Detection of Beef Adulterated with Pork Using a Low-Cost Electronic Nose Based on Colorimetric Sensors
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. General Chemical Analysis
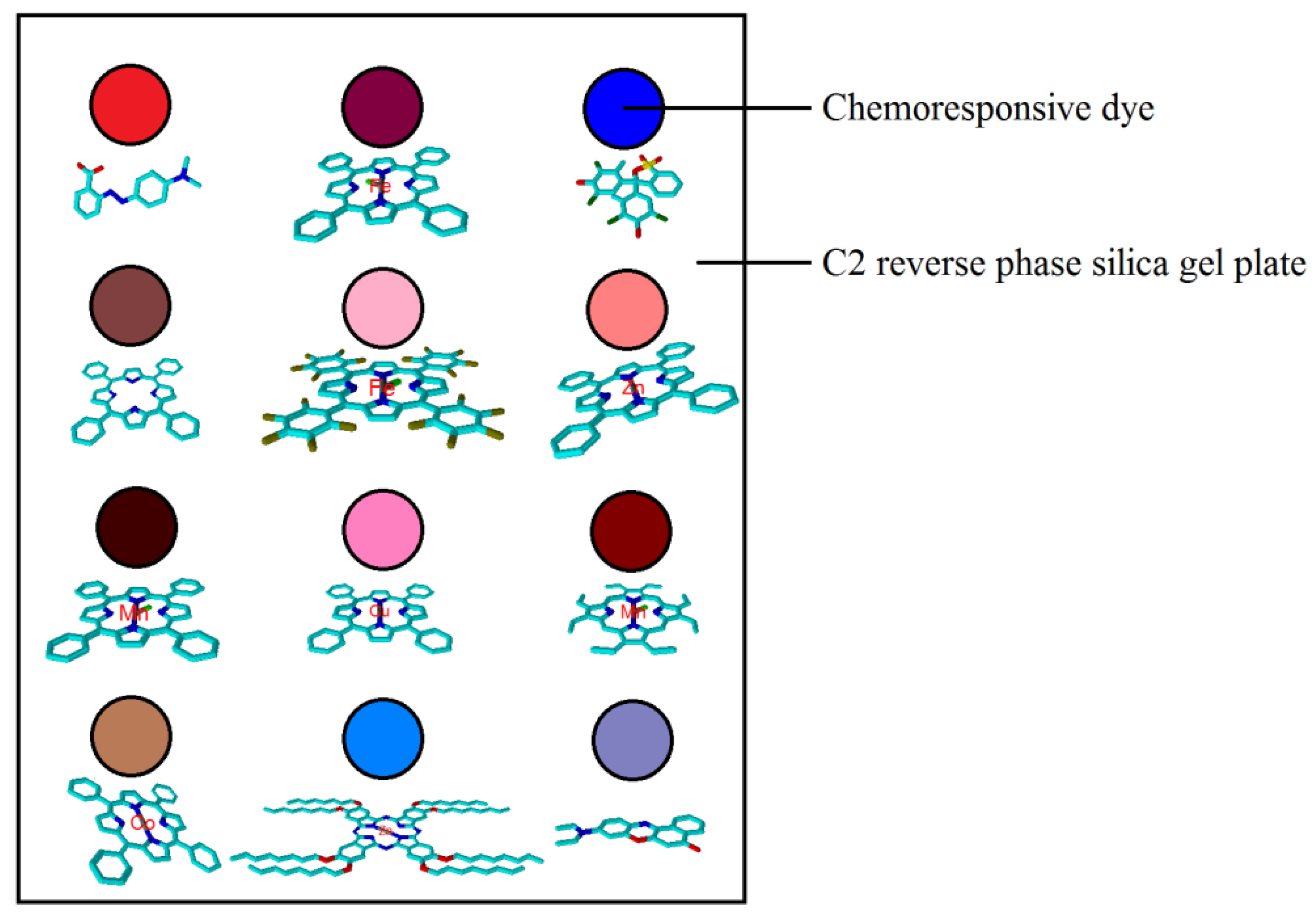
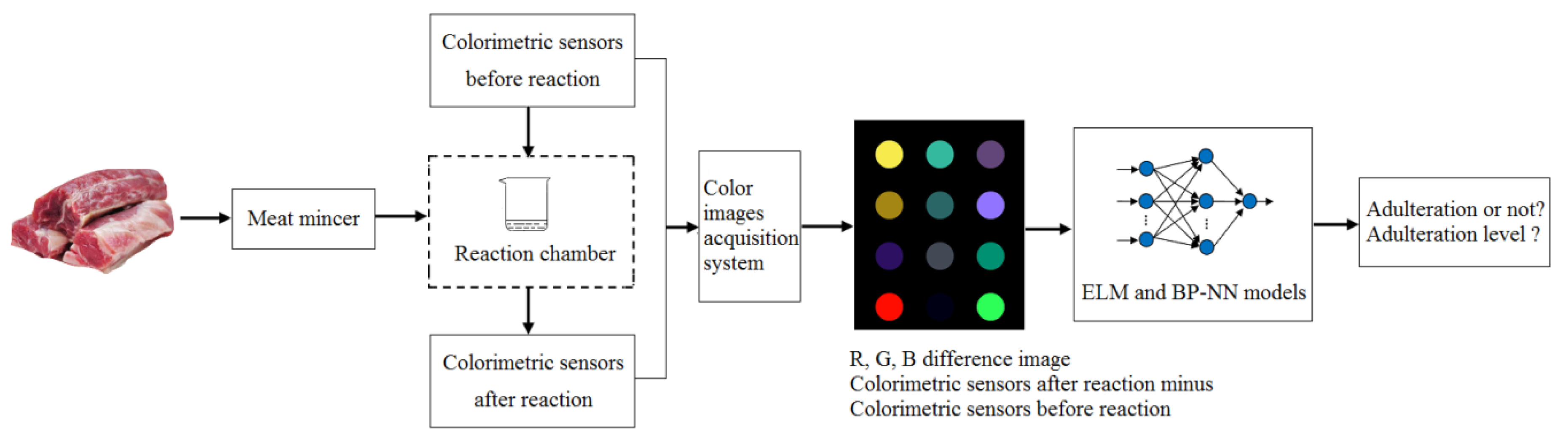
2.3. E-Nose Based on Colorimetric Sensors
- (1)
- manganese(3+),10,12,13,23-tetraphenyl-21H-porphyrin,trichloride;
- (2)
- zinc-2,3,9,10,16,17,23,24-octakis-(octyloxy)-29H,31H-phthalocyanine;
- (3)
- manganese(3+),2,12,13,15,17,18,20,23-octaethyl-21H-porphyrin,trichloride;
- (4)
- 5,10,15,20-Tetraphenyl-21H,23H-porphinecopper;
- (5)
- 5,10,15,20-tetraphenyl-21,22-dihydroporphyrin,zinc;
- (6)
- [5,10,15,20-tetrakis(C6F5)-21H,23H-porphine]Fe(III) chloride;
- (7)
- 5,10,15,20-tetraphenyl-21h,23h-porphine iron(iii) chloride;
- (8)
- 5,10,15,20-Tetraphenyl-21H,23H-porphine Cobalt(II);
- (9)
- 5,10,15,20-tetraphenyl-21H,23H-porphine;
- (10)
- 2-[4-(dimethylamino)phenylazo]benzoic acid;
- (11)
- m-Cresol, 4,4′-(3H-2,1-benzoxathiol-3-ylidene)bis(2,6-dibromo-, S,S-dioxide (8CI);
- (12)
- 9-(Diethylamino)-5H-benzo[a]phenoxazin-5-one.
2.4. Operating Procedure and Feature Extraction
ΔG= |Ga− Gb|
ΔB = |Ba− Bb|
2.5. Multivariate Analysis and Software
2.6. Software
3. Results
3.1. Chemical Analysis Results
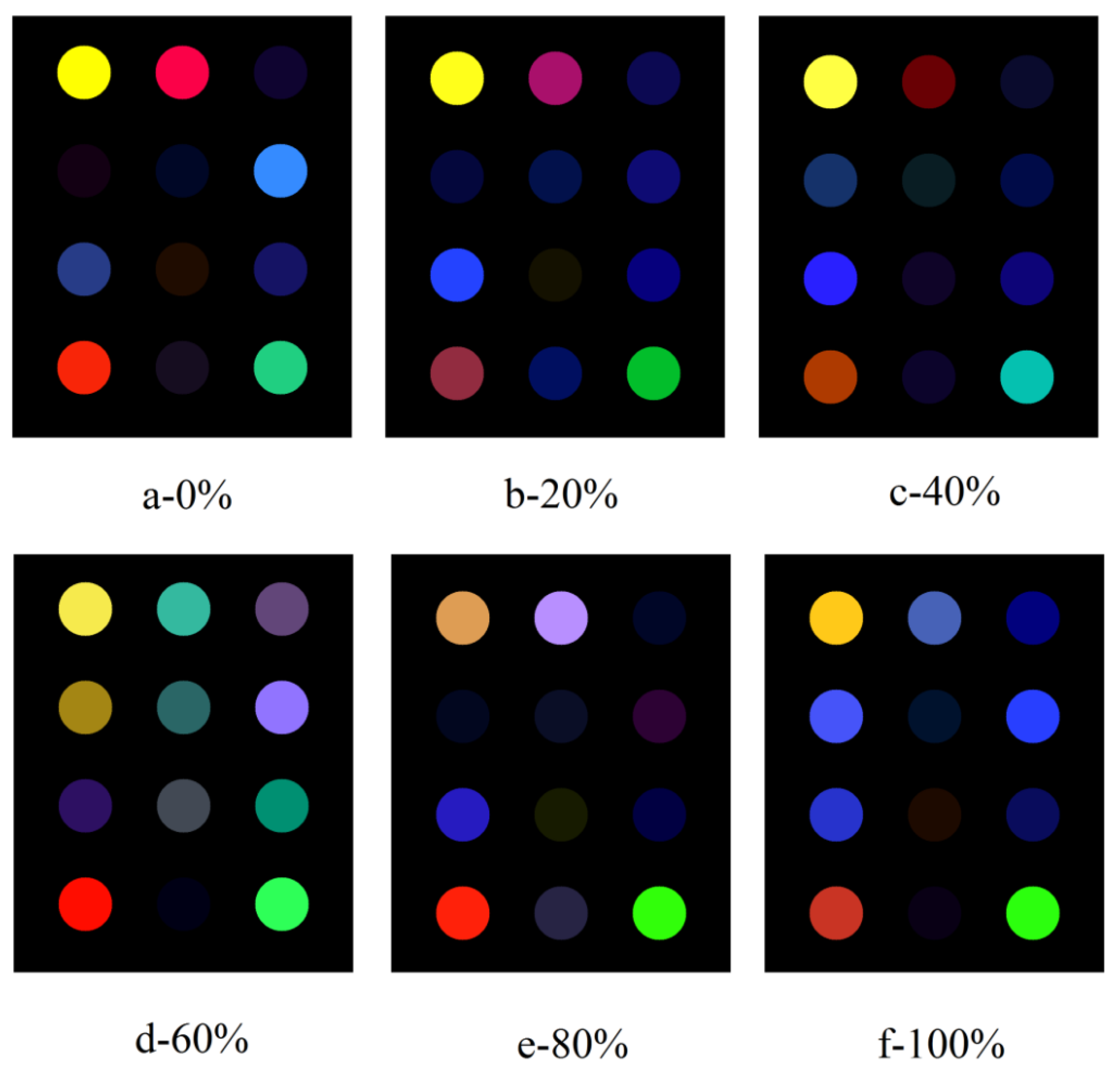
3.2. Sensor Responses and Preprocessing
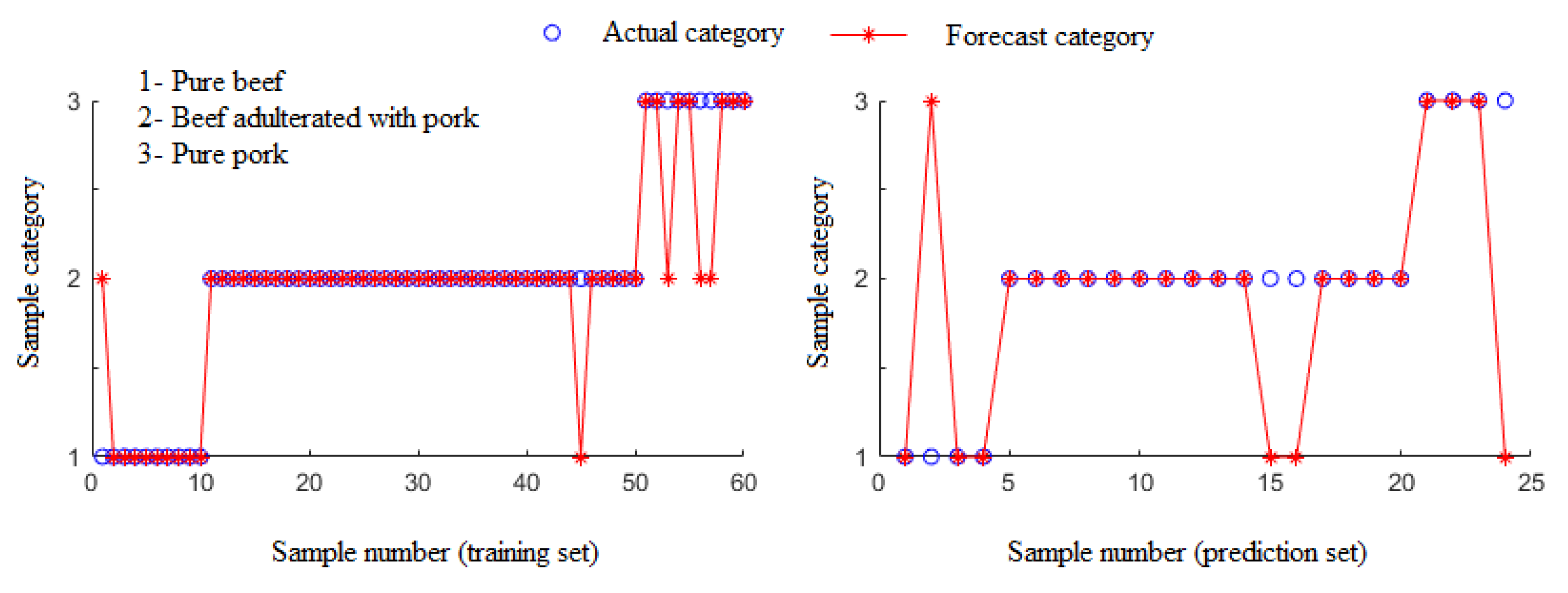
3.3. Identification of the Beef Adulterated with Pork
3.4. Prediction of Adulteration Levels
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Ethical Approval
References
- Sanchez-Sabate, R.; Sabaté, J. Consumer attitudes towards environmental concerns of meat consumption: A systematic review. Int. J. Environ. Res. Public Health 2019, 16, 1220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barai, B.; Nayak, R.; Singhal, R.; Kulkarni, P. Approaches to the detection of meat adulteration. Trends Food Sci. Technol. 1992, 3, 69–72. [Google Scholar] [CrossRef]
- Song, Q.; Chen, Y.; Zhao, L.; Ouyang, H.; Song, J. Monitoring of sausage products sold in Sichuan Province, China: A first comprehensive report on meat species’ authenticity determination. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Gao, X.; Guan, Y. A Novel Isothermal Amplification Method for Detecting Mouse Source Component in Meat. J. AOAC Int. 2019, 102, 872–877. [Google Scholar] [CrossRef] [PubMed]
- Alamprese, C.; Amigo, J.M.; Casiraghi, E.; Engelsen, S.B. Identification and quantification of turkey meat adulteration in fresh, frozen-thawed and cooked minced beef by FT-NIR spectroscopy and chemometrics. Meat Sci. 2016, 121, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Kamruzzaman, M.; Makino, Y.; Oshita, S.; Liu, S. Assessment of visible near-infrared hyperspectral imaging as a tool for detection of horsemeat adulteration in minced beef. Food Bioprocess Technol. 2015, 8, 1054–1062. [Google Scholar] [CrossRef]
- Schmutzler, M.; Beganovic, A.; Böhler, G.; Huck, C.W. Methods for detection of pork adulteration in veal product based on FT-NIR spectroscopy for laboratory, industrial and on-site analysis. Food Control 2015, 57, 258–267. [Google Scholar] [CrossRef]
- López-Maestresalas, A.; Insausti, K.; Jarén, C.; Pérez-Roncal, C.; Urrutia, O.; Beriain, M.J.; Arazuri, S. Detection of minced lamb and beef fraud using NIR spectroscopy. Food Control 2019, 98, 465–473. [Google Scholar] [CrossRef]
- Erkinbaev, C.; Henderson, K.; Paliwal, J. Discrimination of gluten-free oats from contaminants using near infrared hyperspectral imaging technique. Food Control 2017, 80, 197–203. [Google Scholar] [CrossRef]
- Turhan, M.; Balaban, M.O.; Nazan Turhan, K.; Luzuriaga, D. Potential use of electronic nose technique for detection of meat adulteration: Separation of pork-beef mixtures. Fleischwirtsch. Int. 1998, 6, 26–28. [Google Scholar]
- Han, F.; Huang, X.; Teye, E.; Gu, F.; Gu, H. Nondestructive detection of fish freshness during its preservation by combining electronic nose and electronic tongue techniques in conjunction with chemometric analysis. Anal. Methods 2014, 6, 529–536. [Google Scholar] [CrossRef]
- Baietto, M.; Wilson, A.D. Electronic-nose applications for fruit identification, ripeness and quality grading. Sensors 2015, 15, 899–931. [Google Scholar] [CrossRef] [PubMed]
- Di Rosa, A.R.; Leone, F.; Cheli, F.; Chiofalo, V. Fusion of electronic nose, electronic tongue and computer vision for animal source food authentication and quality assessment—A review. J. Food Eng. 2017, 210, 62–75. [Google Scholar] [CrossRef]
- Liu, Q.; Zhao, N.; Zhou, D.; Sun, Y.; Sun, K.; Pan, L.; Tu, K. Discrimination and growth tracking of fungi contamination in peaches using electronic nose. Food Chem. 2018, 262, 226–234. [Google Scholar] [CrossRef]
- Wilson, A.D.; Baietto, M. Applications and advances in electronic-nose technologies. Sensors 2009, 9, 5099–5148. [Google Scholar] [CrossRef]
- Tian, X.; Wang, J.; Cui, S. Analysis of pork adulteration in minced mutton using electronic nose of metal oxide sensors. J. Food Eng. 2013, 119, 744–749. [Google Scholar] [CrossRef]
- Tian, X.; Wang, J.; Ma, Z.; Li, M.; Wei, Z. Combination of an E-Nose and an E-Tongue for Adulteration Detection of Minced Mutton Mixed with Pork. J. Food Qual. 2019, 2019, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Li, L.; Ding, W.; Zhang, D.; Wang, J.; Reed, K.; Zhang, B. Adulterant identification in mutton by electronic nose and gas chromatography-mass spectrometer. Food Control 2019, 98, 431–438. [Google Scholar] [CrossRef]
- Kutsanedzie, F.Y.; Guo, Z.; Chen, Q. Advances in Nondestructive Methods for Meat Quality and Safety Monitoring. Food Rev. Int. 2019, 35, 536–562. [Google Scholar] [CrossRef]
- Rakow, N.A.; Suslick, K.S. A colorimetric sensor array for odour visualization. Nature 2000, 406, 710. [Google Scholar] [CrossRef]
- Suslick, K.S.; Rakow, N.A.; Sen, A. Colorimetric sensor arrays for molecular recognition. Tetrahedron 2004, 60, 11133–11138. [Google Scholar] [CrossRef]
- Han, F.; Huang, X.; Teye, E. Novel prediction of heavy metal residues in fish using a low-cost optical electronic tongue system based on colorimetric sensors array. J. Food Process Eng. 2019, 42, e12983. [Google Scholar] [CrossRef]
- Xiaowei, H.; Xiaobo, Z.; Jiewen, Z.; Jiyong, S.; Zhihua, L.; Tingting, S. Monitoring the biogenic amines in Chinese traditional salted pork in jelly (Yao-meat) by colorimetric sensor array based on nine natural pigments. Int. J. Food Sci. Technol. 2015, 50, 203–209. [Google Scholar] [CrossRef]
- Zhang, C.; Bailey, D.P.; Suslick, K.S. Colorimetric sensor arrays for the analysis of beers: A feasibility study. J. Agric. Food Chem. 2006, 54, 4925–4931. [Google Scholar] [CrossRef]
- Bang, J.H.; Lim, S.H.; Park, E.; Suslick, K.S. Chemically responsive nanoporous pigments: Colorimetric sensor arrays and the identification of aliphatic amines. Langmuir 2008, 24, 13168–13172. [Google Scholar] [CrossRef] [Green Version]
- Lim, S.H.; Musto, C.J.; Park, E.; Zhong, W.; Suslick, K.S. A colorimetric sensor array for detection and identification of sugars. Org. Lett. 2008, 10, 4405. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Hu, W.; Su, J.; Li, H.; Ouyang, Q.; Zhao, J. Nondestructively sensing of total viable count (TVC) in chicken using an artificial olfaction system based colorimetric sensor array. J. Food Eng. 2016, 168, 259–266. [Google Scholar] [CrossRef]
- Huang, X.; Xin, J.; Zhao, J. A novel technique for rapid evaluation of fish freshness using colorimetric sensor array. J. Food Eng. 2011, 105, 632–637. [Google Scholar] [CrossRef]
- Zaragozá, P.; Fuentes, A.; Ruiz-Rico, M.; Vivancos, J.-L.; Fernández-Segovia, I.; Ros-Lis, J.V.; Barat, J.M.; Martínez-Máñez, R. Development of a colorimetric sensor array for squid spoilage assessment. Food Chem. 2015, 175, 315–321. [Google Scholar] [CrossRef]
- Salinas, Y.; Ros-Lis, J.V.; Vivancos, J.-L.; Martínez-Máñez, R.; Marcos, M.D.; Aucejo, S.; Herranz, N.; Lorente, I. Monitoring of chicken meat freshness by means of a colorimetric sensor array. Analyst 2012, 137, 3635–3643. [Google Scholar] [CrossRef] [Green Version]
- Salinas, Y.; Ros-Lis, J.V.; Vivancos, J.-L.; Martínez-Máñez, R.; Marcos, M.D.; Aucejo, S.; Herranz, N.; Lorente, I.; Garcia, E. A novel colorimetric sensor array for monitoring fresh pork sausages spoilage. Food Control 2014, 35, 166–176. [Google Scholar] [CrossRef]
- Zhu, A.; Zhi, W.; Qiu, Y.; Wei, L.; Tian, J.; Pan, Z.; Kang, X.; Gu, W.; Duan, L. Surveillance study of the prevalence and antimicrobial resistance of Salmonella in pork from open markets in Xuzhou, China. Food Control 2019, 98, 474–480. [Google Scholar] [CrossRef]
- Khan, M.R.R.; Kang, B.-H.; Yeom, S.-H.; Kwon, D.-H.; Kang, S.-W. Fiber-optic pulse width modulation sensor for low concentration VOC gas. Sens. Actuators B Chem. 2013, 188, 689–696. [Google Scholar] [CrossRef]
- Chen, Y.; Fu, G.; Zilberman, Y.; Ruan, W.; Ameri, S.K.; Zhang, Y.S.; Miller, E.; Sonkusale, S.R. Low cost smart phone diagnostics for food using paper-based colorimetric sensor arrays. Food Control 2017, 82, 227–232. [Google Scholar] [CrossRef]
- Xiao-wei, H.; Xiao-bo, Z.; Ji-yong, S.; Zhi-hua, L.; Jie-wen, Z. Colorimetric sensor arrays based on chemo-responsive dyes for food odor visualization. Trends Food Sci. Technol. 2018, 81, 90–107. [Google Scholar] [CrossRef]
- Shahidi, F.; Rubin, L.J.; D’Souza, L.A.; Teranishi, R.; Buttery, R.G. Meat flavor volatiles: A review of the composition, techniques of analysis, and sensory evaluation. Crit. Rev. Food Sci. Nutr. 1986, 24, 141–243. [Google Scholar] [CrossRef]
- Chen, Q.; Liu, A.; Zhao, J.; Ouyang, Q.; Sun, Z.; Huang, L. Monitoring vinegar acetic fermentation using a colorimetric sensor array. Sens. Actuators B Chem. 2013, 183, 608–616. [Google Scholar] [CrossRef]
- Han, F.; Huang, X.; Teye, E.; Gu, H. Quantitative analysis of fish microbiological quality using electronic tongue coupled with nonlinear pattern recognition algorithms. J. Food Saf. 2015, 35, 336–344. [Google Scholar] [CrossRef]
- Han, F.; Huang, X.; Teye, E.; Gu, H.; Dai, H.; Yao, L. A nondestructive method for fish freshness determination with electronic tongue combined with linear and non-linear multivariate algorithms. Czech J. Food Sci. 2014, 32, 532–537. [Google Scholar] [CrossRef] [Green Version]
- Huang, G.-B.; Zhu, Q.-Y.; Siew, C.-K. Extreme learning machine: Theory and applications. Neurocomputing 2006, 70, 489–501. [Google Scholar] [CrossRef]
- Lin, H.; Chen, Q.; Zhao, J.; Zhou, P. Determination of free amino acid content in Radix Pseudostellariae using near infrared (NIR) spectroscopy and different multivariate calibrations. J. Pharm. Biomed. Anal. 2009, 50, 803–808. [Google Scholar] [CrossRef] [PubMed]


| Crude Protein | Total Lipid | Total Ash | |
|---|---|---|---|
| beef | 14 ± 0.85a | 34 ± 4.0a | 0.62 ± 0.10a |
| pork | 18 ± 3.0b | 4.3 ± 0.47b | 1.0 ± 0.19b |
| Adulteration Level | 20% | 40% | 60% | 80% | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C1 in Figure 3b | C2 in Figure 3b | C3 in Figure 3b | C1 in Figure 3c | C2 in Figure 3c | C3 in Figure 3c | C1 in Figure 3d | C2 in Figure 3d | C3 in Figure 3d | C1 in Figure 3e | C2 in Figure 3e | C3 in Figure 3e | |
| Red | 7.2 | 4.8 | 0.49 | 12 | 5.1 | 0.55 | 7.9 | 2.0 | 3.4 | 8.1 | 6.8 | 0.031 |
| 0.25 | 0.21 | 0.54 | 1.1 | 0.50 | 0.053 | 5.4 | 1.7 | 4.8 | 0.089 | 0.40 | 1.7 | |
| 1.2 | 0.70 | 0.31 | 2.0 | 0.79 | 0.65 | 1.7 | 2.4 | 0.37 | 1.4 | 0.87 | 0.068 | |
| 4.2 | 0.15 | 0.22 | 8.4 | 0.62 | 0.31 | 8.2 | 0.35 | 1.8 | 9.3 | 1.4 | 1.8 | |
| Green | 7.3 | 0.49 | 0.30 | 11 | 0.024 | 0.53 | 9.3 | 7.4 | 3.0 | 6.7 | 6.1 | 0.33 |
| 0.2 | 0.53 | 0.34 | 2.2 | 1.3 | 0.52 | 5.4 | 4.2 | 4.7 | 0.40 | 0.66 | 0.19 | |
| 1.9 | 0.53 | 0.04 | 1.5 | 0.22 | 0.20 | 0.86 | 3.1 | 5.8 | 1.2 | 1.2 | 0.090 | |
| 1.3 | 0.47 | 5.4 | 2.6 | 0.18 | 8.6 | 0.74 | 0.25 | 10 | 1.5 | 1.6 | 111 | |
| Blue | 0.18 | 0.56 | 0.44 | 1.7 | 0.11 | 1.1 | 3.7 | 7.6 | 5.8 | 2.3 | 6.8 | 1.1 |
| 0.34 | 0.41 | 0.60 | 2.6 | 0.89 | 1.8 | 0.98 | 4.9 | 12 | 0.86 | 1.1 | 1.4 | |
| 1.3 | 0.05 | 0.64 | 6.3 | 1.0 | 3.0 | 4.7 | 4.0 | 5.5 | 5.1 | 0.049 | 1.8 | |
| 0.36 | 0.51 | 0.25 | 0.022 | 1.1 | 4.4 | 0.024 | 1.0 | 4.2 | 0.30 | 1.8 | 0.31 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, F.; Huang, X.; H. Aheto, J.; Zhang, D.; Feng, F. Detection of Beef Adulterated with Pork Using a Low-Cost Electronic Nose Based on Colorimetric Sensors. Foods 2020, 9, 193. https://doi.org/10.3390/foods9020193
Han F, Huang X, H. Aheto J, Zhang D, Feng F. Detection of Beef Adulterated with Pork Using a Low-Cost Electronic Nose Based on Colorimetric Sensors. Foods. 2020; 9(2):193. https://doi.org/10.3390/foods9020193
Chicago/Turabian StyleHan, Fangkai, Xingyi Huang, Joshua H. Aheto, Dongjing Zhang, and Fan Feng. 2020. "Detection of Beef Adulterated with Pork Using a Low-Cost Electronic Nose Based on Colorimetric Sensors" Foods 9, no. 2: 193. https://doi.org/10.3390/foods9020193





