A Systematic Quantitative Determination of the Antimicrobial Efficacy of Grape Seed Extract against Foodborne Bacterial Pathogens
Abstract
1. Introduction
2. Materials and Methods
2.1. Inoculum Preparation
2.2. Preparation of GSE Solution
2.3. Microbial Dynamics in the Presence of GSE
2.4. Statistical Analysis
3. Results and Discussion
3.1. The Impact of GSE on the Microbial Dynamics of L. monocytogenes
3.1.1. The Impact of the Bacterial Initial Inoculum Concentration on the Activity of GSE against L. monocytogenes
3.1.2. The Impact of the Bacterial Growth Phase on the Activity of GSE against L. monocytogenes
3.1.3. The Impact of sigB on the Activity of GSE against L. monocytogenes
3.2. The Impact of GSE on the Microbial Dynamics of Gram-Negative Bacteria
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arenas-Jal, M.; Suñé-Negre, J.M.; Pérez-Lozano, P.; García-Montoya, E. Trends in the food and sports nutrition industry: A review. Crit. Rev. Food Sci. Nutr. 2020, 60, 2405–2421. [Google Scholar] [CrossRef]
- Zhang, Z.-H.; Wang, L.-H.; Zeng, X.-A.; Han, Z.; Brennan, C.S. Non-thermal technologies and its current and future application in the food industry: A review. Int. J. Food Sci. Technol. 2019, 54, 1–13. [Google Scholar] [CrossRef]
- Costello, K.M.; Gutierrez-Merino, J.; Bussemaker, M.; Smet, C.; Van Impe, J.F.; Velliou, E.G. A multi-scale analysis of the effect of complex viscoelastic models on Listeria dynamics and adaptation in co-culture systems. AIChE J. 2019, 66, e16761. [Google Scholar] [CrossRef]
- Goryluk-Salmonowicz, A.; Popowska, M. Factors promoting and limiting antimicrobial resistance in the environment—Existing knowledge gaps. Front. Microbiol. 2022, 13, 992268. [Google Scholar] [CrossRef]
- EFSA (European Food Safety Authority); ECDC (European Centre for Disease Prevention and Control). The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2017. EFSA J. 2017, 17, e05598. [Google Scholar] [CrossRef]
- Landers, T.F.; Cohen, B.; Wittum, T.E.; Larson, E.L. A Review of Antibiotic Use in Food Animals: Perspective, Policy, and Potential. Public Health Rep. 2012, 127, 4–22. [Google Scholar] [CrossRef]
- Ma, F.; Xu, S.; Tang, Z.; Li, Z.; Zhang, L. Use of antimicrobials in food animals and impact of transmission of antimicrobial resistance on humans. Biosaf. Health 2021, 3, 32–38. [Google Scholar] [CrossRef]
- Michael, C.A.; Dominey-Howes, D.; Labbate, M. The Antimicrobial Resistance Crisis: Causes, Consequences, and Management. Front. Public Health 2014, 2, 145. [Google Scholar] [CrossRef]
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; Government of the United Kingdom: London, UK, 2016.
- Teixeira, J.A. Grand Challenges in Sustainable Food Processing. Front. Sustain. Food Syst. 2018, 2, 19. [Google Scholar] [CrossRef]
- Searchinger, T.; Hanson, C.; Ranganathan, J.; Lipinski, B.; Waite, R.; Winterbottom, R.; Dinshaw, A.; Heimlich, R. Creating a Sustainable Food Future: Interim Findings. A Menu of Solutions to Sustainably Feed More than 9 Billion People by 2050; World Resources Institute: Washington, DC, USA, 2013; Volume 130. Available online: http://www.ncbi.nlm.nih.gov/pubmed/13463888 (accessed on 31 January 2023).
- Liu, X.; Liu, R.; Zhao, R.; Wang, J.; Cheng, Y.; Liu, Q.; Wang, Y.; Yang, S. Synergistic Interaction Between Paired Combinations of Natural Antimicrobials Against Poultry-Borne Pathogens. Front. Microbiol. 2022, 13, 811784. [Google Scholar] [CrossRef]
- Quinto, E.J.; Caro, I.; Villalobos-Delgado, L.H.; Mateo, J.; De-Mateo-Silleras, B.; Redondo-Del-Río, M.P. Food Safety through Natural Antimicrobials. Antibiotics 2019, 8, 208. [Google Scholar] [CrossRef]
- Villalobos-Delgado, L.H.; Nevárez-Moorillon, G.V.; Caro, I.; Quinto, E.J.; Mateo, J. Natural antimicrobial agents to improve foods shelf life. In Food Quality and Shelf Life, Galanakis; Elsevier: Amsterdam, The Netherlands, 2019; pp. 125–157. [Google Scholar]
- Gómez, N.C.; Manetsberger, J.; Benomar, N.; Gutiérrez, S.C.; Abriouel, H. Antibacterial and antibiofilm effects of essential oil components, EDTA and HLE disinfectant solution on Enterococcus, Pseudomonas and Staphylococcus sp. multiresistant strains isolated along the meat production chain. Front. Microbiol. 2022, 13, 1014169. [Google Scholar] [CrossRef]
- Sabater, C.; Ruiz, L.; Delgado, S.; Ruas-Madiedo, P.; Margolles, A. Valorization of Vegetable Food Waste and By-Products through Fermentation Processes. Front. Microbiol. 2020, 11, 581997. [Google Scholar] [CrossRef]
- Idris, S.; Ndukwe, G.; Gimba, C. Preliminary phytochemical screening and antimicrobial activity of seed extracts of Persea americana (avocado pear). Bayero J. Pure Appl. Sci. 2010, 2, 173–176. [Google Scholar] [CrossRef]
- Al-Zoreky, N. Antimicrobial activity of pomegranate (Punica granatum L.) fruit peels. Int. J. Food Microbiol. 2009, 134, 244–248. [Google Scholar] [CrossRef]
- Andrade, M.A.; Lima, V.; Silva, A.S.; Vilarinho, F.; Castilho, M.C.; Khwaldia, K.; Ramos, F. Pomegranate and grape by-products and their active compounds: Are they a valuable source for food applications? Trends Food Sci. Technol. 2019, 86, 68–84. [Google Scholar] [CrossRef]
- Agourram, A.; Ghirardello, D.; Rantsiou, K.; Zeppa, G.; Belviso, S.; Romane, A.; Oufdou, K.; Giordano, M. Phenolic Content, Antioxidant Potential, and Antimicrobial Activities of Fruit and Vegetable By-Product Extracts. Int. J. Food Prop. 2013, 16, 1092–1104. [Google Scholar] [CrossRef]
- Abdalla, A.E.; Darwish, S.M.; Ayad, E.H.; El-Hamahmy, R.M. Egyptian mango by-product 2: Antioxidant and antimicrobial activities of extract and oil from mango seed kernel. Food Chem. 2007, 103, 1141–1152. [Google Scholar] [CrossRef]
- Saleem, M.; Saeed, M.T. Potential application of waste fruit peels (orange, yellow lemon and banana) as wide range natural antimicrobial agent. J. King Saud Univ. Sci. 2019, 32, 805–810. [Google Scholar] [CrossRef]
- Pandey, K.B.; Rizvi, S.I. Plant polyphenols as dietary antioxidants in human health and disease. Oxid. Med. Cell. Longev. 2009, 2, 270–278. [Google Scholar] [CrossRef]
- Daglia, M. Polyphenols as antimicrobial agents. Curr. Opin. Biotechnol. 2012, 23, 174–181. [Google Scholar] [CrossRef]
- Chedea, V.S.; Pop, R.M. Total Polyphenols Content and Antioxidant DPPH Assays on Biological Samples, 2nd ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2019; pp. 169–183. [Google Scholar] [CrossRef]
- Martin, M.E.; Grao-Cruces, E.; Millan-Linares, M.C.; Montserrat de la Paz, S. Grape (Vitis vinifera L.) Seed Oil: A Functional Food from the Winemaking Industry. Foods 2020, 9, 1360. [Google Scholar] [CrossRef]
- Oliveira, D.A.; Salvador, A.A.; Smânia, A.; Smânia, E.F.; Maraschin, M.; Ferreira, S.R. Antimicrobial activity and composition profile of grape (Vitis vinifera) pomace extracts obtained by supercritical fluids. J. Biotechnol. 2012, 164, 423–432. [Google Scholar] [CrossRef]
- Özkan, G.; Sagdiç, O.; Baydar, N.G.; Kurumahmutoglu, Z. Antibacterial activities and total phenolic contents of grape pomace extracts. J. Sci. Food Agric. 2004, 84, 1807–1811. [Google Scholar] [CrossRef]
- Anastasiadi, M.; Chorianopoulos, N.G.; Nychas, G.-J.E.; Haroutounian, S.A. Antilisterial Activities of Polyphenol-Rich Extracts of Grapes and Vinification Byproducts. J. Agric. Food Chem. 2008, 57, 457–463. [Google Scholar] [CrossRef]
- Farhadi, K.; Esmaeilzadeh, F.; Hatami, M.; Forough, M.; Molaie, R. Determination of phenolic compounds content and antioxidant activity in skin, pulp, seed, cane and leaf of five native grape cultivars in West Azerbaijan province, Iran. Food Chem. 2016, 199, 847–855. [Google Scholar] [CrossRef]
- Ribeiro, L.F.; Ribani, R.H.; Francisco, T.M.G.; Soares, A.A.; Pontarolo, R.; Haminiuk, C.W.I. Profile of bioactive compounds from grape pomace (Vitis vinifera and Vitis labrusca) by spectrophotometric, chromatographic and spectral analyses. J. Chromatogr. B 2015, 1007, 72–80. [Google Scholar] [CrossRef]
- Zhao, X.; Chen, L.; Wu, J.; He, Y.; Yang, H. Elucidating antimicrobial mechanism of nisin and grape seed extract against Listeria monocytogenes in broth and on shrimp through NMR-based metabolomics approach. Int. J. Food Microbiol. 2019, 319, 108494. [Google Scholar] [CrossRef]
- FDA. Grape Seed Extract (GSE) Notification; U.S. Food and Drug Administration: Silver Spring, MD, USA, 2003.
- Xu, Y.; Burton, S.; Kim, C.; Sismour, E. Phenolic compounds, antioxidant, and antibacterial properties of pomace extracts from four Virginia-grown grape varieties. Food Sci. Nutr. 2016, 4, 125–133. [Google Scholar] [CrossRef]
- Yu, J.; Ahmedna, M. Functional components of grape pomace: Their composition, biological properties and potential applications. Int. J. Food Sci. Technol. 2013, 48, 221–237. [Google Scholar] [CrossRef]
- Lingua, M.S.; Fabani, M.P.; Wunderlin, D.A.; Baroni, M.V. In vivo antioxidant activity of grape, pomace and wine from three red varieties grown in Argentina: Its relationship to phenolic profile. J. Funct. Foods 2016, 20, 332–345. [Google Scholar] [CrossRef]
- Goula, A.; Thymiatis, K.; Kaderides, K. Valorization of grape pomace: Drying behavior and ultrasound extraction of phenolics. Food Bioprod. Process. 2016, 100, 132–144. [Google Scholar] [CrossRef]
- Brewer, M.S. Natural Antioxidants: Sources, Compounds, Mechanisms of Action, and Potential Applications. Compr. Rev. Food Sci. Food Saf. 2011, 10, 221–247. [Google Scholar] [CrossRef]
- Drosou, C.; Kyriakopoulou, K.; Bimpilas, A.; Tsimogiannis, D.; Krokida, M. A comparative study on different extraction techniques to recover red grape pomace polyphenols from vinification byproducts. Ind. Crop. Prod. 2015, 75, 141–149. [Google Scholar] [CrossRef]
- Sagdic, O.; Ozturk, I.; Yilmaz, M.T.; Yetim, H. Effect of Grape Pomace Extracts Obtained from Different Grape Varieties on Microbial Quality of Beef Patty. J. Food Sci. 2011, 76, M515–M521. [Google Scholar] [CrossRef]
- Lončarević, I.; Petrović, J.; Teslić, N.; Nikolić, I.; Maravić, N.; Pajin, B.; Pavlić, B. Cocoa Spread with Grape Seed Oil and Encapsulated Grape Seed Extract: Impact on Physical Properties, Sensory Characteristics and Polyphenol Content. Foods 2022, 11, 2730. [Google Scholar] [CrossRef]
- Silva, V.; Igrejas, G.; Falco, V.; Santos, T.P.; Torres, C.; Oliveira, A.M.; Pereira, J.E.; Amaral, J.S.; Poeta, P. Chemical composition, antioxidant and antimicrobial activity of phenolic compounds extracted from wine industry by-products. Food Control. 2018, 92, 516–522. [Google Scholar] [CrossRef]
- Katalinić, V.; Možina, S.S.; Skroza, D.; Generalić, I.; Abramovič, H.; Miloš, M.; Ljubenkov, I.; Piskernik, S.; Pezo, I.; Terpinc, P.; et al. Polyphenolic profile, antioxidant properties and antimicrobial activity of grape skin extracts of 14 Vitis vinifera varieties grown in Dalmatia (Croatia). Food Chem. 2010, 119, 715–723. [Google Scholar] [CrossRef]
- Adámez, J.D.; Samino, E.G.; Sánchez, E.V.; González-Gómez, D. In vitro estimation of the antibacterial activity and antioxidant capacity of aqueous extracts from grape-seeds (Vitis vinifera L.). Food Control. 2012, 24, 136–141. [Google Scholar] [CrossRef]
- Baydar, N.G.; Özkan, G.; Sagdic, O. Total phenolic contents and antibacterial activities of grape (Vitis vinifera L.) extracts. Food Control. 2004, 15, 335–339. [Google Scholar] [CrossRef]
- Silvan, J.M.; Gutiérrez-Docio, A.; Moreno-Fernandez, S.; Alarcón-Cavero, T.; Prodanov, M.; Martinez-Rodriguez, A.J. Procyanidin-Rich Extract from Grape Seeds as a Putative Tool against Helicobacter pylori. Foods 2020, 9, 1370. [Google Scholar] [CrossRef]
- Corrales, M.; Han, J.H.; Tauscher, B. Antimicrobial properties of grape seed extracts and their effectiveness after incorporation into pea starch films. Int. J. Food Sci. Technol. 2009, 44, 425–433. [Google Scholar] [CrossRef]
- Silván, J.M.; Mingo, E.; Hidalgo, M.; de Pascual-Teresa, S.; Carrascosa, A.V.; Martinez-Rodriguez, A.J. Antibacterial activity of a grape seed extract and its fractions against Campylobacter spp. Food Control. 2013, 29, 25–31. [Google Scholar] [CrossRef]
- Begg, S.L. The role of metal ions in the virulence and viability of bacterial pathogens. Biochem. Soc. Trans. 2019, 47, 77–87. [Google Scholar] [CrossRef]
- Klančnik, A.; Piskernik, S.; Jeršek, B.; Možina, S.S. Evaluation of diffusion and dilution methods to determine the antibacterial activity of plant extracts. J. Microbiol. Methods 2010, 81, 121–126. [Google Scholar] [CrossRef]
- Baydar, N.G.; Sagdic, O.; Ozkan, G.; Cetin, S. Determination of antibacterial effects and total phenolic contents of grape (Vitis vinifera L.) seed extracts. Int. J. Food Sci. Technol. 2006, 41, 799–804. [Google Scholar] [CrossRef]
- Sivarooban, T.; Hettiarachchy, N.S.; Johnson, M.G. Inhibition of Listeria monocytogenes using nisin with grape seed extract on turkey frankfurters stored at 4 and 10 °C. J. Food Prot. 2007, 70, 1017–1020. [Google Scholar] [CrossRef]
- Zhao, X.; Chen, L.; Zhao, L.; He, Y.; Yang, H. Antimicrobial kinetics of nisin and grape seed extract against inoculated Listeria monocytogenes on cooked shrimps: Survival and residual effects. Food Control. 2020, 115, 107278. [Google Scholar] [CrossRef]
- Ahn, J.; Grün, I.U.; Mustapha, A. Antimicrobial and Antioxidant Activities of Natural Extracts in Vitro and in Ground Beef. J. Food Prot. 2004, 67, 148–155. [Google Scholar] [CrossRef]
- Ahn, J.; Grün, I.U.; Mustapha, A. Effects of plant extracts on microbial growth, color change, and lipid oxidation in cooked beef. Food Microbiol. 2007, 24, 7–14. [Google Scholar] [CrossRef]
- Sheng, L.; Olsen, S.A.; Hu, J.; Yue, W.; Means, W.J.; Zhu, M.J. Inhibitory effects of grape seed extract on growth, quorum sensing, and virulence factors of CDC “top-six” non-O157 Shiga toxin producing E. coli. Int. J. Food Microbiol. 2016, 229, 24–32. [Google Scholar] [CrossRef]
- Kao, T.-T.; Tu, H.-C.; Chang, W.-N.; Chen, B.-H.; Shi, Y.-Y.; Chang, T.-C.; Fu, T.-F. Grape seed extract inhibits the growth and pathogenicity of Staphylococcus aureus by interfering with dihydrofolate reductase activity and folate-mediated one-carbon metabolism. Int. J. Food Microbiol. 2010, 141, 17–27. [Google Scholar] [CrossRef]
- Pletnev, P.; Osterman, I.; Sergiev, P.V.; Bogdanov, A.; Dontsova, O. Survival guide: Escherichia coli in the stationary phase. Acta Nat. 2015, 7, 22–33. [Google Scholar] [CrossRef]
- Utratna, M.; Cosgrave, E.; Baustian, C.; Ceredig, R.H.; O’Byrne, C.P. Effects of Growth Phase and Temperature on σB Activity within a Listeria monocytogenes Population: Evidence for RsbV-Independent Activation of σB at Refrigeration Temperatures. BioMed Res. Int. 2014, 2014, 1–11. [Google Scholar] [CrossRef]
- Boura, M.; Keating, C.; Royet, K.; Paudyal, R.; O’Donoghue, B.; O’Byrne, C.P.; Karatzas, K.A.G. Loss of SigB in Listeria monocytogenes Strains EGD-e and 10403S Confers Hyperresistance to Hydrogen Peroxide in Stationary Phase under Aerobic Conditions. Appl. Environ. Microbiol. 2016, 82, 4584–4591. [Google Scholar] [CrossRef]
- O’Byrne, C.P.; Karatzas, K.A. The Role of Sigma B (σB) in the Stress Adaptations of Listeria monocytogenes: Overlaps between Stress Adaptation and Virulence. Adv. Appl. Microbiol. 2008, 65, 115–140. [Google Scholar] [CrossRef]
- Raengpradub, S.; Wiedmann, M.; Boor, K.J. Comparative Analysis of the σ B -Dependent Stress Responses in Listeria monocytogenes and Listeria innocua Strains Exposed to Selected Stress Conditions. Appl. Environ. Microbiol. 2008, 74, 158–171. [Google Scholar] [CrossRef]
- Begley, M.; Hill, C.; Ross, R.P. Tolerance of Listeria monocytogenes to Cell Envelope-Acting Antimicrobial Agents Is Dependent on SigB. Appl. Environ. Microbiol. 2006, 72, 2231–2234. [Google Scholar] [CrossRef]
- Sivarooban, T.; Hettiarachchy, N.S.; Johnson, M.G. Transmission electron microscopy study of Listeria monocytogenes treated with nisin in combination with either grape seed or green tea extract. J. Food Prot. 2008, 71, 2105–2109. [Google Scholar] [CrossRef]
- Fong, I.W.; Engelking, E.R.; Kirby, W.M.M. Relative Inactivation by Staphylococcus aureus of Eight Cephalosporin Antibiotics. Antimicrob. Agents Chemother. 1976, 9, 939–944. [Google Scholar] [CrossRef]
- Bacon, R.T.; Ransom, J.R.; Sofos, J.N.; Kendall, P.A.; Belk, K.E.; Smith, G.C. Thermal Inactivation of Susceptible and Multiantimicrobial-Resistant Salmonella Strains Grown in the Absence or Presence of Glucose. Appl. Environ. Microbiol. 2003, 69, 4123–4128. [Google Scholar] [CrossRef]
- Shen, C.; Sofos, J. Antilisterial Activity of Hops Beta Acids in Broth with or Without Other Antimicrobials. J. Food Sci. 2008, 73, M438–M442. [Google Scholar] [CrossRef]
- Oh, D.-H.; Marshall, D.L. Antimicrobial activity of ethanol, glycerol monolaurate or lactic acid against Listeria monocytogenes. Int. J. Food Microbiol. 1993, 20, 239–246. [Google Scholar] [CrossRef]
- Lambert, R.; Skandamis, P.; Coote, P.; Nychas, G.-J. A study of the minimum inhibitory concentration and mode of action of oregano essential oil, thymol and carvacrol. J. Appl. Microbiol. 2001, 91, 453–462. [Google Scholar] [CrossRef]
- European Food Safety Authority. European Centre for Disease Prevention and Control The European Union One Health 2020 Zoonoses Report. EFSA J. 2021, 19, e06971. [Google Scholar] [CrossRef]
- Velliou, E.; Van Derlinden, E.; Cappuyns, A.; Geeraerd, A.; Devlieghere, F.; Van Impe, J. Heat inactivation of Escherichia coli K12 MG1655: Effect of microbial metabolites and acids in spent medium. J. Therm. Biol. 2012, 37, 72–78. [Google Scholar] [CrossRef]
- Velliou, E.; Noriega, E.; Van Derlinden, E.; Mertens, L.; Boons, K.; Geeraerd, A.; Devlieghere, F.; Van Impe, J. The effect of colony formation on the heat inactivation dynamics of Escherichia coli K12 and Salmonella typhimurium. Food Res. Int. 2013, 54, 1746–1752. [Google Scholar] [CrossRef]
- Costello, K.M.; Smet, C.; Gutierrez-Merino, J.; Bussemaker, M.; Van Impe, J.F.; Velliou, E.G. The impact of food model system structure on the inactivation of Listeria innocua by cold atmospheric plasma and nisin combined treatments. Int. J. Food Microbiol. 2021, 337, 108948. [Google Scholar] [CrossRef]
- Velliou, E.; Van Derlinden, E.; Cappuyns, A.; Aerts, D.; Nikolaidou, E.; Geeraerd, A.; Devlieghere, F.; Van Impe, J. Quantification of the influence of trimethylamine-N-oxide (TMAO) on the heat resistance of Escherichia coli K12 at lethal temperatures. Lett. Appl. Microbiol. 2010, 52, 116–122. [Google Scholar] [CrossRef]
- Costello, K.M.; Gutierrez-Merino, J.; Bussemaker, M.; Ramaioli, M.; Baka, M.; Van Impe, J.F.; Velliou, E.G. Modelling the microbial dynamics and antimicrobial resistance development of Listeria in viscoelastic food model systems of various structural complexities. Int. J. Food Microbiol. 2018, 286, 15–30. [Google Scholar] [CrossRef]
- Velliou, E.G.; Van Derlinden, E.; Cappuyns, A.M.; Goossens, J.; Geeraerd, A.H.; Devlieghere, F.; Van Impe, J.F. Heat adaptation of Escherichia coli K12: Effect of acid and glucose. Procedia Food Sci. 2011, 1, 987–993. [Google Scholar] [CrossRef]
- Velliou, E.; Van Derlinden, E.; Cappuyns, A.; Nikolaidou, E.; Geeraerd, A.; Devlieghere, F.; Van Impe, J. Towards the quantification of the effect of acid treatment on the heat tolerance of Escherichia coli K12 at lethal temperatures. Food Microbiol. 2011, 28, 702–711. [Google Scholar] [CrossRef]
- Jydegaard, A.-M.; Gravesen, A.; Knøchel, S. Growth condition-related response of Listeria monocytogenes 412 to bacteriocin inactivation. Lett. Appl. Microbiol. 2000, 31, 68–72. [Google Scholar] [CrossRef]
- Jaishankar, J.; Srivastava, P. Molecular Basis of Stationary Phase Survival and Applications. Front. Microbiol. 2017, 8, 2000. [Google Scholar] [CrossRef]
- Davis, M.J.; Coote, P.J.; O’Byrne, C.P. Acid tolerance in Listeria monocytogenes: The adaptive acid tolerance response (ATR) and growth-phase-dependent acid resistance. Microbiology 1996, 142, 2975–2982. [Google Scholar] [CrossRef]
- Mendonça, A.F.; Daraba, A. NON-THERMAL PROCESSING|Irradiation. In Encyclopedia of Food Microbiology; Elsevier: Amsterdam, The Netherlands, 2014; Volume 2, pp. 954–961. [Google Scholar] [CrossRef]
- Mañas, P.; Mackey, B.M. Morphological and Physiological Changes Induced by High Hydrostatic Pressure in Exponential- and Stationary-Phase Cells of Escherichia coli: Relationship with Cell Death. Appl. Environ. Microbiol. 2004, 70, 1545–1554. [Google Scholar] [CrossRef]
- Cebrián, G.; Sagarzazu, N.; Pagán, R.; Condón, S.; Mañas, P. Heat and pulsed electric field resistance of pigmented and non-pigmented enterotoxigenic strains of Staphylococcus aureus in exponential and stationary phase of growth. Int. J. Food Microbiol. 2007, 118, 304–311. [Google Scholar] [CrossRef]
- Lu, L.-C.; Chen, Y.-W.; Chou, C.-C. Antibacterial activity of propolis against Staphylococcus aureus. Int. J. Food Microbiol. 2005, 102, 213–220. [Google Scholar] [CrossRef]
- Rocco, A.; Kierzek, A.M.; McFadden, J. Slow Protein Fluctuations Explain the Emergence of Growth Phenotypes and Persistence in Clonal Bacterial Populations. PLoS ONE 2013, 8, e54272. [Google Scholar] [CrossRef]
- Ortuño, C.; Martínez-Pastor, M.T.; Mulet, A.; Benedito, J. Supercritical carbon dioxide inactivation of Escherichia coli and Saccharomyces cerevisiae in different growth stages. J. Supercrit. Fluids 2012, 63, 8–15. [Google Scholar] [CrossRef]
- Zhao, L.; Qin, X.; Wang, Y.; Ling, J.; Shi, W.; Pang, S.; Liao, X. CO2 -assisted high pressure processing on inactivation of Escherichia coli and Staphylococcus aureus. J. CO2 Util. 2017, 22, 53–62. [Google Scholar] [CrossRef]
- Yu, H.; Perni, S.; Shi, J.; Wang, D.; Kong, M.; Shama, G. Effects of cell surface loading and phase of growth in cold atmospheric gas plasma inactivation of Escherichia coli K12. J. Appl. Microbiol. 2006, 101, 1323–1330. [Google Scholar] [CrossRef]
- Coroller, L.; Leguerinel, I.; Mettler, E.; Savy, N.; Mafart, P. General Model, Based on Two Mixed Weibull Distributions of Bacterial Resistance, for Describing Various Shapes of Inactivation Curves. Appl. Environ. Microbiol. 2006, 72, 6493–6502. [Google Scholar] [CrossRef]
- Kearns, D.B.; Losick, R. Cell population heterogeneity during growth of Bacillus subtilis. Genes Dev. 2005, 19, 3083–3094. [Google Scholar] [CrossRef]
- Schottroff, F.; Fröhling, A.; Zunabovic-Pichler, M.; Krottenthaler, A.; Schlüter, O.; Jäger, H. Sublethal Injury and Viable but Non-culturable (VBNC) State in Microorganisms During Preservation of Food and Biological Materials by Non-thermal Processes. Front. Microbiol. 2018, 9, 2773. [Google Scholar] [CrossRef]
- Wu, S.; Yu, P.-L.; Flint, S. Persister cell formation of Listeria monocytogenes in response to natural antimicrobial agent nisin. Food Control. 2017, 77, 243–250. [Google Scholar] [CrossRef]
- Dorey, A.L.; Lee, B.-H.; Rotter, B.; O’Byrne, C.P. Blue Light Sensing in Listeria monocytogenes Is Temperature-Dependent and the Transcriptional Response to It Is Predominantly SigB-Dependent. Front. Microbiol. 2019, 10, 2497. [Google Scholar] [CrossRef]
- Palmer, M.E.; Wiedmann, M.; Boor, K.J. σB and σL Contribute to Listeria monocytogenes 10403S Response to the Antimicrobial Peptides SdpC and Nisin. Foodborne Pathog. Dis. 2009, 6, 1057–1065. [Google Scholar] [CrossRef]
- Guerreiro, D.N.; Arcari, T.; O’Byrne, C.P. The σB-Mediated General Stress Response of Listeria monocytogenes: Life and Death Decision Making in a Pathogen. Front. Microbiol. 2020, 11, 1505. [Google Scholar] [CrossRef]
- Gyawali, R.; Ibrahim, S.A. Natural products as antimicrobial agents. Food Control. 2014, 46, 412–429. [Google Scholar] [CrossRef]
- Deng, Q.; Zhao, Y. Physicochemical, Nutritional, and Antimicrobial Properties of Wine Grape (cv. Merlot) Pomace Extract-Based Films. J. Food Sci. 2011, 76, E309–E317. [Google Scholar] [CrossRef]
- Costello, K.M.; Velliou, E.; Gutierrez-Merino, J.; Smet, C.; El Kadri, H.; Van Impe, J.F.; Bussemaker, M. The effect of ultrasound treatment in combination with nisin on the inactivation of Listeria innocua and Escherichia coli. Ultrason. Sonochemistry 2021, 79, 105776. [Google Scholar] [CrossRef]
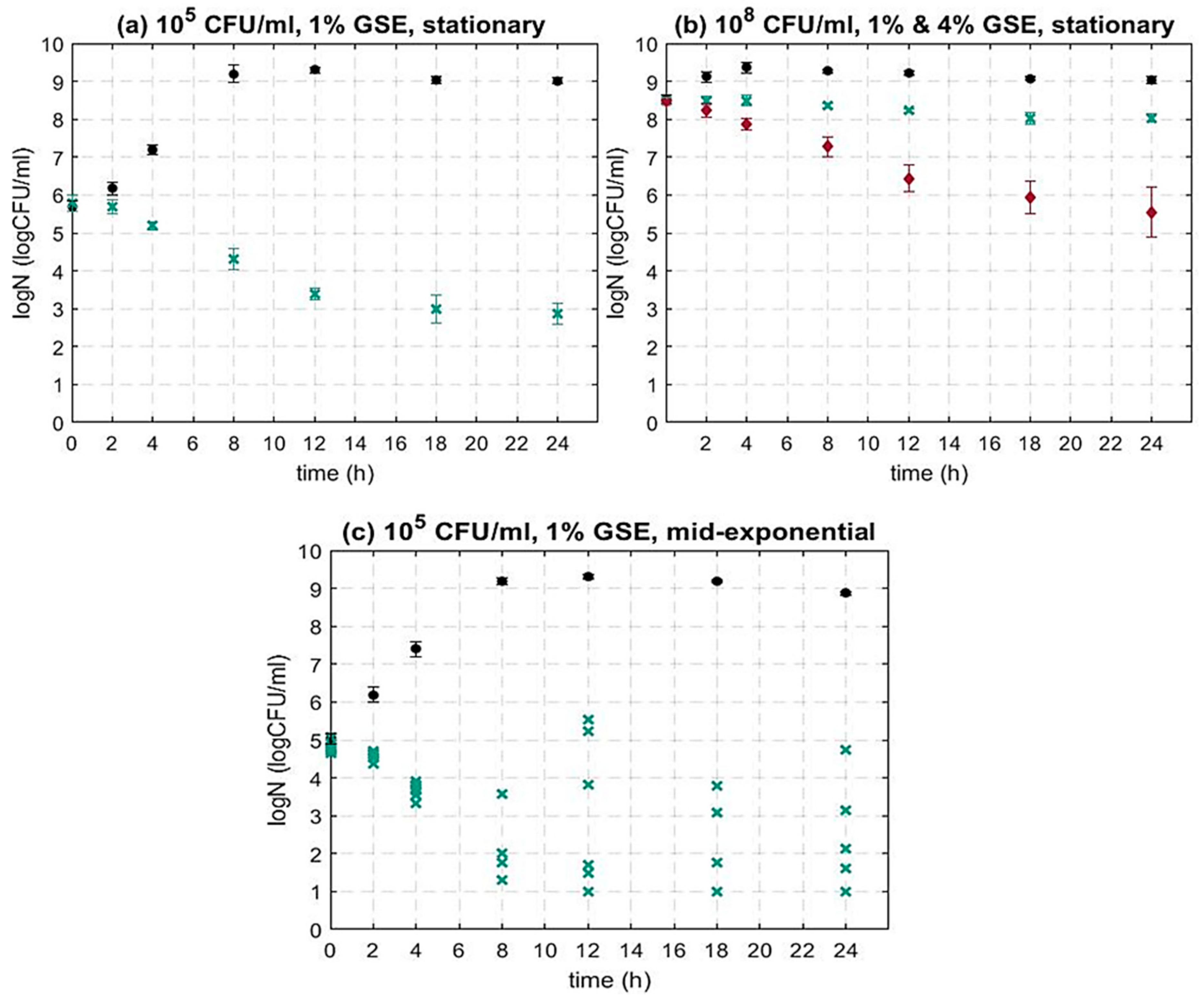
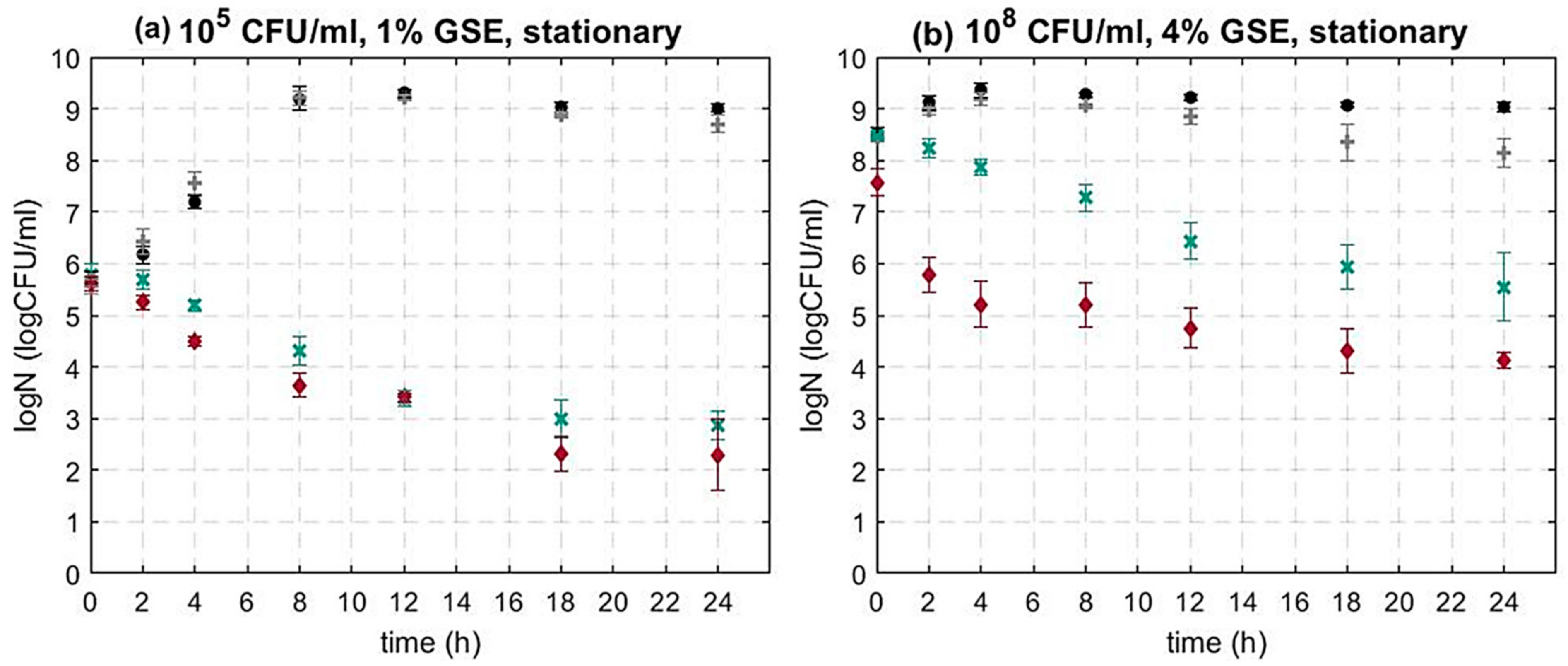
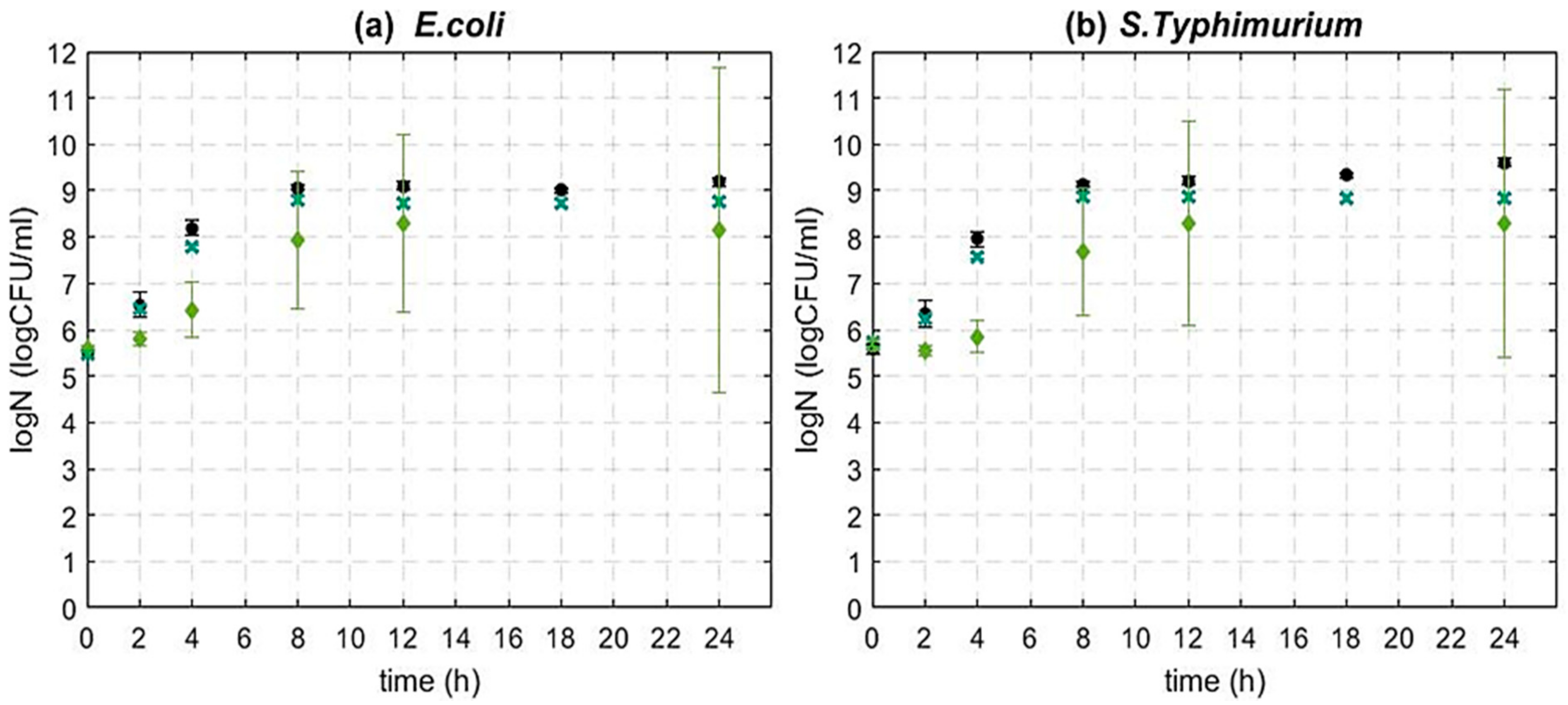
| Microorganism | Growth Phase | Initial Population (CFU/mL) | GSE Concentration (w/v) |
|---|---|---|---|
| L. monocytogenes 10403S WT | Stationary | 105 | 1% |
| 108 | 1% and 4% | ||
| Mid-exponential | 105 | 1% | |
| L. monocytogenes 10403S ΔsigB | Stationary | 105 | 1% |
| 108 | 4% | ||
| E. coli MG1655 | Stationary | 105 | 1% and 4% |
| S. Typhimurium ATCC 14028 | Stationary | 105 | 1% and 4% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kitsiou, M.; Purk, L.; Gutierrez-Merino, J.; Karatzas, K.A.; Klymenko, O.V.; Velliou, E. A Systematic Quantitative Determination of the Antimicrobial Efficacy of Grape Seed Extract against Foodborne Bacterial Pathogens. Foods 2023, 12, 929. https://doi.org/10.3390/foods12050929
Kitsiou M, Purk L, Gutierrez-Merino J, Karatzas KA, Klymenko OV, Velliou E. A Systematic Quantitative Determination of the Antimicrobial Efficacy of Grape Seed Extract against Foodborne Bacterial Pathogens. Foods. 2023; 12(5):929. https://doi.org/10.3390/foods12050929
Chicago/Turabian StyleKitsiou, Melina, Lisa Purk, Jorge Gutierrez-Merino, Kimon Andreas Karatzas, Oleksiy V. Klymenko, and Eirini Velliou. 2023. "A Systematic Quantitative Determination of the Antimicrobial Efficacy of Grape Seed Extract against Foodborne Bacterial Pathogens" Foods 12, no. 5: 929. https://doi.org/10.3390/foods12050929
APA StyleKitsiou, M., Purk, L., Gutierrez-Merino, J., Karatzas, K. A., Klymenko, O. V., & Velliou, E. (2023). A Systematic Quantitative Determination of the Antimicrobial Efficacy of Grape Seed Extract against Foodborne Bacterial Pathogens. Foods, 12(5), 929. https://doi.org/10.3390/foods12050929





