Wearable 12-Lead ECG Acquisition Using a Novel Deep Learning Approach from Frank or EASI Leads with Clinical Validation
Abstract
:1. Introduction
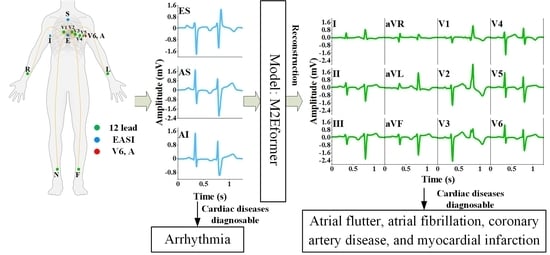
- For the first time, a deep learning-based ECG reconstruction network is presented, which deeply extracts latent cardiac information from EASI leads and reconstructs a standard 12-lead ECG consistent with the diagnostic practices of cardiac experts. This provides a feasible approach to the application of wearable ECG for clinical diagnosis.
- We propose a 2D-ECGblock module for the reconstruction network that transforms time-domain signals into multiperiod 2D tensors based on spectral energy. This module simultaneously extracts dependent information from both within-cycle and between-cycle components in the ECG. Additionally, we designed the ProbDecoder module, which employs a sparse attention mechanism to achieve ECG reconstruction in a residual-like manner.
- We conducted a clinical diagnostic validation study of 25 cases using a 12-lead ECG reconstructed from EASI leads. Next, focusing on four cardiac conditions, namely, atrial fibrillation, atrial flutter, coronary artery disease, and myocardial infarction, which require 12-lead ECG monitoring, three experts were invited to participate in a double-blind diagnostic experiment comparing the reconstructed 12-lead ECG with standard recorded ones. The overall consistency coefficient reached 96%.
2. Materials and Methods
2.1. Databases
2.2. Multichannel 2D-Variation ECG Reconstruction Network (M2Eformer)
2.3. Evaluation
3. Results
3.1. A Comparison of Training Results
3.2. ECG Reconstruction Effect Comparison
3.3. EASI Leads to 12-Lead ECG
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Benjamin, E.J.; Muntner, P.; Alonso, A.; Bittencourt, M.S.; Callaway, C.W.; Carson, A.P.; Chamberlain, A.M.; Chang, A.R.; Cheng, S.; Das, S.R. Heart disease and stroke statistics—2019 update: A report from the American Heart Association. Circulation 2019, 139, e56–e528. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Yang, M.; Di, J.; Xing, Y.; Li, Y.; Li, J. Wearable ECG: History, key technologies and future challenges. Chin. J. Biomed. Eng. 2019, 38, 641–652. [Google Scholar]
- Zhang, X.; Jiang, M.; Polat, K.; Alhudhaif, A.; Hemanth, J.; Wu, W. Detection of Atrial Fibrillation from Variable-Duration ECG Signal Based on Time-Adaptive Densely Network and Feature Enhancement Strategy. IEEE J. Biomed. Health Inform. 2022, 27, 944–955. [Google Scholar] [CrossRef] [PubMed]
- Holter, N.J. New method for heart studies: Continuous electrocardiography of active subjects over long periods is now practical. Science 1961, 134, 1214–1220. [Google Scholar] [CrossRef]
- Rajanna, R.R.; Sriraam, N.; Prakash, V.; Vittala, P.R.; Arun, U.; Sahoo, S. External cardiac loop recorders: Functionalities, diagnostic efficacy, challenges and opportunities. IEEE Rev. Biomed. Eng. 2021, 15, 273–292. [Google Scholar] [CrossRef]
- Liu, H.; Gao, T.; Liu, Z.; Shu, M. FGSQA-Net: A Weakly Supervised Approach to Fine-grained Electrocardiogram Signal Quality Assessment. IEEE J. Biomed. Health Inform. 2023, 27, 3844–3855. [Google Scholar] [CrossRef]
- Gladstone, D.J.; Wachter, R.; Schmalstieg-Bahr, K.; Quinn, F.R.; Hummers, E.; Ivers, N.; Marsden, T.; Thornton, A.; Djuric, A.; Suerbaum, J. Screening for atrial fibrillation in the older population: A randomized clinical trial. JAMA Cardiol. 2021, 6, 558–567. [Google Scholar] [CrossRef]
- Perez, M.V.; Mahaffey, K.W.; Hedlin, H.; Rumsfeld, J.S.; Garcia, A.; Ferris, T.; Balasubramanian, V.; Russo, A.M.; Rajmane, A.; Cheung, L. Large-scale assessment of a smartwatch to identify atrial fibrillation. N. Engl. J. Med. 2019, 381, 1909–1917. [Google Scholar] [CrossRef] [PubMed]
- Teferra, M.N.; Hobbs, D.A.; Clark, R.A.; Reynolds, K.J. Electronic-textile 12-lead equivalent diagnostic electrocardiogram based on the EASI lead placement. IEEE Sens. 2022, 22, 5994–6001. [Google Scholar] [CrossRef]
- Fang, B.; Chen, J.; Liu, Y.; Wang, W.; Wang, K.; Singh, A.K.; Lv, Z. Dual-channel neural network for atrial fibrillation detection from a single lead ECG wave. IEEE J. Biomed. Health Inform. 2021, 27, 2296–2305. [Google Scholar] [CrossRef] [PubMed]
- Rajakariar, K.; Koshy, A.N.; Sajeev, J.K.; Nair, S.; Roberts, L.; Teh, A.W. Accuracy of a smartwatch based single-lead electrocardiogram device in detection of atrial fibrillation. Heart 2020, 106, 665–670. [Google Scholar] [CrossRef]
- Himmelreich, J.C.; Karregat, E.P.; Lucassen, W.A.; van Weert, H.C.; de Groot, J.R.; Handoko, M.L.; Nijveldt, R.; Harskamp, R.E. Diagnostic accuracy of a smartphone-operated, single-lead electrocardiography device for detection of rhythm and conduction abnormalities in primary care. Ann. Fam. Med. 2019, 17, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Frank, E. An accurate, clinically practical system for spatial vectorcardiography. Circulation 1956, 13, 737–749. [Google Scholar] [CrossRef] [PubMed]
- Frank, E. General theory of heart-vector projection. Circ. Res. 1954, 2, 258–270. [Google Scholar] [CrossRef]
- Levkov, C.L. Orthogonal electrocardiogram derived from the limb and chest electrodes of the conventional 12-lead system. Med. Biol. Eng. Comput. 1987, 25, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Dower, G.E.; Yakush, A.; Nazzal, S.B.; Jutzy, R.V.; Ruiz, C.E. Deriving the 12-lead electrocardiogram from four (EASI) electrodes. J. Electrocardiol. 1988, 21, S182–S187. [Google Scholar] [CrossRef] [PubMed]
- Nelwan, S.P.; Kors, J.A.; Meij, S.H.; van Bemmel, J.H.; Simoons, M.L. Reconstruction of the 12-lead electrocardiogram from reduced lead sets. J. Electrocardiol. 2004, 37, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Feild, D.Q.; Feldman, C.L.; Hor, B.M. Improved EASI coefficients: Their derivation, values, and performance. J. Electrocardiol. 2002, 35, 23–33. [Google Scholar] [CrossRef]
- Li, S.F.; Jing, Y.; Guo, S.; Yu, A.X.; Zhu, T.; Li, Z.J. A comparative analysis of Cabrera and Wilson lead electrocardiogram in normal subjects. Clin. Meta-Anal. 2010, 25, 1149–1150. [Google Scholar]
- Drew, B.J.; Pelter, M.M.; Wung, S.-F.; Adams, M.G.; Taylor, C.; Evans, G.T., Jr.; Foster, E. Accuracy of the EASI 12-lead electrocardiogram compared to the standard 12-lead electrocardiogram for diagnosing multiple cardiac abnormalities. J. Electrocardiol. 1999, 32, 38–47. [Google Scholar] [CrossRef]
- Nelwan, S.P.; Kors, J.A.; Crater, S.W.; Meij, S.H.; van Dam, T.B.; Simoons, M.L.; Krucoff, M.W. Simultaneous comparison of 3 derived 12-lead electrocardiograms with standard electrocardiogram at rest and during percutaneous coronary occlusion. J. Electrocardiol. 2008, 41, 230–237. [Google Scholar] [CrossRef]
- Schreck, D.; Brotea, C.; Shah, S. Derivation of the 12-lead electrocardiogram using abstract factor analysis and simplex optimization. Int. J. Bioelectromagn. 2002, 4, 337–338. [Google Scholar]
- Schreck, D.M.; Fishberg, R.D. Derivation of the 12-lead electrocardiogram and 3-lead vectorcardiogram. Am. J. Emerg. Med. 2013, 31, 1183–1190. [Google Scholar] [CrossRef] [PubMed]
- Feild, D.Q.; Zhou, S.H.; Helfenbein, E.D.; Gregg, R.E.; Lindauer, J.M. Technical challenges and future directions in lead reconstruction for reduced-lead systems. J. Electrocardiol. 2008, 41, 466–473. [Google Scholar] [CrossRef]
- Trobec, R.; Tomašić, I. Synthesis of the 12-lead electrocardiogram from differential leads. IEEE Trans. Inf. Technol. Biomed. 2011, 15, 615–621. [Google Scholar] [CrossRef] [PubMed]
- Mulyadi, I.H.; Supriyanto, E. Segment-specific EASI coefficients for improving accuracy of derived 12-lead electrocardiography. In Proceedings of the 2018 International Conference on Applied Engineering, Batam, Indonesia, 3–4 October 2018; pp. 1–4. [Google Scholar]
- Atoui, H.; Fayn, J.; Rubel, P. A neural network approach for patient-specific 12-lead ECG synthesis in patient monitoring environments. In Proceedings of the Computers in Cardiology, Chicago, IL, USA, 19–22 September 2004; pp. 161–164. [Google Scholar]
- Smith, G.H.; Van den Heever, D.J.; Swart, W. The reconstruction of a 12-lead electrocardiogram from a reduced lead set using a focus time-delay neural network. Acta Cardiol. Sin. 2021, 37, 47. [Google Scholar]
- Zhou, H.; Zhang, S.; Peng, J.; Zhang, S.; Li, J.; Xiong, H.; Zhang, W. Informer: Beyond efficient transformer for long sequence time-series forecasting. In Proceedings of the AAAI Conference on Artificial Intelligence, Vancouver, BC, Canada, 2–9 February 2021; pp. 11106–11115. [Google Scholar]
- Liu, S.; Yu, H.; Liao, C.; Li, J.; Lin, W.; Liu, A.X.; Dustdar, S. Pyraformer: Low-complexity pyramidal attention for long-range time series modeling and forecasting. In Proceedings of the International Conference on Learning Representations, Virtual, 3–7 May 2021. [Google Scholar]
- Wu, H.; Xu, J.; Wang, J.; Long, M. Autoformer: Decomposition transformers with auto-correlation for long-term series forecasting. Adv. Neural Inf. Process. Syst. 2021, 34, 22419–22430. [Google Scholar]
- Zhou, T.; Ma, Z.; Wen, Q.; Wang, X.; Sun, L.; Jin, R. Fedformer: Frequency enhanced decomposed transformer for long-term series forecasting. In Proceedings of the International Conference on Machine Learning, Baltimore MD, USA, 17 July 2022; pp. 27268–27286. [Google Scholar]
- Bousseljot, R.; Kreiseler, D.; Schnabel, A. Nutzung der EKG-Signaldatenbank CARDIODAT der PTB über das Internet. Biomed. Eng./Biomed. Tech. 1995, 40, 317–318. [Google Scholar] [CrossRef]
- Wu, H.; Hu, T.; Liu, Y.; Zhou, H.; Wang, J.; Long, M. Timesnet: Temporal 2d-variation modeling for general time series analysis. arXiv 2022, arXiv:2210.02186. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Wang, J. An intelligent computer-aided approach for atrial fibrillation and atrial flutter signals classification using modified bidirectional LSTM network. Inf. Sci. 2021, 574, 320–332. [Google Scholar] [CrossRef]
- Almutairi, H.; Hassan, G.M.; Datta, A. Classification of Obstructive Sleep Apnoea from single-lead ECG signals using convolutional neural and Long Short Term Memory networks. Biomed. Signal Process. Control 2021, 69, 102906. [Google Scholar] [CrossRef]
- Andersen, R.S.; Peimankar, A.; Puthusserypady, S. A deep learning approach for real-time detection of atrial fibrillation. Expert Syst. Appl. 2019, 115, 465–473. [Google Scholar] [CrossRef]










| Training | Validation | Test | Total | |
|---|---|---|---|---|
| Healthy controls | 35 | 11 | 11 | 57 |
| Myocardial infarction | 127 | 43 | 43 | 213 |
| Bundle branch block | 7 | 2 | 2 | 11 |
| Myocardial hypertrophy | 6 | 2 | 2 | 10 |
| Valvular heart disease | 2 | 1 | 1 | 4 |
| Cardiomyopathy | 4 | 1 | 1 | 6 |
| Total | 183 | 61 | 61 | 305 |
| Age ± Std | Training | Test | Total | |
|---|---|---|---|---|
| Healthy controls | 26.2 ± 7.2 | 8 | 2 | 10 |
| Atrial flutter | 73 | 12 | 3 | 15 |
| Atrial tachycardia | 76.5 ± 7.5 | |||
| Myocardial infarction | 66.2 ± 7.9 | |||
| Coronary heart disease | 59.3 ± 17.6 | |||
| Total | 50.5 ± 22.2 | 20 | 5 | 25 |
| Model | Best Epoch/Min Loss | |||||||
|---|---|---|---|---|---|---|---|---|
| I | II | III | aVF | V2 | V3 | V5 | V6 | |
| M2Eformer | 64/0.0027 | 98/0.0009 | 97/0.0022 | 98/0.0008 | 55/0.0229 | 74/0.0215 | 66/0.0050 | 85/0.0016 |
| T-Transformer | 12/0.0028 | 70/0.0010 | 91/0.0024 | 95/0.0009 | 93/0.0236 | 10/0.0219 | 94/0.0054 | 66/0.0027 |
| Transformer | 18/0.0027 | 51/0.0010 | 97/0.0038 | 70/0.0009 | 28/0.0243 | 10/0.0215 | 15/0.0049 | 89/0.0016 |
| Model | I | II | III | aVR | aVL | aVF | V1 | V2 | V3 | V4 | V5 | V6 | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M2Eformer (ours) | 0.8465 | 0.9588 | 0.7817 | 0.8921 | 0.8447 | 0.8321 | 0.9105 | 0.8930 | 0.9420 | 0.8641 | 0.8554 | 0.9215 | 0.8785 |
| M2Eformer (ours) | 0.8441 | 0.9207 | 0.7496 | 0.8534 | 0.7875 | 0.8647 | 0.8932 | 0.9052 | 0.9220 | 0.8409 | 0.8326 | 0.8814 | 0.8579 |
| Transformer | 0.8568 | 0.9063 | 0.6705 | 0.7983 | 0.7036 | 0.8748 | 0.8865 | 0.9133 | 0.9217 | 0.7883 | 0.7895 | 0.9004 | 0.8342 |
| LSTM | 0.6573 | 0.6357 | 0.4625 | 0.6181 | 0.6273 | 0.6366 | 0.7009 | 0.5212 | 0.5600 | 0.5341 | 0.5612 | 0.6926 | 0.6006 |
| LSR | 0.8507 | 0.8266 | 0.5891 | 0.9224 | 0.6456 | 0.6741 | 0.9015 | 0.8835 | 0.9516 | 0.8552 | 0.8594 | 0.9621 | 0.8268 |
| Linear | 0.8087 | 0.9485 | 0.6381 | 0.8642 | 0.6155 | 0.8534 | 0.9068 | 0.8795 | 0.9440 | 0.7759 | 0.6735 | 0.9220 | 0.8192 |
| Model | I | II | III | aVR | aVL | aVF | V1 | V2 | V3 | V4 | V5 | V6 | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M2Eformer (ours) | 0.0399 | 0.0215 | 0.0395 | 0.0266 | 0.0370 | 0.0241 | 0.0619 | 0.1044 | 0.0815 | 0.0730 | 0.0629 | 0.0406 | 0.0511 |
| M2Eformer (ours) | 0.0401 | 0.0260 | 0.0368 | 0.0316 | 0.0465 | 0.0252 | 0.0632 | 0.0972 | 0.0879 | 0.0827 | 0.0688 | 0.0465 | 0.0544 |
| Transformer | 0.0380 | 0.0265 | 0.0430 | 0.0343 | 0.0474 | 0.0241 | 0.0638 | 0.1003 | 0.0933 | 0.1045 | 0.0635 | 0.0457 | 0.0570 |
| LSTM | 0.0479 | 0.0576 | 0.0507 | 0.0488 | 0.0373 | 0.0464 | 0.0789 | 0.1410 | 0.1403 | 0.1147 | 0.0853 | 0.0620 | 0.0759 |
| LSR | 0.0352 | 0.0422 | 0.0491 | 0.0234 | 0.0401 | 0.0487 | 0.0517 | 0.0980 | 0.0655 | 0.0756 | 0.0570 | 0.0311 | 0.0515 |
| Linear | 0.0516 | 0.0212 | 0.0470 | 0.0322 | 0.0478 | 0.0257 | 0.0432 | 0.0753 | 0.0582 | 0.0954 | 0.0997 | 0.0358 | 0.0528 |
| Data | Cardiologist 1 | Cardiologist 2 | Cardiologist 3 | Overall Agreement (OvA) | ||||
|---|---|---|---|---|---|---|---|---|
| CS/AS | PoC | CS/AS | PoC | CS/AS | PoC | CS/AS | PoC | |
| Healthy controls | 9/11 | 81.8% | 9/11 | 81.8% | 10/11 | 90.9% | 9/11 | 81.8% |
| Myocardial infarction | 41/43 | 95.3% | 41/43 | 95.3% | 39/43 | 90.7% | 43/43 | 100% |
| Dysrhythmia | 2/2 | 100% | 2/2 | 100% | 2/2 | 100% | 2/2 | 100% |
| Bundle branch block | 2/2 | 100% | 2/2 | 100% | 2/2 | 100% | 2/2 | 100% |
| Myocardial hypertrophy | 1/1 | 100% | 1/1 | 100% | 1/1 | 100% | 1/1 | 100% |
| Valvular heart disease | 1/1 | 100% | 1/1 | 100% | 0/1 | 0% | 1/1 | 100% |
| Cardiomyopathy | 1/1 | 100% | 1/1 | 100% | 1/1 | 100% | 1/1 | 100% |
| Total | 57/61 | 93.4% | 57/61 | 93.4% | 55/61 | 90.2% | 59/61 | 96.7% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, F.; Zhong, D.; Liu, J.; Xu, T.; Shen, Q.; Wang, W.; Zhu, S.; Li, J. Wearable 12-Lead ECG Acquisition Using a Novel Deep Learning Approach from Frank or EASI Leads with Clinical Validation. Bioengineering 2024, 11, 293. https://doi.org/10.3390/bioengineering11030293
Fu F, Zhong D, Liu J, Xu T, Shen Q, Wang W, Zhu S, Li J. Wearable 12-Lead ECG Acquisition Using a Novel Deep Learning Approach from Frank or EASI Leads with Clinical Validation. Bioengineering. 2024; 11(3):293. https://doi.org/10.3390/bioengineering11030293
Chicago/Turabian StyleFu, Fan, Dacheng Zhong, Jiamin Liu, Tianxiang Xu, Qin Shen, Wei Wang, Songsheng Zhu, and Jianqing Li. 2024. "Wearable 12-Lead ECG Acquisition Using a Novel Deep Learning Approach from Frank or EASI Leads with Clinical Validation" Bioengineering 11, no. 3: 293. https://doi.org/10.3390/bioengineering11030293