Isolation and Characterization of High-Ethanol-Tolerance Lactic Acid Bacteria from Australian Wine
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of Highly Ethanol Tolerant Bacteria from Wine
2.2. Bacterial DNA Extraction
2.3. Bacterial Species Identification
2.4. Amplified Fragment Length Polymorphism (AFLP) Analysis of O. oeni
2.5. Growth Assay in MRS-AJ
2.6. Ethanol Tolerance Assay
2.7. Statistical Analysis
3. Results
3.1. Isolation and Characterization of High Ethanol Tolerant Strains
3.2. Genotyping of O. oeni Strains by AFLP Analysis
3.3. Ethanol Tolerance Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ribéreau-Gayon, P.; Dubourdieu, D.; Done che, B.; Lonvaud-Funel, A. Handbook of Enology: The Microbiology of Wine and Vinifications, 2nd ed.; Wiley and Sons: Chichester, UK, 2006; pp. 145–149. [Google Scholar]
- Le Marrec, C.; Bon, E.; Lonvaud-Funel, A. Tolerance to high osmolality of the lactic acid bacterium Oenococcus oeni and identification of potential osmoprotectants. Int. J. Food Microbiol. 2007, 115, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Lonvaud-Funel, A. Lactic acid bacteria in the quality improvement and depreciation of wine. Antonie Van Leeuwenhoek 1999, 76, 317–331. [Google Scholar] [CrossRef] [PubMed]
- Cañas, P.M.I.; Pérez, P.R.; Prieto, S.S.; Herreros, M.L.P. Ecological study of lactic acid microbiota isolated from Tempranillo wines of Castilla-La Mancha. J. Biosci. Bioeng. 2009, 108, 220–224. [Google Scholar] [CrossRef]
- Ruiz, P.; Izquierdo, P.M.; Seseña, S.; Palop, M.L.I. Analysis of lactic acid bacteria populations during spontaneous malolactic fermentation of Tempranillo wines at five wineries during two consecutive vintages. Food Control 2010, 21, 70–75. [Google Scholar] [CrossRef]
- Du Toit, M.; Engelbrecht, L.; Lerm, E.; Krieger-Weber, S. Lactobacillus: The Next Generation of Malolactic Fermentation Starter Cultures—An Overview. Food Bioprocess Technol. 2011, 4, 876–906. [Google Scholar] [CrossRef]
- Li, H.; Wang, H.; Yuan, C.; Wang, S. Enology, 1st ed.; Science Publishing House: Beijing, China, 2000; pp. 108–114. [Google Scholar]
- Renouf, V.; Delaherche, A.; Claisse, O.; Lonvaud-Funel, A. Correlation between indigenous Oenococcus oeni strain resistance and the presence of genetic markers. J. Ind. Microbiol. Biotechnol. 2008, 35, 27–33. [Google Scholar] [CrossRef] [PubMed]
- De Orduña, R.M. Climate change associated effects on grape and wine quality and production. Food Res. Int. 2010, 43, 1844–1855. [Google Scholar] [CrossRef]
- Pilone, G.J.; Rankine, B.C.; Pilone, D.A. Inhibiting malo-lactic fermentation n in Australian dry red wines by adding fumaric acid. Am. J. Enol. Vitic. 1974, 25, 99. [Google Scholar]
- Jin, G.; Wang, H.; Zhang, C.; Li, C.; Du, L.; Grbin, P.R.; Li, H. Characterization and amino acid metabolism performances of indigenous Oenococcus oeni isolated from Chinese wines. Eur. Food Res. Technol. 2014, 238, 597–605. [Google Scholar] [CrossRef]
- Harrigan, W.F.; McCance, M.E. Laboratory Methods in Microbiology; Academic Press: London, UK, 1966; pp. 8–144. [Google Scholar]
- Zapparoli, G.; Torriani, S.; Pesente, P.; Dellaglio, F. Design and evaluation of malolactic enzyme gene targeted primers for rapid identification and detection of Oenococcus oeni in wine. Lett. Appl. Microbiol. 1998, 27, 243–246. [Google Scholar] [CrossRef]
- Sohier, D.; Coulon, J.; Lonvaud-Funel, A. Molecular identification of Lactobacillus hilgardii and genetic relatedness with Lactobacillus brevis. Int. J. Syst. Evol. Microbiol. 1999, 49, 1075–1081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nei, M.; Li, W.H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 1979, 76, 5269–5273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davidson, P.M.; Branen, A.L. Antimicrobials in Foods; Marcel Dekker: New York, NY, USA, 1993; pp. 25–29. [Google Scholar]
- Cappello, M.; Stefani, D.; Grieco, F.; Logrieco, A.; Zapparoli, G. Genotyping by Amplified Fragment Length Polymorphism and malate metabolism performances of indigenous Oenococcus oeni strains isolated from Primitivo wine. Int. J. Food Microbiol. 2008, 127, 241–245. [Google Scholar] [CrossRef] [PubMed]
- López, I.; Tenorio, C.; Zarazaga, M.; Dizy, M.; Torres, C.; Ruiz-Larrea, F. Evidence of mixed wild populations of Oenococcus oeni strains during wine spontaneous malolactic fermentations. Eur. Food Res. Technol. 2007, 226, 215–223. [Google Scholar] [CrossRef]
- Guerrini, S.; Bastianini, A.; Blaiotta, G.; Granchi, L.; Moschetti, G.; Coppola, S.; Romano, P.; Vincenzini, M. Phenotypic and genotypic characterization of Oenococcus oeni strains isolated from Italian wines. Int. J. Food Microbiol. 2002, 83, 1–14. [Google Scholar] [CrossRef]
- Lechiancole, T.; Blaiotta, G.; Messina, D.; Fusco, V.; Villani, F.; Salzano, G. Evaluation of intra-specific diversities in Oenococcus oeni through analysis of genomic and expressed DNA. Syst. Appl. Microbiol. 2006, 29, 375–381. [Google Scholar] [CrossRef]
- Vigentini, I.; Picozzi, C.; Tirelli, A.; Giugni, A.; Foschino, R. Survey on indigenous Oenococcus oeni strains isolated from red wines of Valtellina, a cold climate wine-growing Italian area. Int. J. Food Microbiol. 2009, 136, 123–128. [Google Scholar] [CrossRef]
- Giammanco, G.M.; Mammina, C.; Romani, C.; Luzzi, I.; Dionisi, A.M.; Nastasi, A. Evaluation of a modified single-enzyme amplified fragment length polymorphism (SE-AFLP) technique for subtyping Salmonella enterica serotype Enteritidis. Res. Microbiol. 2007, 158, 10–17. [Google Scholar] [CrossRef]
- Dimitrov, Z.P.; Minkova, S.; Michaylova, M. Comparative evaluation of three molecular typing methods in their applicability to differentiate Lactobacillus strains with human origin. World J. Microbiol. Biotechnol. 2008, 24, 1305–1312. [Google Scholar] [CrossRef]
- Vos, P.; Hogers, R.; Bleeker, M.; Reijans, M.; Van De Lee, T.; Hornes, M.; Frijters, A.; Pot, J.; Peleman, J.; Kuiper, M. AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res. 1995, 23, 4407–4414. [Google Scholar] [CrossRef] [Green Version]
- Salminen, S.; Isolauri, E.; Salminen, E. Clinical uses of probiotics for stabilizing the gut mucosal barrier: Successful strains and future challenges. Antonie van Leeuwenhoek 1996, 70, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Rico, A.; Ortiz-Barredo, A.; Ritter, E.; Murillo, J. Genetic characterization of Erwinia amylovora strains by amplified fragment length polymorphism. J. Appl. Microbiol. 2004, 96, 302–310. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; García, M.; Levisohn, S.; Savelkoul, P.; Leiting, V.; Lysnyansky, I.; Ley, D.H.; Kleven, S.H. Differentiation of Mycoplasma gallisepticum Strains Using Amplified Fragment Length Polymorphism and Other DNA-Based Typing Methods. Avian Dis. 2005, 49, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Janssen, P.; Coopman, R.; Huys, G.; Swings, J.; Bleeker, M.; Vos, P.; Zabeau, M.; Kersters, K. Evaluation of the DNA fingerprinting method AFLP as a new tool in bacterial taxonomy. Microbiology 1996, 142 Pt 7, 1881–1893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- G-Alegría, E.; López, I.; Ruiz, J.I.; Sáenz, J.; Fernández, E.; Zarazaga, M.; Dizy, M.; Torres, C.; Ruiz-Larrea, F. High tolerance of wild Lactobacillus plantarum and Oenococcus oeni strains to lyophilisation and stress environmental conditions of acid pH and ethanol. FEMS Microbiol. Lett. 2004, 230, 53–61. [Google Scholar] [CrossRef]
- Mtshali, P.; Divol, B.; Van Rensburg, P.; du Toit, M. Genetic screening of wine-related enzymes in Lactobacillus species isolated from South African wines. J. Appl. Microbiol. 2010, 108, 1389–1397. [Google Scholar] [CrossRef] [PubMed]
- Bou, M.; Krieger, S. Alcohol-Tolerant Malolactic Strains for the Maturation of Wines with Average or High pH. U.S. Patent US-2006153822-A1, 13 July 2006. [Google Scholar]
- Fumi, M.D.; Krieger Weber, S.; Deleris Bou, M.; Silva, A.; Dutoit, M. A new generation of malolactic starter cultures for high-pH wine. In Proceedings of the International IVIF Congress 2010, WB3 Microorganisms-Malolactic Fermentation, Stuttgart, Germany, 24–26 March 2010. [Google Scholar]
- Iorizzo, M.; Testa, B.; Lombardi, S.J.; García-Ruiz, A.; Muñoz-González, C.; Bartolomé, B.; Moreno-Arribas, M.V. Selection and technological potential of Lactobacillus plantarum bacteria suitable for wine malolactic fermentation and grape aroma release. LWT 2016, 73, 557–566. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Li, S.; Zhao, H.; Gu, P.; Chen, Y.; Zhang, B.; Zhu, B. Acetaldehyde released by Lactobacillus plantarum enhances accumulation of pyranoanthocyanins in wine during malolactic fermentation. Food Res. Int. 2018, 108, 254–263. [Google Scholar] [CrossRef] [Green Version]
- Gold, R.S.; Meagher, M.M.; Hutkins, R.; Conway, T. Ethanol tolerance and carbohydrate metabolism in lactobacilli. J. Ind. Microbiol. Biotechnol. 1992, 10, 45–54. [Google Scholar] [CrossRef]
- Jones, G.V. Climate variability and change: Influences on viticulture and wine production. In Proceedings of the 4th International Conferences of the South African Society for Enology and Viticulture, Cape Town, South Africa, 28–30 July 2009. [Google Scholar]
- Iacumin, L.; Comi, G.; Cantoni, C.; Cocolin, L. Ecology and dynamics of coagulase-negative cocci isolated from naturally fermented Italian sausages. Syst. Appl. Microbiol. 2006, 29, 480–486. [Google Scholar] [CrossRef]
- Wainwright, M.; Alharbi, S.; Wickramasinghe, N. How do microorganisms reach the stratosphere? Int. J. Astrobiol. 2006, 5, 13–15. [Google Scholar] [CrossRef] [Green Version]
- Bansal, E.; Garg, A.; Bhatia, S.; Attri, A.K.; Chander, J. Spectrum of microbial flora in diabetic foot ulcers. Indian J. Pathol. Microbiol. 2008, 51, 204–208. [Google Scholar] [CrossRef] [PubMed]
- Cogen, A.L.; Nizet, V.; Gallo, R.L. Skin microbiota: A source of disease or defence? Br. J. Dermatol. 2008, 158, 442–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sivadon, V.; Rottman, M.; Chaverot, S.; Quincampoix, J.-C.; Avettand, V.; de Mazancourt, P.; Bernard, L.; Trieu-Cuot, P.; Féron, J.-M.; Lortat-Jacob, A.; et al. Use of Genotypic Identification by sodA Sequencing in a Prospective Study to Examine the Distribution of Coagulase-Negative Staphylococcus Species among Strains Recovered during Septic Orthopedic Surgery and Evaluate Their Significance. J. Clin. Microbiol. 2005, 43, 2952–2954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, J.; Quan, L.-H.; Heu, S.; Jung, K.S.; Han, S.-W.; Moon, E.; Roh, E. A new antimicrobial substance produced by Staphylococcus pasteuri isolated from vegetables. Food Sci. Biotechnol. 2014, 23, 983–990. [Google Scholar] [CrossRef]
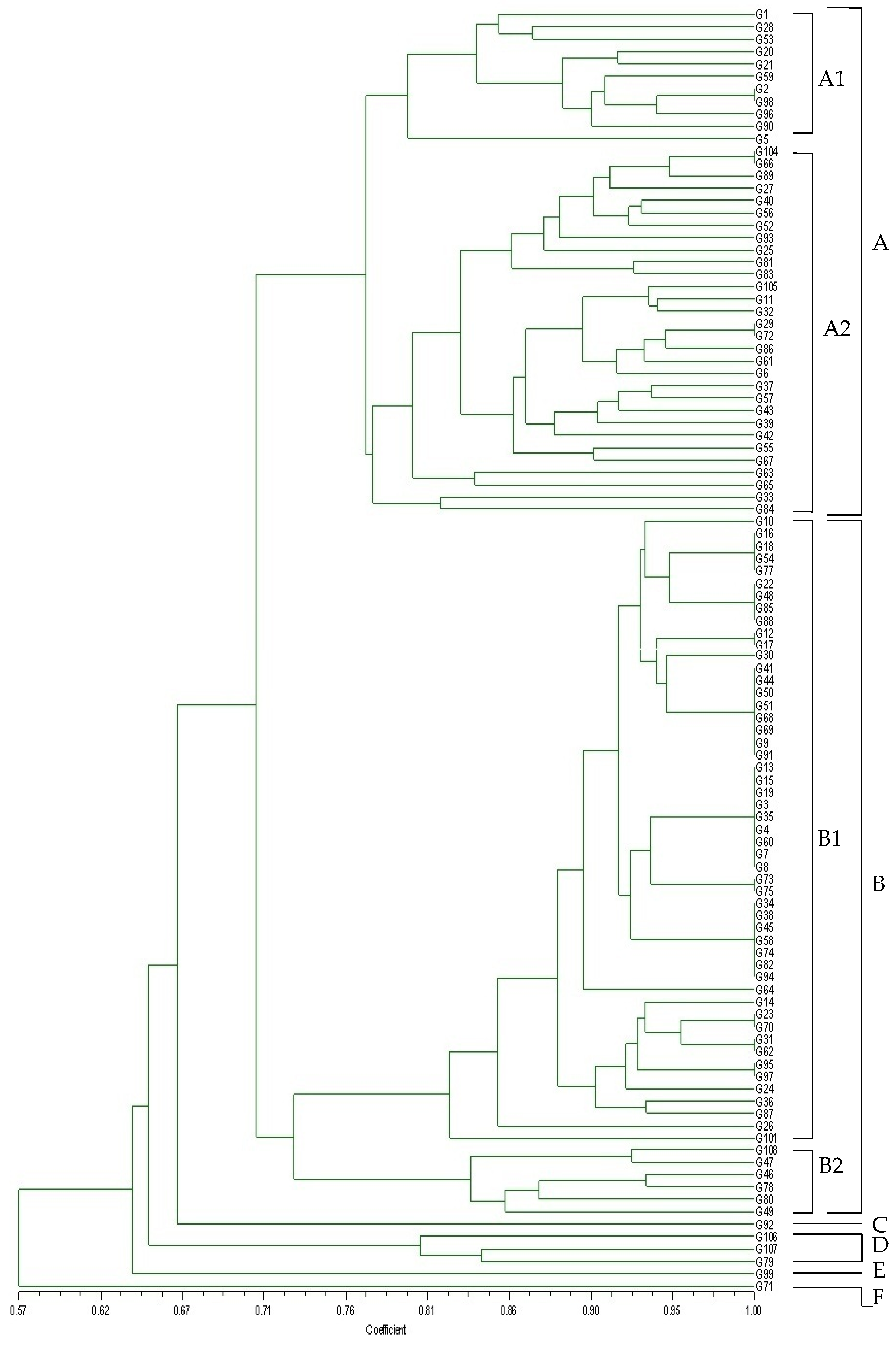
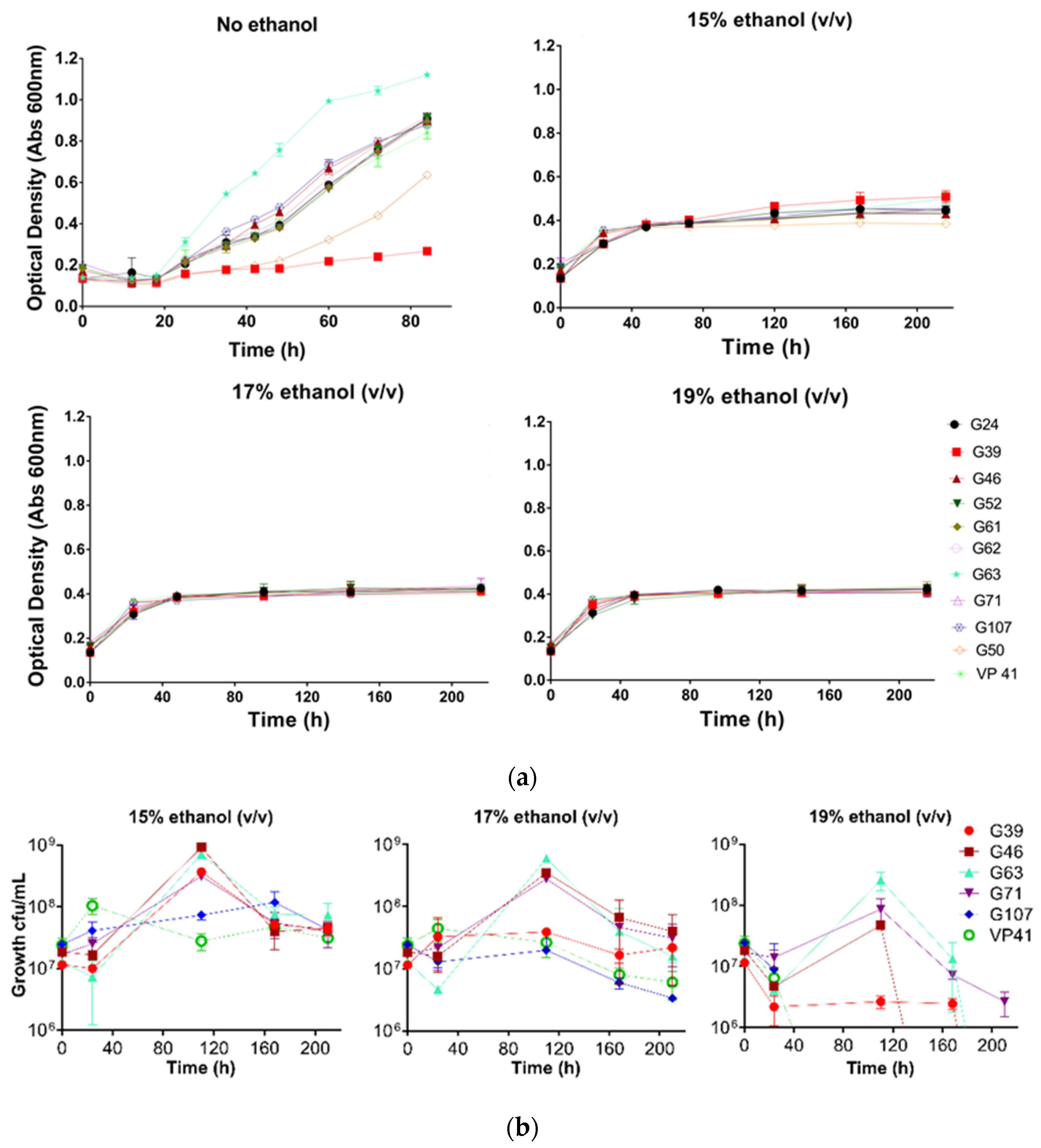
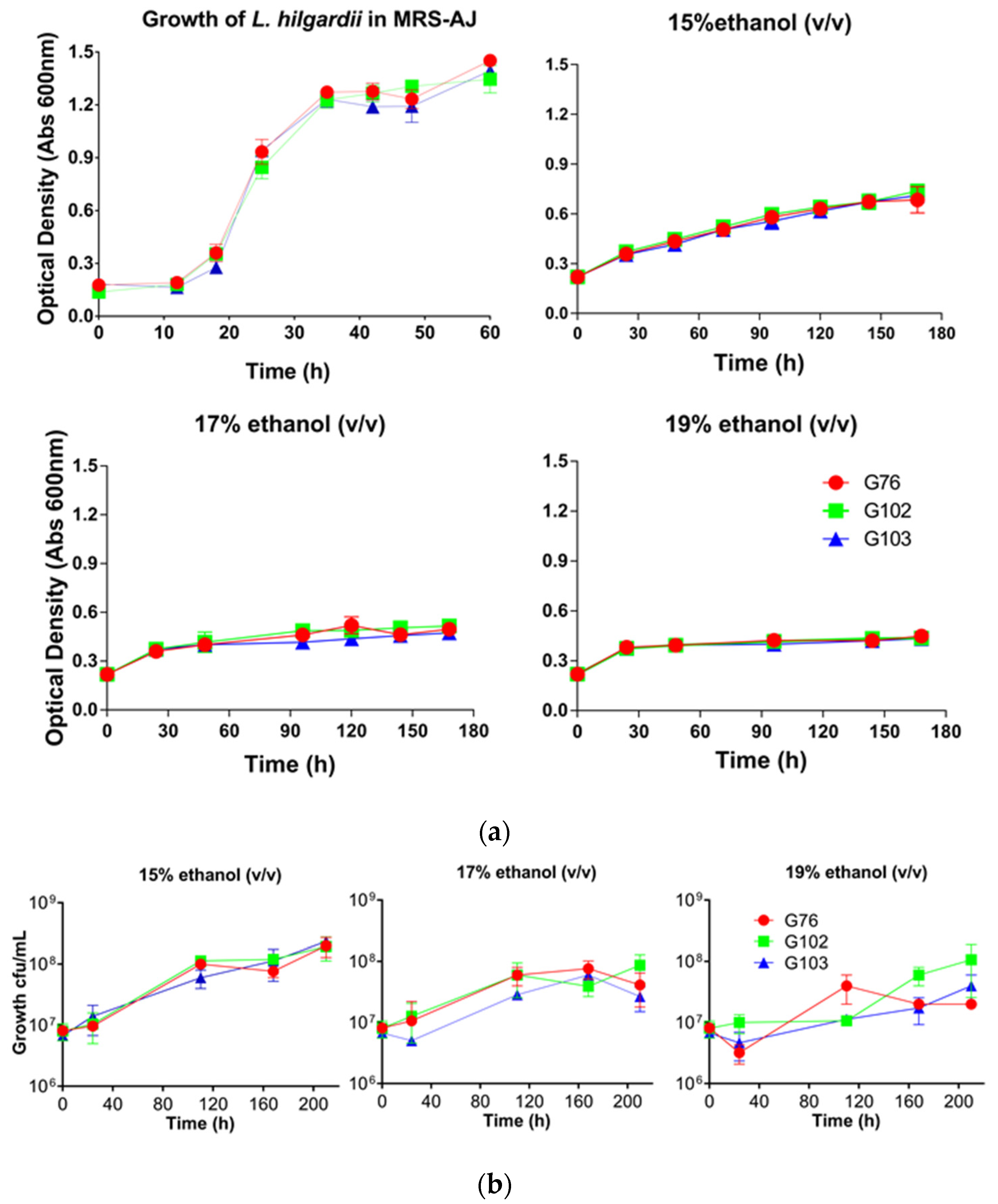
| Origin | Barrel | Alcohol Content (v/v) | pH | Malic Acid (g/L) | Isolates |
|---|---|---|---|---|---|
| Barossa | AU2 | 16.8 | 3.76 | 1.50 | G1, G2, G3, G89 |
| Barossa | AU4 | 16.8 | 3.76 | 1.63 | G4, G5, G6, G7, G8, G9, G10, G11, G12, G13, G14, G15, G16, G17, G18, G19, G20, G21, G22, G23, G24, G25, G26, G27, G28, G29, G30, G31, G32, G33, G34, G35, G36, G37, G38, G39, G40, G41, G42, G43, G90, G91, G92, G93, G94, G95, G96, G97, G100, G101, G102, G103 |
| Barossa | AU6 | 16.9 | 3.71 | 1.50 | G84, G85, G86, G87, G88, G98, G99, G104, G105 |
| Barossa | AU12 | 17.0 | 3. 78 | 1.56 | G44, G45, G46, G47, G48, G49, G50, G51, G52, G53, G54, G55, G56, G57, G58, G59, G60, G61, G62, G63, G64, G65, G66, G67, G68, G69, G70, G71, G72, G73, G74, G75, G76, G77, G78, G79, G80, G81, G82, G83, G106, G107, G108 |
| Identification | Number of Isolates | 16S rRNA Sequencing Similarity (%) |
|---|---|---|
| Oenococcus oeni | 104 | 99.42–100.00 |
| Lactobacillus hilgardii | 3 | 99.03–100.00 |
| Staphylococcus pasteuri | 1 | 99.46 |
| Primer Pair | Sequences of Selective Primers | Fragment No. |
|---|---|---|
| MC-HT FAM | MC 5′-GATGAGTCCTGAGTAAC-3′ HT 5′-GACTGCGTACCAGCTTT-3′ | 57–81 |
| MA-HT FAM | MA 5′-GATGAGTCCTGAGTAAA-3′ HT 5′-GACTGCGTACCAGCTTT-3′ | 76–85 |
| MT-HT FAM | MT 5′-GATGAGTCCTGAGTAAT-3′ HT 5′-GACTGCGTACCAGCTTT-3′ | 70–85 |
| MG-HT FAM | MG 5′-GATGAGTCCTGAGTAAG-3′ HT 5′-GACTGCGTACCAGCTTT-3′ | 57–64 |
| MA-HC FAM | MA 5′-GATGAGTCCTGAGTAAA-3′ HC 5′-GACTGCGTACCAGCTTC-3′ | 54–62 |
| MT-HC FAM | MT 5′-GATGAGTCCTGAGTAAT-3′ HC 5′-GACTGCGTACCAGCTTC-3′ | 52/74 |
| MC-HC FAM | MC 5′-GATGAGTCCTGAGTAAC-3′ HC 5′-GACTGCGTACCAGCTTC-3′′ | 57–75 |
| MG-HC FAM | MG 5′-GATGAGTCCTGAGTAAG-3′ HC 5′-GACTGCGTACCAGCTTC-3′ | 63–66 |
| HG-MA HEX | HG 5′-GACTGCGTACCAGCTTG-3′ MA 5′-GATGAGTCCTGAGTAAA-3 | 52–78 |
| HG-MT HEX | HG 5′-GACTGCGTACCAGCTTG-3′ MT 5′-GATGAGTCCTGAGTAAT-3 | 41–49 |
| HA-MT HEX | HA 5′-GACTGCGTACCAGCTTA-3′ MT 5′-GATGAGTCCTGAGTAAT-3 | 34–49 |
| Primer | O. oeni | L. hilgardii | ||||
|---|---|---|---|---|---|---|
| G13 | G22 | G63 | G76 | G102 | G103 | |
| MA-HT FAM | 99.60% ± 0.37 | 99.28% ± 0.19 | 99.71% ± 0.21 | 95.03% ± 0.37 | 96.17% ± 0.87 | 97.6% ± 0.93 |
| MT-HT FAM | 100% | 99.70% ± 0.22 | 99.69% ± 0.40 | 95.65% ± 0.59 | 95.52% ± 0.61 | 96.58% ± 0.20 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, G.; Jiranek, V.; Hayes, A.M.; Grbin, P.R. Isolation and Characterization of High-Ethanol-Tolerance Lactic Acid Bacteria from Australian Wine. Foods 2022, 11, 1231. https://doi.org/10.3390/foods11091231
Jin G, Jiranek V, Hayes AM, Grbin PR. Isolation and Characterization of High-Ethanol-Tolerance Lactic Acid Bacteria from Australian Wine. Foods. 2022; 11(9):1231. https://doi.org/10.3390/foods11091231
Chicago/Turabian StyleJin, Gang, Vladimir Jiranek, Aaron Mark Hayes, and Paul R. Grbin. 2022. "Isolation and Characterization of High-Ethanol-Tolerance Lactic Acid Bacteria from Australian Wine" Foods 11, no. 9: 1231. https://doi.org/10.3390/foods11091231






