The Role of Microfluidics for Organ on Chip Simulations
Abstract
:1. Introduction
2. Organ Simulation Is Better with Microfluidics
3. Limitations of Organ Simulation on Microfluidics
4. Microfluidics Devices Accompanied with Supporting Technologies for Better Simulation
4.1. 3D Bioprinting
4.2. Biosensors
4.3. Multi-Electrode Array (MEA)
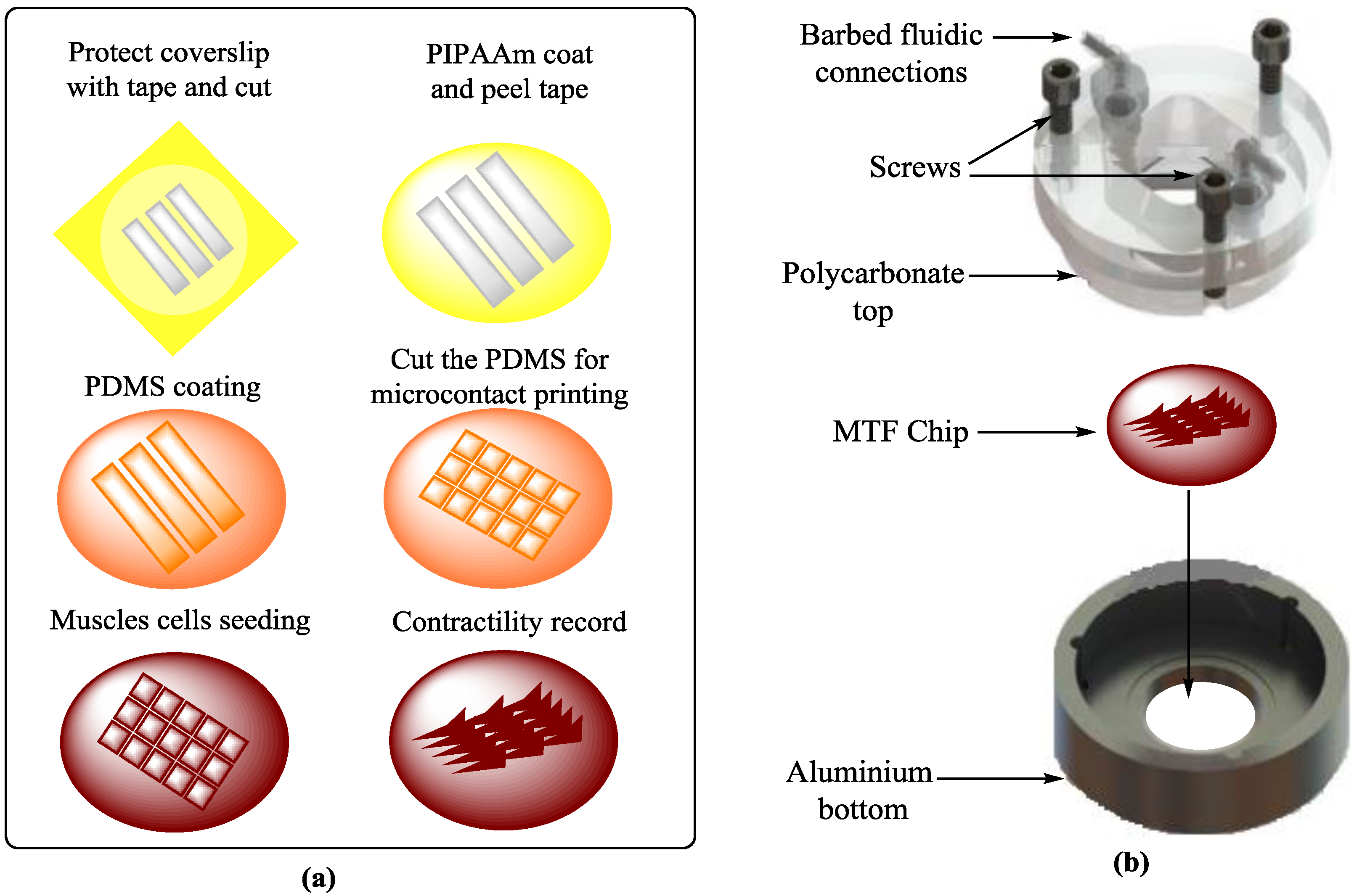
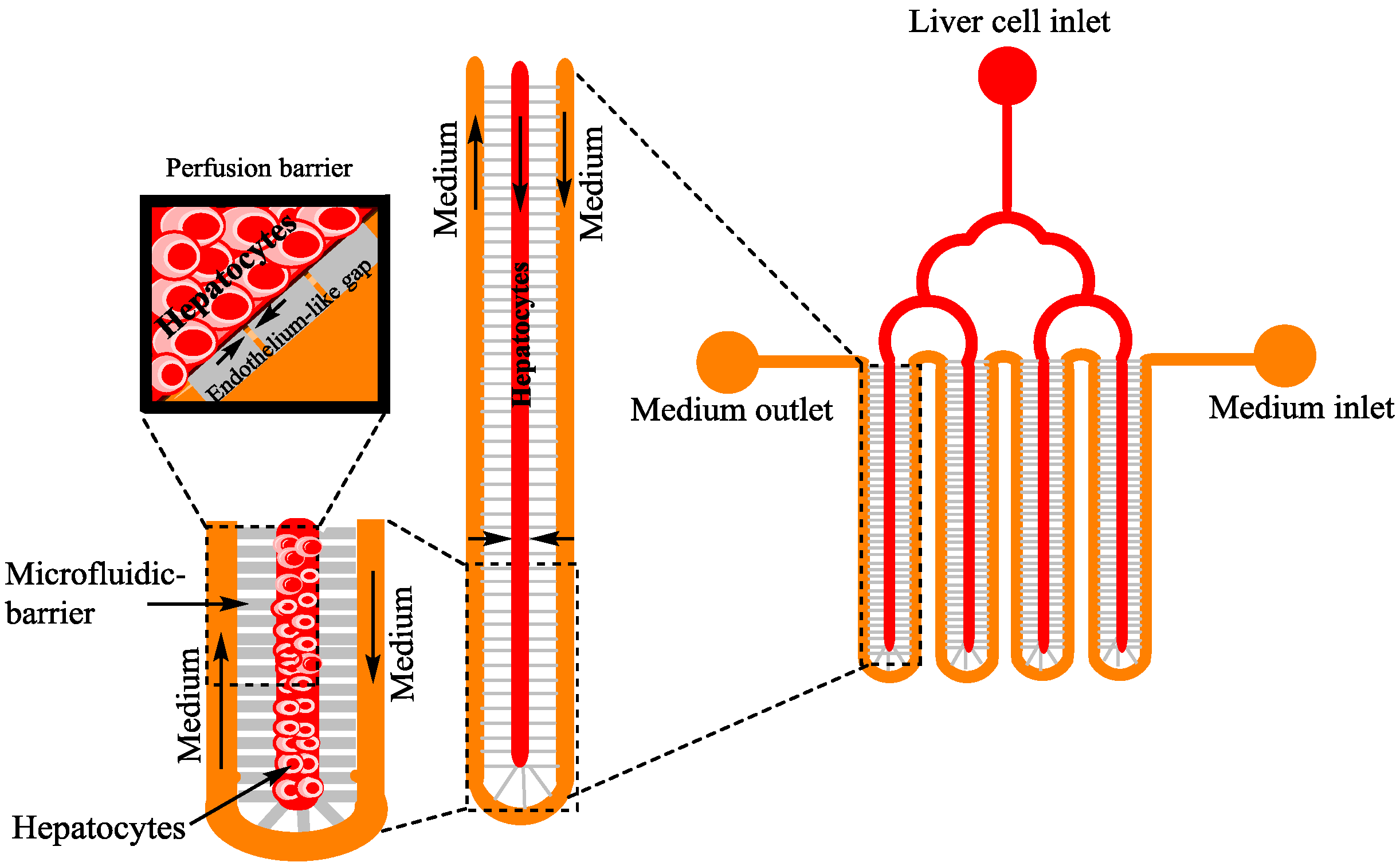
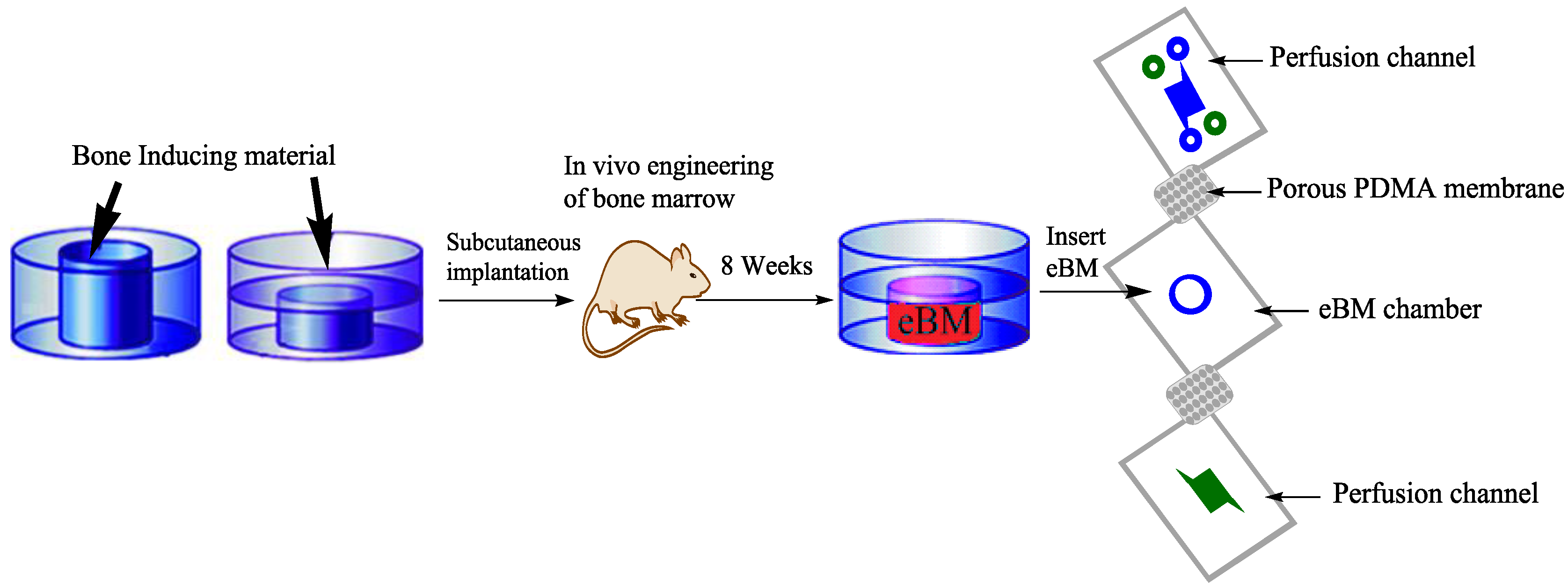
4.4. Tissue Engineering (TE)
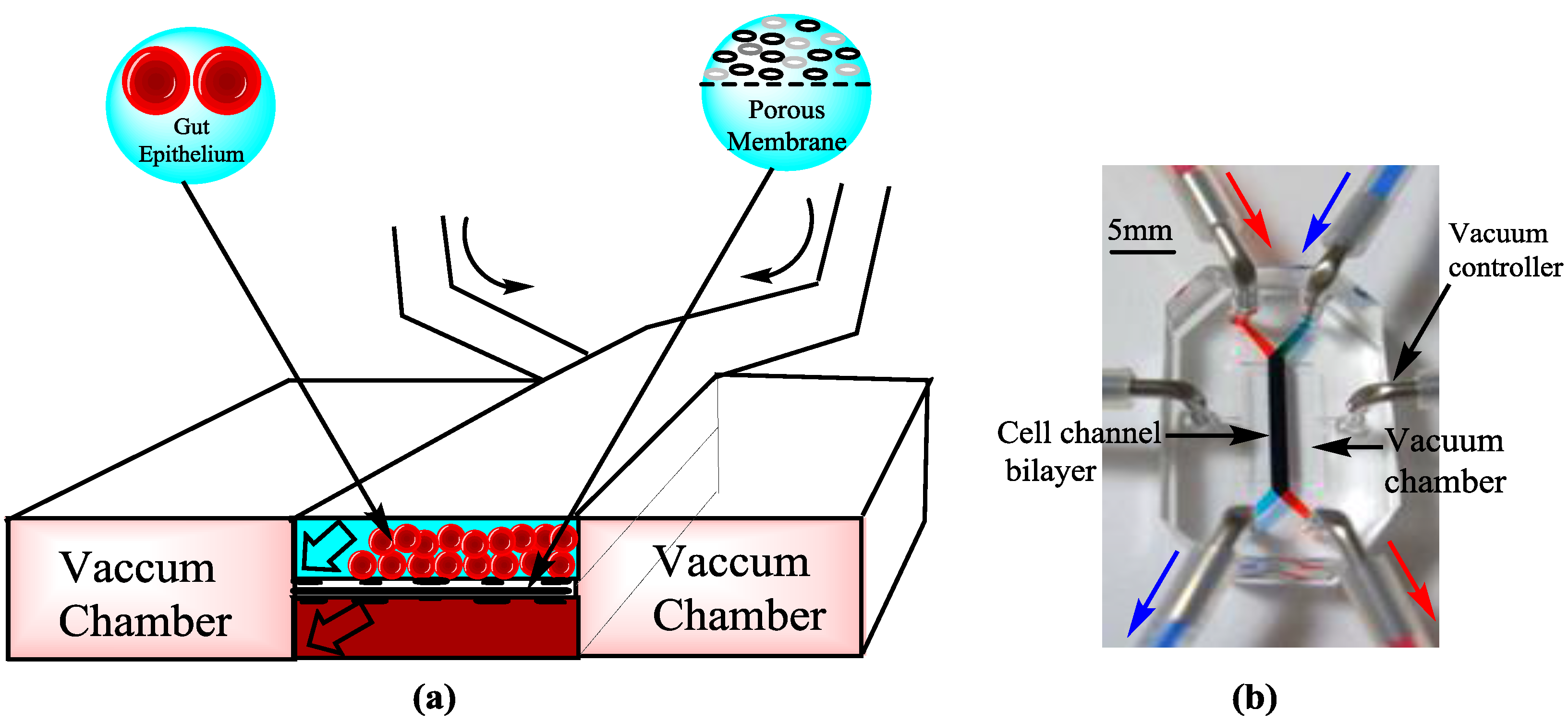
4.5. Omics and Microbiome
5. Future Opportunities and Challenges
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Mehling, M.; Tay, S. Microfluidic cell culture. Curr. Opin. Biotechnol. 2014, 25, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Volpatti, L.R.; Yetisen, A.K. Commercialization of microfluidic devices. Trends Biotechnol. 2014, 32, 347–350. [Google Scholar] [CrossRef] [PubMed]
- Wikswo, J.P.; Curtis, E.L.; Eagleton, Z.E.; Evans, B.C.; Kole, A.; Hofmeister, L.H.; Matloff, W.J. Scaling and systems biology for integrating multiple organs-on-a-chip. Lab Chip 2013, 13, 3496–3511. [Google Scholar] [CrossRef] [PubMed]
- Greek, R.; Menache, A. Systematic reviews of animal models: Methodology versus epistemology. Int. J. Med. Sci. 2013, 10, 206–221. [Google Scholar] [CrossRef] [PubMed]
- Mammoto, T.; Mammoto, A.; Ingber, D.E. Mechanobiology and developmental control. Annu. Rev. Cell Dev. Biol. 2013, 29, 27–61. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, S.N.; Ingber, D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014, 32, 760–772. [Google Scholar] [CrossRef] [PubMed]
- Huh, D.; Leslie, D.C.; Matthews, B.D.; Fraser, J.P.; Jurek, S.; Hamilton, G.A.; Thorneloe, K.S.; McAlexander, M.A.; Ingber, D.E. A human disease model of drug toxicity-induced pulmonary edema in a lung-on-a-chip microdevice. Sci. Transl. Med. 2012, 4, 159ra147. [Google Scholar] [CrossRef] [PubMed]
- Gao, D.; Liu, H.X.; Jiang, Y.Y.; Lin, J.M. Recent developments in microfluidic devices for in vitro cell culture for cell-biology research. TrAC Trends Anal. Chem. 2012, 35, 150–164. [Google Scholar] [CrossRef]
- Muranen, T.; Selfors, L.M.; Worster, D.T.; Iwanicki, M.P.; Song, L.; Morales, F.C.; Gao, S.; Mills, G.B.; Brugge, J.S. Inhibition of PI3K/mTOR leads to adaptive resistance in matrix-attached cancer cells. Cancer Cell 2012, 21, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Mroue, R.; Bissell, M.J. Three-dimensional cultures of mouse mammary epithelial cells. Methods Mol. Biol. 2013, 945, 221–250. [Google Scholar] [PubMed]
- Sato, T.; Clevers, H. Growing self-organizing mini-guts from a single intestinal stem cell: Mechanism and applications. Science 2013, 340, 1190–1194. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, M.A.; Renner, M.; Martin, C.A.; Wenzel, D.; Bicknell, L.S.; Hurles, M.E.; Homfray, T.; Penninger, J.M.; Jackson, A.P.; Knoblich, J.A. Cerebral organoids model human brain development and microcephaly. Nature 2013, 501, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Prentice-Mott, H.V.; Chang, C.H.; Mahadevan, L.; Mitchison, T.J.; Irimia, D.; Shah, J.V. Biased migration of confined neutrophil-like cells in asymmetric hydraulic environments. Proc. Natl. Acad. Sci. USA 2013, 110, 21006–21011. [Google Scholar] [CrossRef] [PubMed]
- Xiao, R.R.; Zeng, W.J.; Li, Y.T.; Zou, W.; Wang, L.; Pei, X.F.; Xie, M.; Huang, W.H. Simultaneous generation of gradients with gradually changed slope in a microfluidic device for quantifying axon response. Anal. Chem. 2013, 85, 7842–7850. [Google Scholar] [CrossRef] [PubMed]
- Cimetta, E.; Cannizzaro, C.; James, R.; Biechele, T.; Moon, R.T.; Elvassore, N.; Vunjak-Novakovic, G. Microfluidic device generating stable concentration gradients for long term cell culture: Application to Wnt3a regulation of beta-catenin signaling. Lab Chip 2010, 10, 3277–3283. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.C.; Liao, W.H.; Chen, Y.H.; Wu, C.Y.; Tung, Y.C. A microfluidic cell culture array with various oxygen tensions. Lab Chip 2013, 13, 3239–3245. [Google Scholar] [CrossRef] [PubMed]
- Seidi, A.; Kaji, H.; Annabi, N.; Ostrovidov, S.; Ramalingam, M.; Khademhosseini, A. A microfluidic-based neurotoxin concentration gradient for the generation of an in vitro model of Parkinson’s disease. Biomicrofluidics 2011, 5, 22214. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Lee, L.P. Non-invasive microfluidic gap junction assay. Integr. Biol. 2010, 2, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, W.; Lim, S.; Jeon, J. Vasculature-on-a-chip for in vitro disease models. Bioengineering 2017, 4, 8. [Google Scholar] [CrossRef]
- Huh, D.; Matthews, B.D.; Mammoto, A.; Montoya-Zavala, M.; Hsin, H.Y.; Ingber, D.E. Reconstituting organ-level lung functions on a chip. Science 2010, 328, 1662–1668. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Huh, D.; Hamilton, G.; Ingber, D.E. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip 2012, 12, 2165–2174. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Ingber, D.E. Gut-on-a-chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr. Biol. 2013, 5, 1130–1140. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, A.; Goss, J.A.; Cho, A.; McCain, M.L.; Parker, K.K. Microfluidic heart on a chip for higher throughput pharmacological studies. Lab Chip 2013, 13, 3599–3608. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Sim, W.Y.; Min, B.H.; Yang, S.S.; Khademhosseini, A.; Kaplan, D.L. Chip-based comparison of the osteogenesis of human bone marrow- and adipose tissue-derived mesenchymal stem cells under mechanical stimulation. PLoS ONE 2012, 7, e46689. [Google Scholar] [CrossRef] [PubMed]
- Takayama, S.; Ostuni, E.; LeDuc, P.; Naruse, K.; Ingber, D.E.; Whitesides, G.M. Subcellular positioning of small molecules. Nature 2001, 411, 1016. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.J.; Hung, P.J.; Lee, L.P. An artificial liver sinusoid with a microfluidic endothelial-like barrier for primary hepatocyte culture. Biotechnol. Bioeng. 2007, 97, 1340–1346. [Google Scholar] [CrossRef] [PubMed]
- Tumarkin, E.; Tzadu, L.; Csaszar, E.; Seo, M.; Zhang, H.; Lee, A.; Peerani, R.; Purpura, K.; Zandstra, P.W.; Kumacheva, E. High-throughput combinatorial cell co-culture using microfluidics. Integr. Biol. 2011, 3, 653–662. [Google Scholar] [CrossRef] [PubMed]
- Esch, M.B.; Sung, J.H.; Yang, J.; Yu, C.; Yu, J.; March, J.C.; Shuler, M.L. On chip porous polymer membranes for integration of gastrointestinal tract epithelium with microfluidic ‘body-on-a-chip’ devices. Biomed. Microdevices 2012, 14, 895–906. [Google Scholar] [CrossRef] [PubMed]
- Baker, B.M.; Trappmann, B.; Stapleton, S.C.; Toro, E.; Chen, C.S. Microfluidics embedded within extracellular matrix to define vascular architectures and pattern diffusive gradients. Lab Chip 2013, 13, 3246–3252. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.H.; Stapleton, S.C.; Yang, M.T.; Cha, S.S.; Choi, C.K.; Galie, P.A.; Chen, C.S. Biomimetic model to reconstitute angiogenic sprouting morphogenesis in vitro. Proc. Natl. Acad. Sci. USA 2013, 110, 6712–6717. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Yan, J.J.; Shin, Y.; Jeon, J.J.; Won, J.; Jeong, H.E.; Kamm, R.D.; Kim, Y.J.; Chung, S. A versatile assay for monitoring in vivo-like transendothelial migration of neutrophils. Lab Chip 2012, 12, 3861–3865. [Google Scholar] [CrossRef] [PubMed]
- Khanal, G.; Chung, K.; Solis-Wever, X.; Johnson, B.; Pappas, D. Ischemia/reperfusion injury of primary porcine cardiomyocytes in a low-shear microfluidic culture and analysis device. Analyst 2011, 136, 3519–3526. [Google Scholar] [CrossRef] [PubMed]
- Tsantoulas, C.; Farmer, C.; Machado, P.; Baba, K.; McMahon, S.B.; Raouf, R. Probing functional properties of nociceptive axons using a microfluidic culture system. PLoS ONE 2013, 8, e80722. [Google Scholar] [CrossRef] [PubMed]
- Douville, N.J.; Tung, Y.C.; Li, R.; Wang, J.D.; El-Sayed, M.E.; Takayama, S. Fabrication of two-layered channel system with embedded electrodes to measure resistance across epithelial and endothelial barriers. Anal. Chem. 2010, 82, 2505–2511. [Google Scholar] [CrossRef] [PubMed]
- Jang, K.J.; Suh, K.Y. A multi-layer microfluidic device for efficient culture and analysis of renal tubular cells. Lab Chip 2010, 10, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Booth, R.; Kim, H. Characterization of a microfluidic in vitro model of the blood-brain barrier (uBBB). Lab Chip 2012, 12, 1784–1792. [Google Scholar] [CrossRef] [PubMed]
- Whitesides, G.M. The origins and the future of microfluidics. Nature 2006, 442, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Sackmann, E.K.; Fulton, A.L.; Beebe, D.J. The present and future role of microfluidics in biomedical research. Nature 2014, 507, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Van de Stolpe, A.; Den Toonder, J. Workshop meeting report organs-on-chips: Human disease models. Lab Chip 2013, 13, 3449–3470. [Google Scholar] [CrossRef] [PubMed]
- Valencia, P.M.; Farokhzad, O.C.; Karnik, R.; Langer, R. Microfluidic technologies for accelerating the clinical translation of nanoparticles. Nat. Nanotechnol. 2012, 7, 623–629. [Google Scholar] [CrossRef] [PubMed]
- Chrimes, A.F.; Khoshmanesh, K.; Stoddart, P.R.; Mitchell, A.; Kalantar-Zadeh, K. Microfluidics and raman microscopy: Current applications and future challenges. Chem. Soc. Rev. 2013, 42, 5880–5906. [Google Scholar] [CrossRef] [PubMed]
- Pati, F.; Gantelius, J.; Svahn, H.A. 3D bioprinting of tissue/organ models. Angew. Chem. Int. Ed. 2016, 55, 4650–4665. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.W.; Lee, J.S.; Cho, D.W. Computer-aided multiple-head 3D printing system for printing of heterogeneous organ/tissue constructs. Sci. Rep. 2016, 6, 21685. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.S.; Yue, K.; Aleman, J.; Mollazadeh-Moghaddam, K.; Bakht, S.M.; Yang, J.Z.; Jia, W.T.; Dell'Erba, V.; Assawes, P.; Shin, S.R.; et al. 3D bioprinting for tissue and organ fabrication. Ann. Biomed. Eng. 2017, 45, 148–163. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, N.; Urrios, A.; Kang, S.; Folch, A. The upcoming 3D-printing revolution in microfluidics. Lab Chip 2016, 16, 1720–1742. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.-G.; Lee, H.; Cho, D.-W. 3D printing of organs-on-chips. Bioengineering 2017, 4, 10. [Google Scholar] [CrossRef]
- Bhise, N.S.; Manoharan, V.; Massa, S.; Tamayol, A.; Ghaderi, M.; Miscuglio, M.; Lang, Q.; Shrike Zhang, Y.; Shin, S.R.; Calzone, G.; et al. A liver-on-a-chip platform with bioprinted hepatic spheroids. Biofabrication 2016, 8, 014101. [Google Scholar] [CrossRef] [PubMed]
- Knowlton, S.; Tasoglu, S. A bioprinted liver-on-a-chip for drug screening applications. Trends Biotechnol. 2016, 34, 681–682. [Google Scholar] [CrossRef] [PubMed]
- Ware, B.R.; Khetani, S.R. Engineered liver platforms for different phases of drug development. Trends Biotechnol. 2017, 35, 172–183. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Cho, D.W. One-step fabrication of an organ-on-a-chip with spatial heterogeneity using a 3D bioprinting technology. Lab Chip 2016, 16, 2618–2625. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.S.; Arneri, A.; Bersini, S.; Shin, S.R.; Zhu, K.; Goli-Malekabadi, Z.; Aleman, J.; Colosi, C.; Busignani, F.; Dell’Erba, V.; et al. Bioprinting 3D microfibrous scaffolds for engineering endothelialized myocardium and heart-on-a-chip. Biomaterials 2016, 110, 45–59. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Arneri, A.; Dell’Erba, V.; Shin, S.; Aleman, J.; Busignani, F.; Bersini, S.; Dokmeci, M.R.; Annabi, N.; Moretti, M.; et al. Bioprinting the heart: Applications in tissue fabrication and organs-on-a-chip. Tissue Eng. Part A 2015, 21, S257–S258. [Google Scholar]
- Duan, B. State-of-the-art review of 3D bioprinting for cardiovascular tissue engineering. Ann. Biomed. Eng. 2017, 45, 195–209. [Google Scholar] [CrossRef] [PubMed]
- Homan, K.A.; Kolesky, D.B.; Skylar-Scott, M.A.; Herrmann, J.; Obuobi, H.; Moisan, A.; Lewis, J.A. Bioprinting of 3D convoluted renal proximal tubules on perfusable chips. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.N.; Lancaster, K.Z.; Hogue, I.B.; Meng, F.; Kong, Y.L.; Enquist, L.W.; McAlpine, M.C. Correction: 3D printed nervous system on a chip. Lab Chip 2016, 16, 1946. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.C.; Zhang, Y.S.; Akpek, A.; Shin, S.R.; Khademhosseini, A. 4D bioprinting: The next-generation technology for biofabrication enabled by stimuli-responsive materials. Biofabrication 2017, 9, 012001. [Google Scholar] [CrossRef] [PubMed]
- Moral-Vico, J.; Barallat, J.; Abad, L.; Olive-Monllau, R.; Munoz-Pascual, F.X.; Galan Ortega, A.; del Campo, F.J.; Baldrich, E. Dual chronoamperometric detection of enzymatic biomarkers using magnetic beads and a low-cost flow cell. Biosens. Bioelectron. 2015, 69, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.Y.; Loke, W.K.; Nguyen, N.T.; Tan, S.N.; Tay, N.B.; Wang, W.; Ng, S.H. Lab-on-a-chip for rapid electrochemical detection of nerve agent sarin. Biomed. Microdevices 2014, 16, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.; Klauke, N.; Sedgwick, H.; Smith, G.L.; Cooper, J.M. Metabolic monitoring of the electrically stimulated single heart cell within a microfluidic platform. Lab Chip 2006, 6, 1424–1431. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.S.; Peng, S.W.; Cheng, J.Y. In vitro electrical-stimulated wound-healing chip for studying electric field-assisted wound-healing process. Biomicrofluidics 2012, 6, 34117. [Google Scholar] [CrossRef] [PubMed]
- Huval, R.M.; Miller, O.H.; Curley, J.L.; Fan, Y.W.; Hall, B.J.; Moore, M.J. Microengineered peripheral nerve-on-a-chip for preclinical physiological testing. Lab Chip 2015, 15, 2221–2232. [Google Scholar] [CrossRef] [PubMed]
- Rattanarat, P.; Teengam, P.; Siangproh, W.; Ishimatsu, R.; Nakano, K.; Chailapakul, O.; Imato, T. An electrochemical compact disk-type microfluidics platform for use as an enzymatic biosensor. Electroanalysis 2015, 27, 703–712. [Google Scholar] [CrossRef]
- Lafleur, J.P.; Jonsson, A.; Senkbeil, S.; Kutter, J.P. Recent advances in lab-on-a-chip for biosensing applications. Biosens. Bioelectron. 2016, 76, 213–233. [Google Scholar] [CrossRef] [PubMed]
- Dastider, S.G.; Barizuddin, S.; Yuksek, N.S.; Dweik, M.; Almasri, M.F. Efficient and rapid detection of salmonella using microfluidic impedance based sensing. J. Sens. 2015, 2015. [Google Scholar] [CrossRef]
- Prill, S.; Bavli, D.; Levy, G.; Ezra, E.; Schmalzlin, E.; Jaeger, M.S.; Schwarz, M.; Duschl, C.; Cohen, M.; Nahmias, Y. Real-time monitoring of oxygen uptake in hepatic bioreactor shows CYP450-independent mitochondrial toxicity of acetaminophen and amiodarone. Arch. Toxicol. 2016, 90, 1181–1191. [Google Scholar] [CrossRef] [PubMed]
- Bavli, D.; Prill, S.; Ezra, E.; Levy, G.; Cohen, M.; Vinken, M.; Vanfleteren, J.; Jaeger, M.; Nahmias, Y. Real-time monitoring of metabolic function in liver-on-chip microdevices tracks the dynamics of mitochondrial dysfunction. Proc. Natl. Acad. Sci. USA 2016, 113, E2231–E2240. [Google Scholar] [CrossRef] [PubMed]
- Kopparthy, V.; Tangutooru, S.; Guilbeau, E. Label free detection of l-glutamate using microfluidic based thermal biosensor. Bioengineering 2015, 2, 2–14. [Google Scholar] [CrossRef]
- Chen, H.; Huang, J.; Fam, D.; Tok, A. Horizontally aligned carbon nanotube based biosensors for protein detection. Bioengineering 2016, 3, 23. [Google Scholar] [CrossRef]
- Wilmer, M.J.; Ng, C.P.; Lanz, H.L.; Vulto, P.; Suter-Dick, L.; Masereeuw, R. Kidney-on-a-chip technology for drug-induced nephrotoxicity screening. Trends Biotechnol. 2016, 34, 156–170. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Islas-Robles, A.; Nicolini, A.M.; Monks, T.J.; Yoon, J.Y. In situ, dual-mode monitoring of organ-on-a-chip with smartphone-based fluorescence microscope. Biosens. Bioelectron. 2016, 86, 697–705. [Google Scholar] [CrossRef] [PubMed]
- McConnell, E.R.; McClain, M.A.; Ross, J.; Lefew, W.R.; Shafer, T.J. Evaluation of multi-well microelectrode arrays for neurotoxicity screening using a chemical training set. Neurotoxicology 2012, 33, 1048–1057. [Google Scholar] [CrossRef] [PubMed]
- De Groot, M.W.; Dingemans, M.M.; Rus, K.H.; De Groot, A.; Westerink, R.H. Characterization of calcium responses and electrical activity in differentiating mouse neural progenitor cells in vitro. Toxicol. Sci. 2014, 137, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Wainger, B.J.; Kiskinis, E.; Mellin, C.; Wiskow, O.; Han, S.S.; Sandoe, J.; Perez, N.P.; Williams, L.A.; Lee, S.; Boulting, G.; et al. Intrinsic membrane hyperexcitability of amyotrophic lateral sclerosis patient-derived motor neurons. Cell Rep. 2014, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Muratore, C.R.; Rice, H.C.; Srikanth, P.; Callahan, D.G.; Shin, T.; Benjamin, L.N.; Walsh, D.M.; Selkoe, D.J.; Young-Pearse, T.L. The familial Alzheimer’s disease APPV717I mutation alters APP processing and Tau expression in iPSC-derived neurons. Hum. Mol. Genet. 2014, 23, 3523–3536. [Google Scholar] [CrossRef] [PubMed]
- Sidorov, V.Y.; Samson, P.C.; Sidorova, T.N.; Davidson, J.M.; Lim, C.C.; Wikswo, J.P. I-Wire Heart-on-a-Chip I: Three-dimensional cardiac tissue constructs for physiology and pharmacology. Acta Biomater. 2017, 48, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Marsano, A.; Conficconi, C.; Lemme, M.; Occhetta, P.; Gaudiello, E.; Votta, E.; Cerino, G.; Redaelli, A.; Rasponi, M. Beating heart on a chip: A novel microfluidic platform to generate functional 3D cardiac microtissues. Lab Chip 2016, 16, 599–610. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.L.; Zhao, L.P.; Yang, J.; Wan, X.P.; Hu, N.; Yeh, L.H.; Joo, S.W.; Qian, S.Z. Low-voltage pulsed electric field sterilization on a microfluidic chip. Electroanalysis 2013, 25, 1301–1309. [Google Scholar] [CrossRef]
- Lee, S.A.; No da, Y.; Kang, E.; Ju, J.; Kim, D.S.; Lee, S.H. Spheroid-based three-dimensional liver-on-a-chip to investigate hepatocyte-hepatic stellate cell interactions and flow effects. Lab Chip 2013, 13, 3529–3537. [Google Scholar] [CrossRef] [PubMed]
- Huh, D.; Hamilton, G.A.; Ingber, D.E. From 3D cell culture to organs-on-chips. Trends Cell Biol. 2011, 21, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Zhao, L.; Zhou, E.M.; Xu, J.; Shen, S.; Wang, J. On-chip construction of liver lobule-like microtissue and its application for adverse drug reaction assay. Anal. Chem. 2016, 88, 1719–1727. [Google Scholar] [CrossRef] [PubMed]
- Li, C.Y.; Stevens, K.R.; Schwartz, R.E.; Alejandro, B.S.; Huang, J.H.; Bhatia, S.N. Micropatterned cell-cell interactions enable functional encapsulation of primary hepatocytes in hydrogel microtissues. Tissue Eng. Part A 2014, 20, 2200–2212. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.A.; Underhill, G.H.; Bhatia, S.N. Multiplexed, high-throughput analysis of 3D microtissue suspensions. Integr. Biol. 2010, 2, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Torisawa, Y.S.; Spina, C.S.; Mammoto, T.; Mammoto, A.; Weaver, J.C.; Tat, T.; Collins, J.J.; Ingber, D.E. Bone marrow-on-a-chip replicates hematopoietic niche physiology in vitro. Nat. Methods 2014, 11, 663–669. [Google Scholar] [CrossRef] [PubMed]
- Chiu, L.L.Y.; Chu, Z.; Radisic, M.; Mozafari, M. Tissue engineering. In Reference Module in Materials Science and Materials Engineering; Elsevier: Tehran, Iran, 2017. [Google Scholar]
- Torisawa, Y.S.; Mammoto, T.; Jiang, E.; Jiang, A.; Mammoto, A.; Watters, A.L.; Bahinski, A.; Ingber, D.E. Modeling hematopoiesis and responses to radiation countermeasures in a bone marrow-on-a-chip. Tissue Eng. Part C Methods 2016, 22, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Doryab, A.; Amoabediny, G.; Salehi-Najafabadi, A. Advances in pulmonary therapy and drug development: Lung tissue engineering to lung-on-a-chip. Biotechnol. Adv. 2016, 34, 588–596. [Google Scholar] [CrossRef] [PubMed]
- Schroer, A.K.; Shotwell, M.S.; Sidorov, V.Y.; Wikswo, J.P.; Merryman, W.D. I-Wire heart-on-a-Chip II: Biomechanical analysis of contractile, three-dimensional cardiomyocyte tissue constructs. Acta Biomater. 2017, 48, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Hamid, Q.; Wang, C.Y.; Zhao, Y.; Snyder, J.; Sun, W. Fabrication of biological microfluidics using a digital microfabrication system. J. Manuf. Sci. Eng.Trans. ASME 2014, 136, 061001. [Google Scholar] [CrossRef]
- Wadsworth, M.H.; Hughes, T.K.; Shalek, A.K. Marrying microfluidics and microwells for parallel, high-throughput single-cell genomics. Genome Biol. 2015, 16, 129. [Google Scholar] [CrossRef] [PubMed]
- Kotz, K.T.; Xiao, W.; Miller-Graziano, C.; Qian, W.J.; Russom, A.; Warner, E.A.; Moldawer, L.L.; De, A.; Bankey, P.E.; Petritis, B.O.; et al. Clinical microfluidics for neutrophil genomics and proteomics. Nat. Med. 2010, 16, 1042–1142. [Google Scholar] [CrossRef] [PubMed]
- Han, X.L.; Yang, K.; Gross, R.W. Microfluidics-based electrospray ionization enhances the intrasource separation of lipid classes and extends identification of individual molecular species through multi-dimensional mass spectrometry: Development of an automated high-throughput platform for shotgun lipidomics. Rapid Commun. Mass Spectrom. 2008, 22, 2115–2124. [Google Scholar] [PubMed]
- Kim, H.J.; Lee, J.; Choi, J.H.; Bahinski, A.; Ingber, D.E. Co-culture of living microbiome with microengineered human intestinal villi in a gut-on-a-chip microfluidic device. JoVE J. Vis. Exp. 2016, 114. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Li, H.; Collins, J.J.; Ingber, D.E. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc. Natl. Acad. Sci. USA 2016, 113, E7–E15. [Google Scholar] [CrossRef] [PubMed]
- Mertz, L. Omics tech, gut-on-a-chip, and bacterial engineering new approaches for treating inflammatory bowel diseases. IEEE Pulse 2016, 7, 9–12. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Hamon, M.; Jambovane, S. Microfluidics for antibiotic susceptibility and toxicity testing. Bioengineering 2016, 3, 25. [Google Scholar] [CrossRef]
- Oleaga, C.; Bernabini, C.; Smith, A.S.T.; Srinivasan, B.; Jackson, M.; McLamb, W.; Platt, V.; Bridges, R.; Cai, Y.Q.; Santhanam, N.; et al. Multi-organ toxicity demonstration in a functional human in vitro system composed of four organs. Sci. Rep. UK 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Sung, J.H.; Shuler, M.L. Prevention of air bubble formation in a microfluidic perfusion cell culture system using a microscale bubble trap. Biomed. Microdevices 2009, 11, 731–738. [Google Scholar] [CrossRef] [PubMed]
- Feinberg, A.W.; Ripplinger, C.M.; van der Meer, P.; Sheehy, S.P.; Domian, I.; Chien, K.R.; Parker, K.K. Functional differences in engineered myocardium from embryonic stem cell-derived versus neonatal cardiomyocytes. Stem. Cell Rep. 2013, 1, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Kroon, E.; Martinson, L.A.; Kadoya, K.; Bang, A.G.; Kelly, O.G.; Eliazer, S.; Young, H.; Richardson, M.; Smart, N.G.; Cunningham, J.; et al. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat. Biotechnol. 2008, 26, 443–452. [Google Scholar] [CrossRef] [PubMed]
- Spater, D.; Abramczuk, M.K.; Buac, K.; Zangi, L.; Stachel, M.W.; Clarke, J.; Sahara, M.; Ludwig, A.; Chien, K.R. A HCN4+ cardiomyogenic progenitor derived from the first heart field and human pluripotent stem cells. Nat. Cell Biol. 2013, 15, 1098–1106. [Google Scholar] [CrossRef] [PubMed]
- van Laake, L.W.; Qian, L.; Cheng, P.; Huang, Y.; Hsiao, E.C.; Conklin, B.R.; Srivastava, D. Reporter-based isolation of induced pluripotent stem cell-and embryonic stem cell-derived cardiac progenitors reveals limited gene expression variance. Circ. Res. 2010, 107, 340–347. [Google Scholar] [CrossRef] [PubMed]
- Shan, J.; Schwartz, R.E.; Ross, N.T.; Logan, D.J.; Thomas, D.; Duncan, S.A.; North, T.E.; Goessling, W.; Carpenter, A.E.; Bhatia, S.N. Identification of small molecules for human hepatocyte expansion and iPS differentiation. Nat. Chem. Biol. 2013, 9, 514–520. [Google Scholar] [CrossRef] [PubMed]
- Domansky, K.; Leslie, D.C.; McKinney, J.; Fraser, J.P.; Sliz, J.D.; Hamkins-Indik, T.; Hamilton, G.A.; Bahinski, A.; Ingber, D.E. Clear castable polyurethane elastomer for fabrication of microfluidic devices. Lab Chip 2013, 13, 3956–3964. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, B.; Kolli, A.R.; Esch, M.B.; Abaci, H.E.; Shuler, M.L.; Hickman, J.J. TEER measurement techniques for in vitro barrier model systems. J. Lab. Autom. 2015, 20, 107–126. [Google Scholar] [CrossRef] [PubMed]
- Wikswo, J.P.; Block, F.E., 3rd; Cliffel, D.E.; Goodwin, C.R.; Marasco, C.C.; Markov, D.A.; McLean, D.L.; McLean, J.A.; McKenzie, J.R.; Reiserer, R.S.; et al. Engineering challenges for instrumenting and controlling integrated organ-on-chip systems. IEEE Trans. Biomed. Eng. 2013, 60, 682–690. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aziz, A.U.R.; Geng, C.; Fu, M.; Yu, X.; Qin, K.; Liu, B. The Role of Microfluidics for Organ on Chip Simulations. Bioengineering 2017, 4, 39. https://doi.org/10.3390/bioengineering4020039
Aziz AUR, Geng C, Fu M, Yu X, Qin K, Liu B. The Role of Microfluidics for Organ on Chip Simulations. Bioengineering. 2017; 4(2):39. https://doi.org/10.3390/bioengineering4020039
Chicago/Turabian StyleAziz, Aziz Ur Rehman, Chunyang Geng, Mengjie Fu, Xiaohui Yu, Kairong Qin, and Bo Liu. 2017. "The Role of Microfluidics for Organ on Chip Simulations" Bioengineering 4, no. 2: 39. https://doi.org/10.3390/bioengineering4020039




