Sun J, Ge Y, Gu X, Li R, Ma W, Jin G. Identification and Characterization of Malolactic Bacteria Isolated from the Eastern Foothills of Helan Mountain in China. Foods. 2022; 11(16):2455. https://doi.org/10.3390/foods11162455
Chicago/Turabian StyleSun, Jingxian, Yuzi Ge, Xiaobo Gu, Ruyi Li, Wen Ma, and Gang Jin. 2022. "Identification and Characterization of Malolactic Bacteria Isolated from the Eastern Foothills of Helan Mountain in China" Foods 11, no. 16: 2455. https://doi.org/10.3390/foods11162455






