Encapsulation of a Lactic Acid Bacteria Cell-Free Extract in Liposomes and Use in Cheddar Cheese Ripening
Abstract
:1. Introduction
2. Results and Discussion
2.1. Lactococcus lactis subsp. lactis 303 CFE Manufacture and Encapsulation in Liposomes
| Sample | Total cell counts (CFU/mL or CFE/g) * | PepX (mM AMC/min·mL or CFE/g) * | LDH (units/mL or CFE/g) * |
|---|---|---|---|
| In RSM after inoculation | 5.0 × 106 b | nd | nd |
| In RSM after growth | 1.1 × 1010 c | 3.77 ± 0.01 a | 0.00 ± 0.00 a |
| After microfluidization (4000 psi) | 9.0 × 109 c | 25.20 ± 0.01 b | 1.31 ± 0.14 b |
| Freeze-dried microfluidized cells ** | 4.2 × 1010 c | 208.69 ± 14.75 c | 13.8 ± 0.37 c |
| Freeze-dried CFE ** | 0 a | 1333.1 ± 116.07 d | nd |
| Zeta potential | Total activity | Unbound | Encapsulated | |||||
|---|---|---|---|---|---|---|---|---|
| PepX activity * | % total activity | PepX activity * | % total activity | PepX activity * | % total activity | |||
| Prolipo S | −17.0 | 822.1 ± 273.1 | 100.0 | 610.4 ± 216.1 | 74.2 a | 162.2 ± 33.4 | 19.7 b | |
| Prolipo Duo | −39.3 | 3275.0 ± 1073.8 | 100.0 | 2448.4 ± 905.6 | 74.8 a | 459.4 ± 144.5 | 14.0 b | |
2.2. Influence of CFE and Encapsulated CFE on the Composition and Water Activity of the Cheeses
| Moisture (% w/w) | Fat (% w/w) | Protein (% w/w) | Salt (% w/w) | Water activity | pH | |
|---|---|---|---|---|---|---|
| Cheese 1 | 39.19 ± 1.03 a | 29.63 ± 0.68 a | 24.95 ± 0.62 a | 1.79 ± 0.23 a | 0.967 ± 0.002 a | 5.12 ± 0.11 a |
| Cheese 2 | 39.29 ± 0.96 a | 29.78 ± 0.65 a | 24.87 ± 0.44 a | 1.72 ± 0.17 a | 0.972 ± 0.005 a | 5.08 ± 0.07 a |
| Cheese 3 | 39.31 ± 0.83 a | 29.67 ± 0.68 a | 24.82 ± 0.32 a | 1.74 ± 0.12 a | 0.969 ± 0.002 a | 5.12 ± 0.09 a |
| Cheese 4 | 38.68 ± 0.89 a | 29.92 ± 0.58 a | 25.19 ± 0.61 a | 1.95 ± 0.22 a | 0.970± 0.002 a | 5.16 ± 0.12 a |
2.3. Enumeration of Starter and Non-Starter Lactic Acid Bacteria and Enzyme Activities during Cheese Ripening
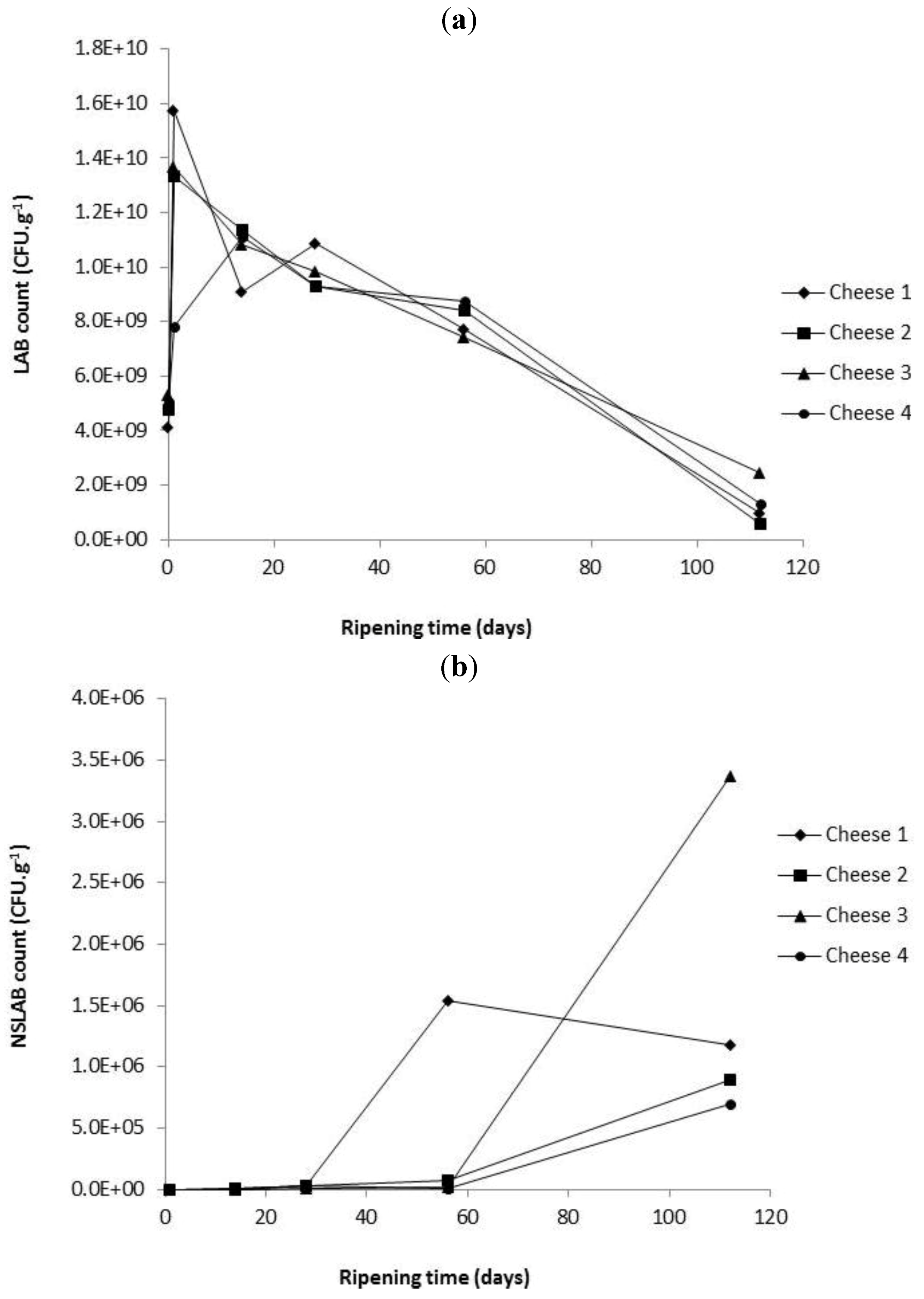
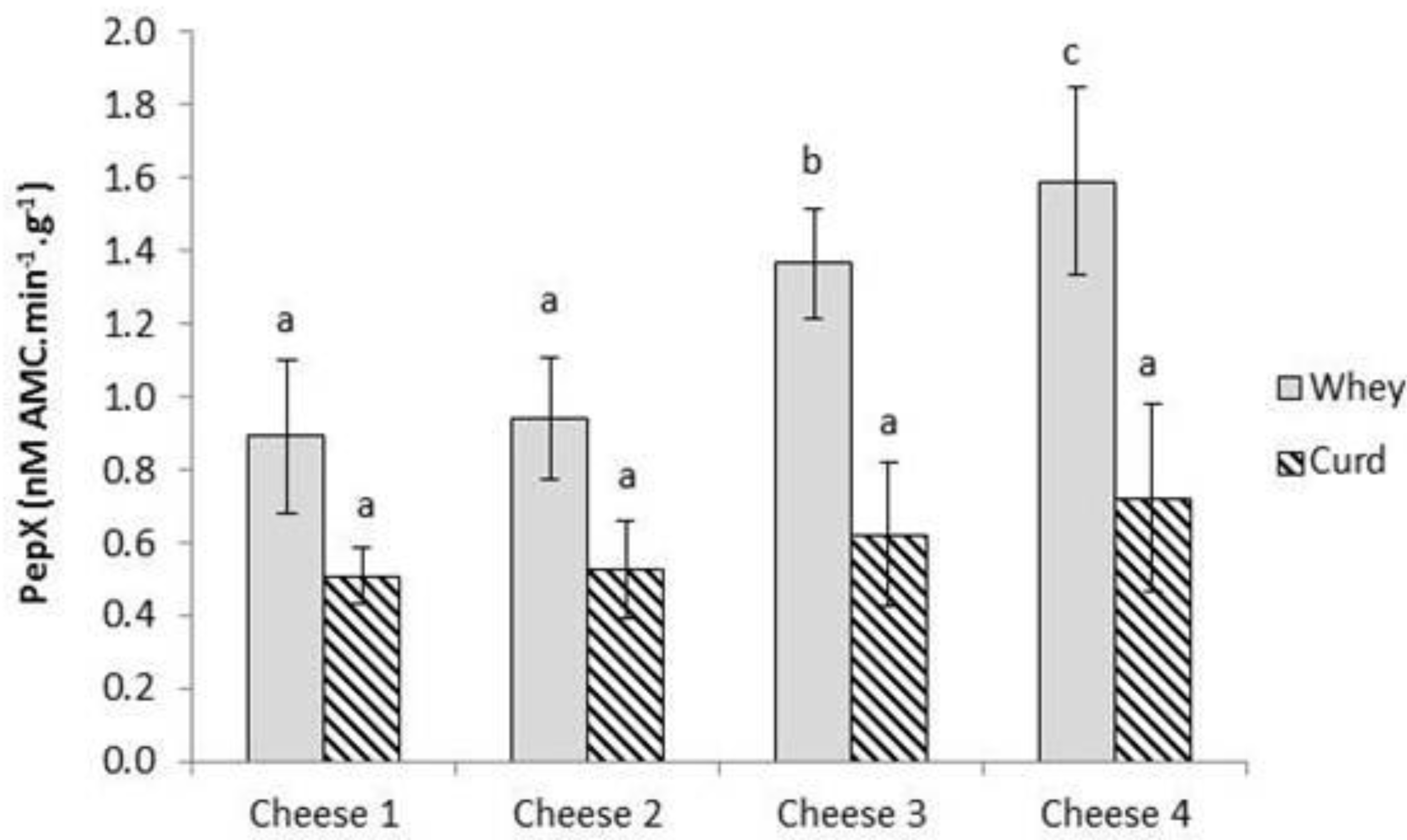
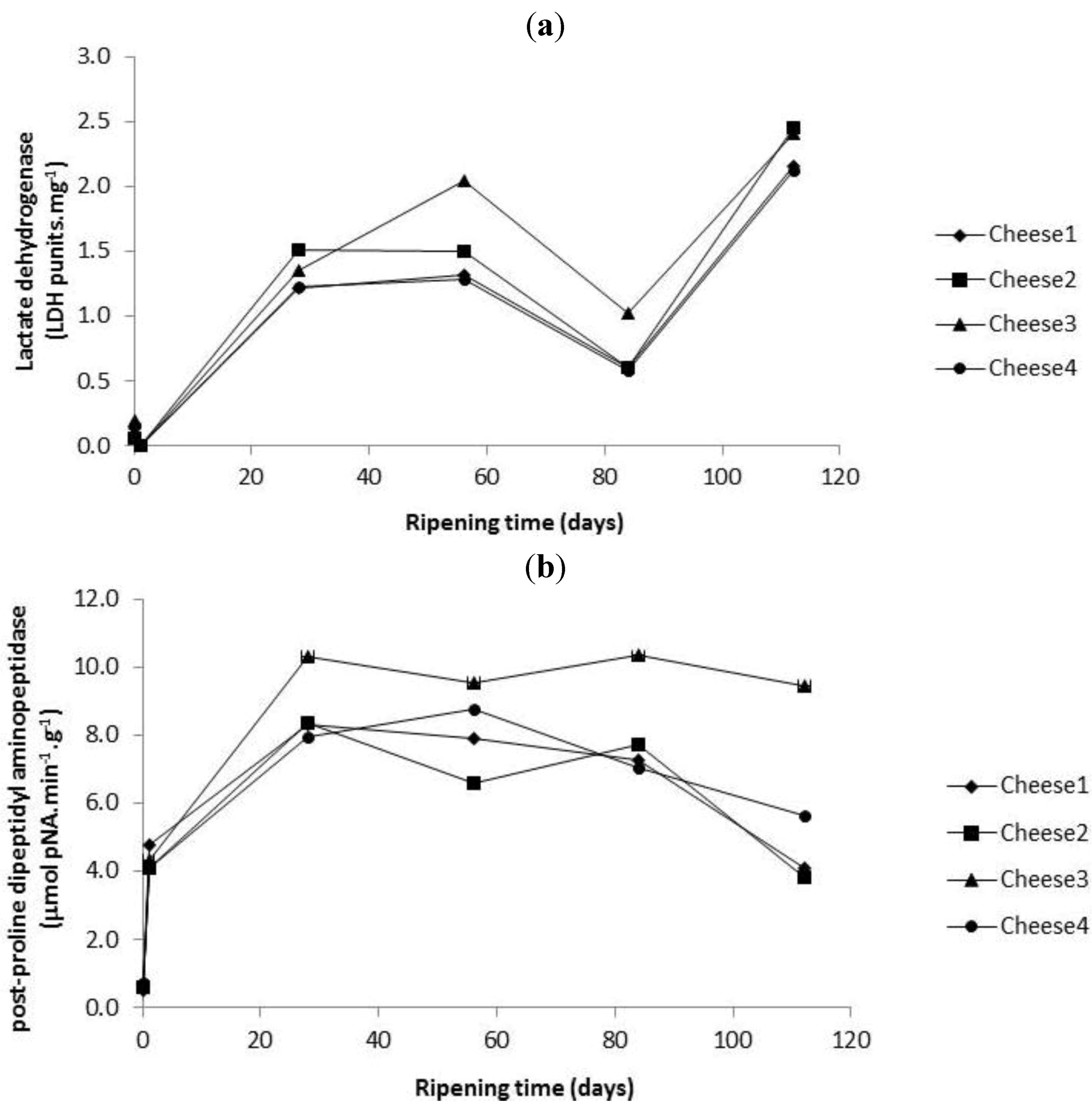
2.4. Evolution of Proteolysis in Cheeses over Ripening
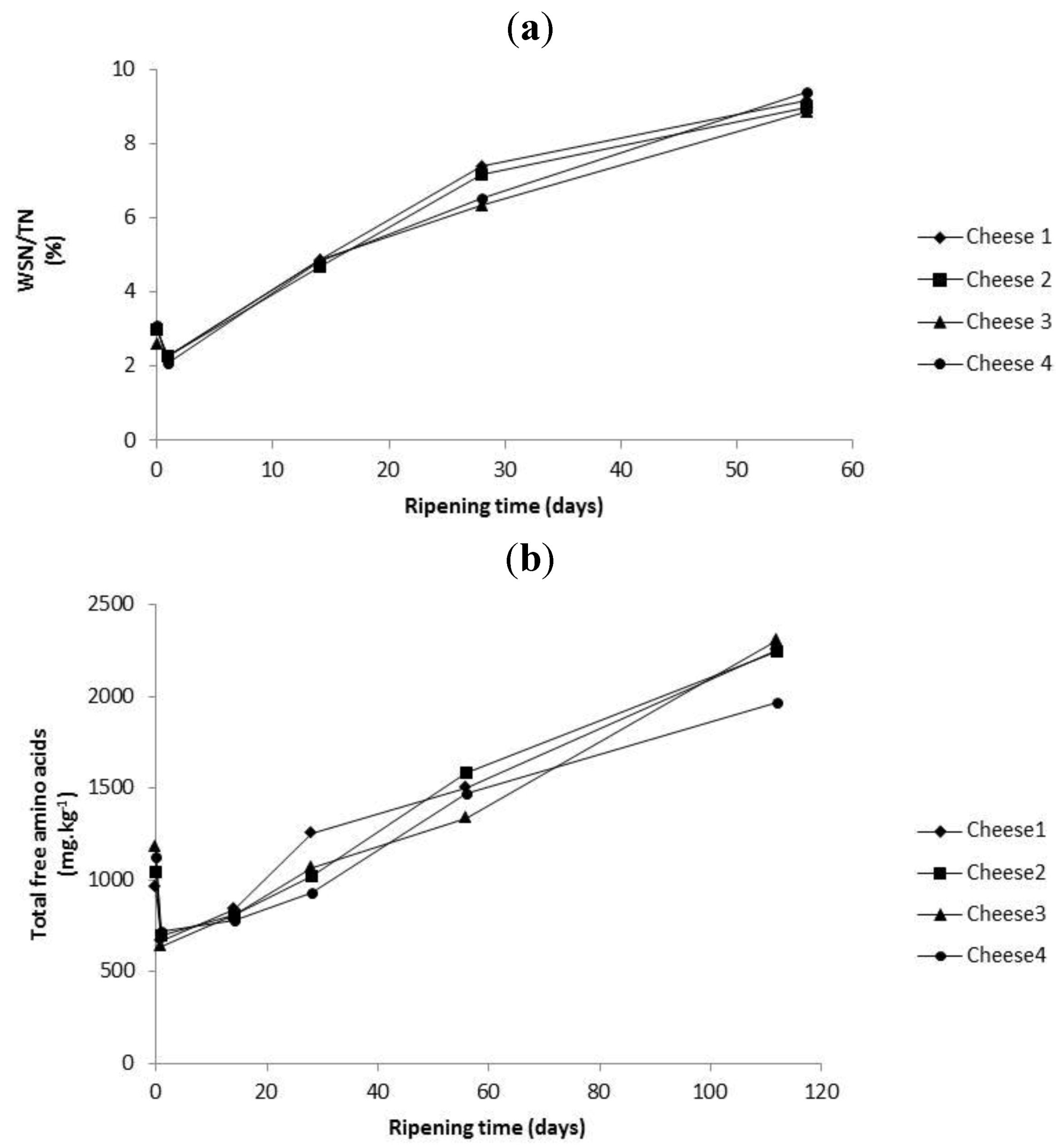
2.5. Cryo SEM of Liposomes within Cheese Curd up to 28 Days of Ripening
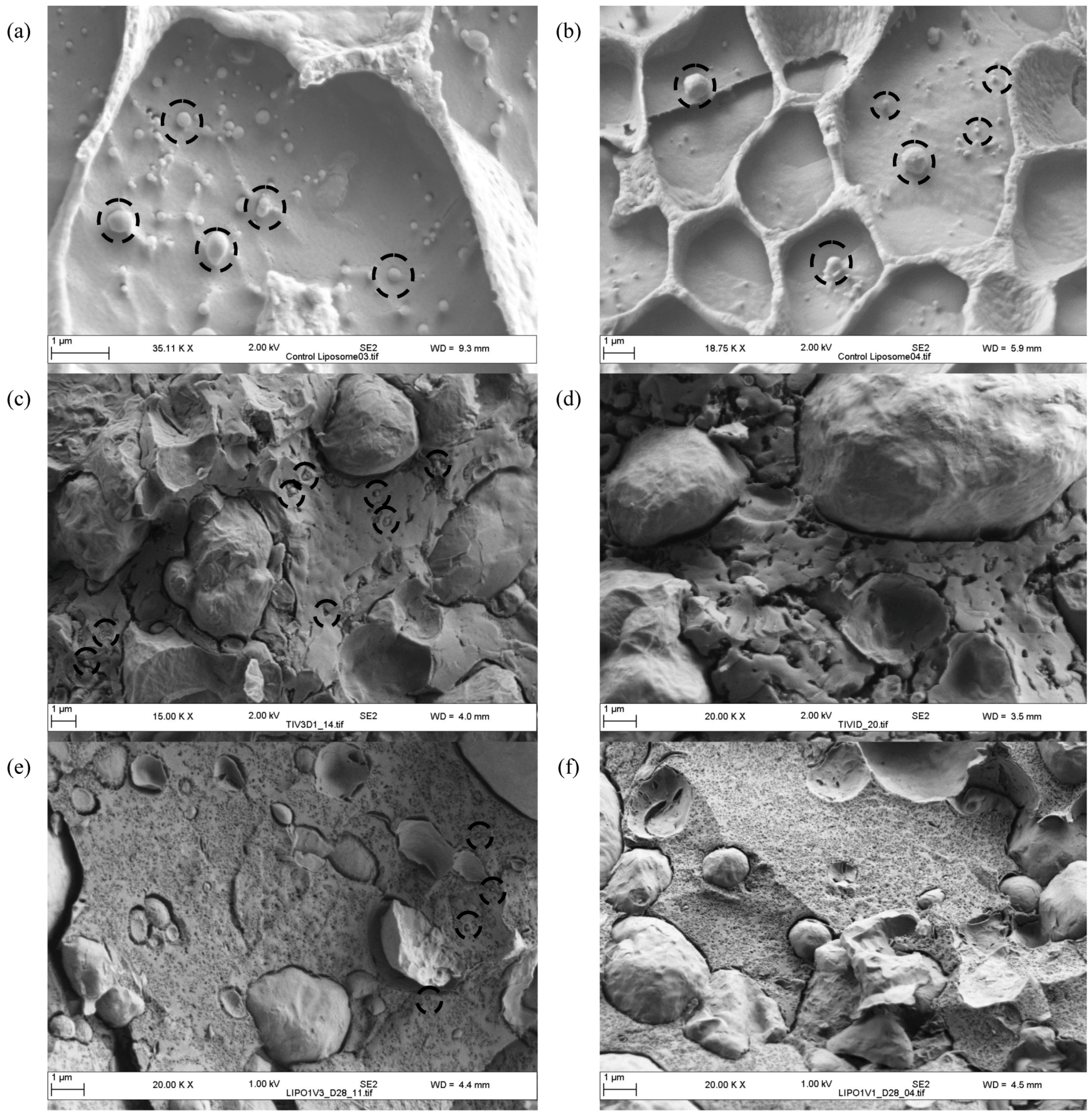
2.6. Descriptive Sensory Analysis and Volatile Profiles of Cheeses at Day 112
| Volatiles | Concentration of volatiles (µg/kg) in the different cheese samples * | |||
|---|---|---|---|---|
| Cheese 1 | Cheese 2 | Cheese 3 | Cheese 4 | |
| Methanethiol | 2.18 a | 3.80 a | 3.86 a | 3.02 a |
| Ethyl alcohol | 196.78 b | 347.96 a, b | 350.35 a | 248.24 a, b |
| Propanol | 355.71 a, b | 373.39 a, b | 55.58 b | 686.59 a |
| Carbon disulfide | 24.73 a | 15.69 a | 21.22 a | 17.24 a |
| 2-Butanone | 641.36 a | 654.36 a | 400.35 a | 977.05 a |
| 2-Butanol, (R)- | 736.40 a | 776.54 a | 1597.63 a | 1096.35 a |
| 2-Pentanone | 13.95 b | 28.24 b | 86.34 a | 33.04 b |
| Acetoin | 39.43 a | 148.11 a | 134.13 a | 54.22 a |
| Methyl butanoate | 5.60 a | 7.17 a | 9.02 a | 6.26 a |
| Ethyl butanoate | 42.24 b | 374.76 a | 145.41 a, b | 184.72 a, b |
| Pentanoic acid | 3.94 a | 6.17 a | 7.47 a | 5.20 a |
| 2-Heptanone | 17.63 b | 38.24 a | 25.44 a, b | 26.69 a, b |
| Heptanal | 1.87 b | 4.32 a | 2.88 a, b | 2.75 a, b |
| 2,6-Dimethyl-pyrazine | 0.00 b | 0.00 b | 0.00 b | 0.23 a |
| Methyl hexanoate | 5.47 a | 7.95 a | 8.46 a | 5.85 a |
| Dimethyl trisulfide | 10.25 a | 20.27 a | 14.98 a | 8.90 a |
| Ethyl hexanoate | 7.13 a | 11.79 a | 11.22 a | 9.38 a |
| Benzeneacetaldehyde | 2.50 a | 5.13 a | 5.05 a | 2.54 a |
| 2-Nonanone | 2.95 b | 6.63 a | 3.99 a, b | 4.84 a, b |
| Nonanal | 5.73 a | 16.30 a | 4.04 a | 8.73 a |
| 2-Ethyl-hexanoic acid | 0.73 a | 1.07 a | 1.42 a | 0.68 a |
| Methyl octanoate | 0.96 a | 1.11 a | 0.93 a | 0.70 a |
| Hexanoic acid | 10.17 a | 14.85 a | 9.16 a | 7.48 a |
| 2,5-Dihydro-3-methyl-furan | 4.35 a | 5.45 a | 4.20 a | 2.87 a |
| Ethyl octanoate | 0.65 a | 1.26 a | 1.18 a | 1.15 a |
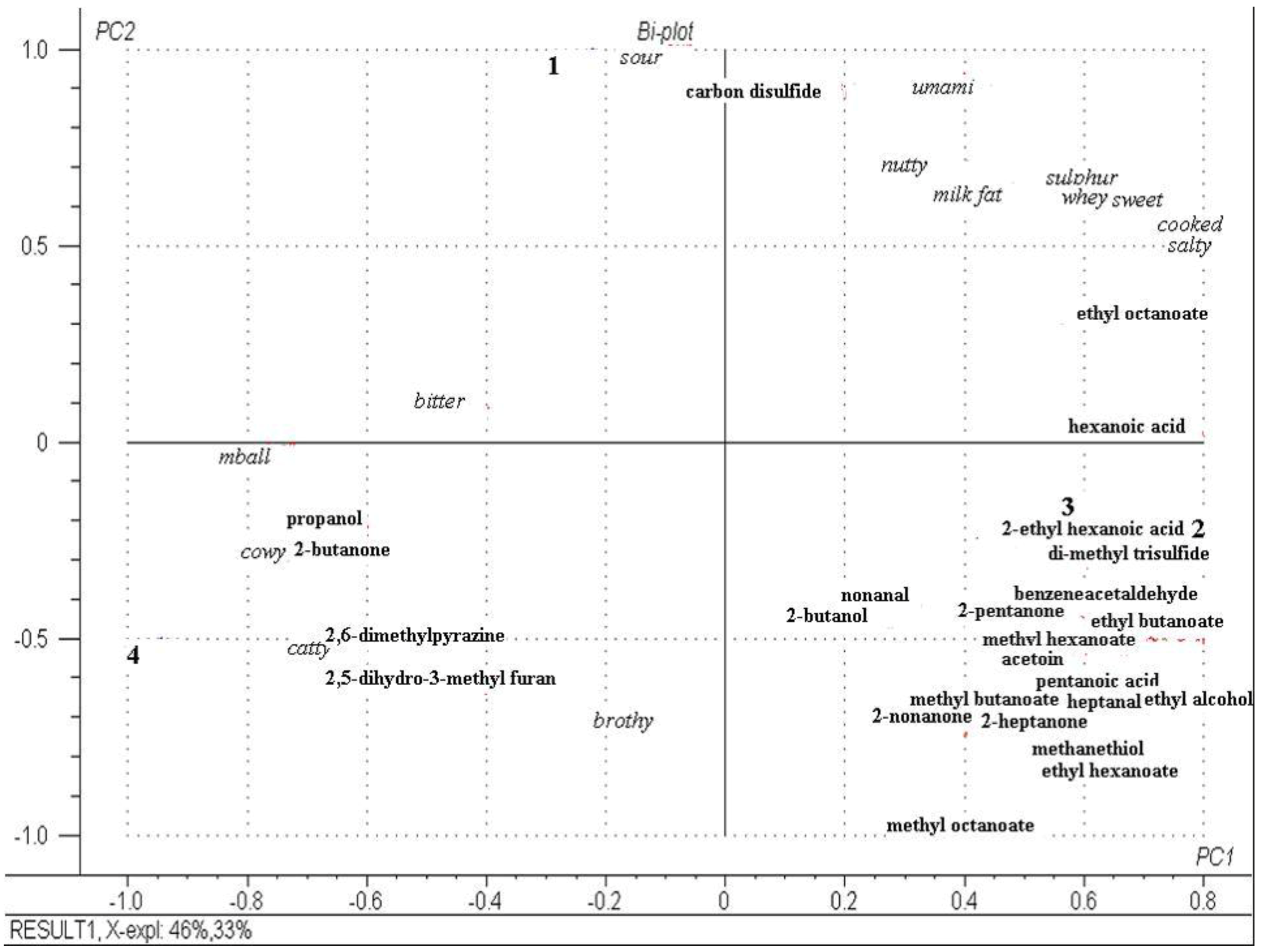
3. Experimental Section
3.1. Preparation of Lactococcus lactis subsp. lactis 303 CFE
3.2. Preparation of Lactococcus lactis subsp. lactis 303 CFE
3.3. Encapsulation of CFE in Liposomes
3.4. Cheese Manufacture
3.5. Determination of the Enzyme Activity
3.5.1. Determination of Enzyme Activities in the CFE Samples
3.5.2. Determination of PepX Activity in the Liposomes
3.5.3. Determination of Enzyme Activities in the Cheese Samples
3.6. Microbial Analyses
3.7. Physicochemical Analysis of the Cheese Samples
3.7.1. Cheese Composition at Day 14
3.7.2. Determination of Individual Free Amino Acids
3.7.3. Measurement of the Water Activity of the Cheese Samples
3.8. Cryogenic Scanning Cryogenic-Scanning Electron Micrography
3.9. Sensory Evaluation and Volatiles
3.10. Statistical Analyses
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Wilkinson, M.G.; Kilcawley, K.N. Mechanisms of incorporation and release of enzymes into cheese during ripening. Int. Dairy J. 2005, 15, 817–830. [Google Scholar] [CrossRef]
- El Soda, M.; Pandian, S. Recent developments in accelerated cheddar cheese ripening. J. Dairy Sci. 1991, 74, 2317–2335. [Google Scholar] [CrossRef]
- Azarnia, S.; Robert, N.; Lee, B. Biotechnological methods to accelerate cheddar cheese ripening. Crit. Rev. Biotechnol. 2006, 26, 121–143. [Google Scholar] [CrossRef]
- Kailasapathy, K.; Lam, S.H. Application of encapsulated enzymes to accelerate cheese ripening. Int. Dairy J. 2005, 15, 929–939. [Google Scholar] [CrossRef]
- Azarnia, S.; Lee, B.H.; St-Gelais, D.; Champagne, C.P.; Kilcawley, K.N. Effect of free or encapsulated recombinant aminopeptidase of Lactobacillus rhamnosus S93 on acceleration of cheddar cheese ripening. Food Biotechnol. 2010, 24, 135–149. [Google Scholar] [CrossRef]
- Thompson, A.K.; Singh, H. Preparation of liposomes from milk fat globule membrane phospholipids using a microfluidizer. J. Dairy Sci. 2006, 89, 410–419. [Google Scholar] [CrossRef]
- Lariviere, B.; Soda, M.E.; Soucy, Y.; Trepanier, G.; Paquin, P.; Vuillemard, J.C. Microfluidized liposomes for the acceleration of cheese ripening. Int. Dairy J. 1991, 1, 111–124. [Google Scholar] [CrossRef]
- Kheadr, E.E.; Vuillemard, J.C.; El-Deeb, S.A. Accelerated cheddar cheese ripening with encapsulated proteinases. Int. J. Food Sci. Technol. 2000, 35, 483–495. [Google Scholar] [CrossRef]
- Dufour, P.; Vuillemard, J.C.; Laloy, E.; Simard, R.E. Characterization of enzyme immobilization in liposomes prepared from proliposomes. J. Microencapsul. 1996, 13, 185–194. [Google Scholar] [CrossRef]
- Kheadr, E.E.; Vuillemard, J.C.; El-Deeb, S.A. Acceleration of Cheddar cheese lipolysis by using liposome-entrapped lipases. J. Dairy Sci. 2002, 67, 485–492. [Google Scholar]
- Kheadr, E.E.; Vuillemard, J.C.; El-Deeb, S.A. Impact of liposome-encapsulated enzyme cocktails on the cheddar cheese ripening. Food Res. Int. 2003, 36, 241–252. [Google Scholar] [CrossRef]
- Laloy, E.; Vuillemard, J.C.; Simard, R.E. Characterization of liposomes and their effect on the properties of Cheddar cheese during ripening. Lait 1998, 78, 401–412. [Google Scholar] [CrossRef]
- Picon, A.; Serrano, C.; Gaya, P.; Medina, M.; Nunez, M. The effect of liposomes-encapsulated cryopsins on Manchego cheese ripening. J. Dairy Sci. 1996, 79, 1699–1705. [Google Scholar] [CrossRef]
- Nongonierma, A.B.; Abrlova, M.; Fenelon, M.A.; Kilcawley, K.N. Evaluation of two food grade proliposomes to encapsulate an extract of a commercial enzyme preparation by microfluidization. J. Agric. Food Chem. 2009, 57, 3291–3297. [Google Scholar]
- Jackson, L.S.; Lee, K. Microencapsulation and the food industry. Lebensm. Wiss. Technol. 1991, 24, 289–297. [Google Scholar]
- Zeisig, R.; Cammerer, B. Single Core Encapsulation. Liposomes in Food Industry. In Microencapsulation of Food Ingredients, 1st; Vilstrup, P., Ed.; Leatherhead Publishing: Surrey, UK, 2001; pp. 101–119. [Google Scholar]
- Kim, H.-H.Y.; Baianu, I.C. Novel liposome microencapsulation techniques for food applications. Trends Food Sci. Technol. 1991, 2, 55–61. [Google Scholar] [CrossRef]
- Wilkinson, M.G. Acceleration of Cheese Ripening. In Cheese: Chemistry, Physics and Microbiology; Fox, P.F., Ed.; Chapman and Hall: London, UK, 1993; pp. 523–553. [Google Scholar]
- Connor, J.; Yatvin, M.B.; Huang, L. pH-Sensitive liposomes: Acid-induced liposome fusion. Proc. Natl. Acad. Sci. 1984, 81, 1715–1718. [Google Scholar] [CrossRef]
- Bainville, C.; Vuillemard, J.C.; Lacroix, C. Comparison of different methods for fortifying Cheddar cheese with vitamin D. Int. Dairy J. 2000, 10, 375–382. [Google Scholar] [CrossRef]
- Lawrence, R.C.; Gilles, J.; Creamer, L.K.; Crow, V.L.; Heap, H.A.; Honoré, C.G.; Johnston, K.A.; Samal, P.K. Cheddar Cheese and Related Dry-Salted Cheese Varieties. In Cheese: Chemistry, Physics and Microbiology, 3rd; Fox, P.F., McSweeny, P.L.H., Cogan, T.M., Guinee, T.P., Eds.; Elsevier Academic Press: London, UK, 2004; pp. 71–102. [Google Scholar]
- O’Donovan, C.M.; Wilkinson, M.G.; Guinee, T.P.; Fox, P.F. An investigation of the autolytic properties of three lactococcal strains during cheese ripening. Int. Dairy J. 1996, 6, 1149–1165. [Google Scholar] [CrossRef]
- Crow, V.L.; Coolbear, T.; Gopal, P.K.; Martley, F.G.; McKay, L.L.; Riepe, H. The role of autolysis of lactic acid bacteria in the ripening of cheese. Int. Dairy J. 1995, 5, 855–875. [Google Scholar] [CrossRef]
- McSweeney, P.L.H.; Sousa, M.J. Biochemical pathways for the production of flavour compounds in cheeses during ripening: A review. Lait 2000, 80, 293–324. [Google Scholar] [CrossRef]
- Singh, T.K.; Drake, M.A.; Cadwallader, K.R. Flavor of Cheddar cheese: A chemical and sensory perspective. Compr. Rev. Food Sci. Food Saf. 2003, 2, 166–189. [Google Scholar] [CrossRef]
- Kilcawley, K.N.; Nongonierma, A.B.; Hannon, J.A.; Doolan, I.A.; Wilkinson, M.G. Evaluation of commercial enzyme systems to accelerate Cheddar cheese ripening. Int. Dairy J. 2012, 26, 50–57. [Google Scholar] [CrossRef]
- Drake, M.A.; McIngvale, S.C.; Gerard, P.D.; Cadwallader, K.R.; Civille, G.V. Development of a descriptive language for Cheddar cheese. J. Food Sci. 2001, 66, 1422–1427. [Google Scholar] [CrossRef]
- Young, N.D.; Drake, M.; Lopetcharat, K.; McDaniel, M.R. Preference mapping of cheddar cheese with varying maturity levels. J. Dairy Sci. 2004, 87, 11–19. [Google Scholar] [CrossRef]
- Laridi, R.; Kheadr, E.E.; Benech, R.-O.; Vuillemard, J.C.; Lacroix, C.; Fliss, I. Liposome encapsulated nisin Z: Optimisation, stability and release during milk fermentation. Int. Dairy J. 2003, 13, 325–336. [Google Scholar] [CrossRef]
- Wilkinson, M.G.; Guinee, T.P.; O’Callaghan, D.M.; Fox, P.F. Autolysis and proteolysis in different strains of starter bacteria during Cheddar cheese ripening. J. Dairy Res. 1994, 61, 249–262. [Google Scholar] [CrossRef]
- Habibi-Najafi, M.B.; Lee, B.H. Proline-specific peptidases of Lactobacillus casei subspecies. J. Dairy Sci. 1994, 77, 385–392. [Google Scholar] [CrossRef]
- Kilcawley, K.N.; Wilkinson, M.G.; Fox, P.F. Determination of key enzyme activities in commercial peptidase and lipase preparations from microbial or animal sources. Enzym. Microb. Technol. 2002, 31, 310–320. [Google Scholar] [CrossRef]
- Cogan, T.M.; O’Dowd, M.; Mellerick, D. Effects of pH and sugar on acetoin production from citrate by Leuconostoc lactis. Appl. Environ. Microbiol. 1981, 41, 1–8. [Google Scholar]
- Hickey, D.K.; Kilcawley, K.N.; Beresford, T.P.; Sheehan, E.M.; Wilkinson, M.G. The influence of a seasonal milk supply on the biochemical and sensory properties of Cheddar cheese. Int. Dairy J. 2006, 16, 679–690. [Google Scholar] [CrossRef]
- Kuchroo, C.N.; Fox, P.F. Soluble nitrogen in cheese: Comparison of extraction procedures. Milchwiss. 1982, 37, 331–335. [Google Scholar]
- Gee, V.L.; Carey, M.A.; Auty, M.A.E. A new cryo-STEM method for imaging food colloids in the scanning electron microscope. Scanning 2010, 32, 150–154. [Google Scholar] [CrossRef]
- Drake, M.A.; Civille, G.V. Flavor lexicons. Compr. Rev. Food Sci. Food Saf. 2003, 2, 33–40. [Google Scholar] [CrossRef]
- Meilgaard, M.; Civille, G.; Carr, B. Descriptive Analysis Techniques. In Sensory Evaluation Techniques; CRC Press: New York, NY, USA, 1999; pp. 173–183. [Google Scholar]
- Carunchiawhetstine, M.E.; Karagul-Yuceer, Y.; Avsar, Y.K.; Drake, M.A. Identification and quantification of character aroma components in fresh chevre-style goat cheese. J. Food Sci. 2003, 68, 2441–2447. [Google Scholar] [CrossRef]
- Drake, M.A.; Yates, M.D.; Gerard, P.D.; Delahunty, C.M.; Sheehan, E.M.; Turnbull, R.P.; Dodds, T.M. Comparison of differences between lexicons for descriptive analysis of Cheddar cheese flavour in Ireland, New Zealand, and the United States of America. Int. Dairy J. 2005, 15, 473–483. [Google Scholar] [CrossRef]
- Kang, E.J.; Smith, T.J.; Drake, M.A. Alternative bleaching methods for Cheddar cheese whey. J. Food Sci. 2012, 77, C818–C823. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Nongonierma, A.B.; Abrlova, M.; Kilcawley, K.N. Encapsulation of a Lactic Acid Bacteria Cell-Free Extract in Liposomes and Use in Cheddar Cheese Ripening. Foods 2013, 2, 100-119. https://doi.org/10.3390/foods2010100
Nongonierma AB, Abrlova M, Kilcawley KN. Encapsulation of a Lactic Acid Bacteria Cell-Free Extract in Liposomes and Use in Cheddar Cheese Ripening. Foods. 2013; 2(1):100-119. https://doi.org/10.3390/foods2010100
Chicago/Turabian StyleNongonierma, Alice Beebyaanda, Magdalena Abrlova, and Kieran Noel Kilcawley. 2013. "Encapsulation of a Lactic Acid Bacteria Cell-Free Extract in Liposomes and Use in Cheddar Cheese Ripening" Foods 2, no. 1: 100-119. https://doi.org/10.3390/foods2010100






