The Popeye Domain Containing Genes and Their Function in Striated Muscle
Abstract
:1. Introduction
2. Structural Elements of the Popeye Domain Containing Proteins
3. Evolution of the Popeye Domain Containing Proteins
4. The POPDC Gene Family Encodes Membrane Proteins Predominantly Expressed in Skeletal Muscle and Heart
5. Muscle Regeneration Is Retarded in Popdc1 Mouse Mutants
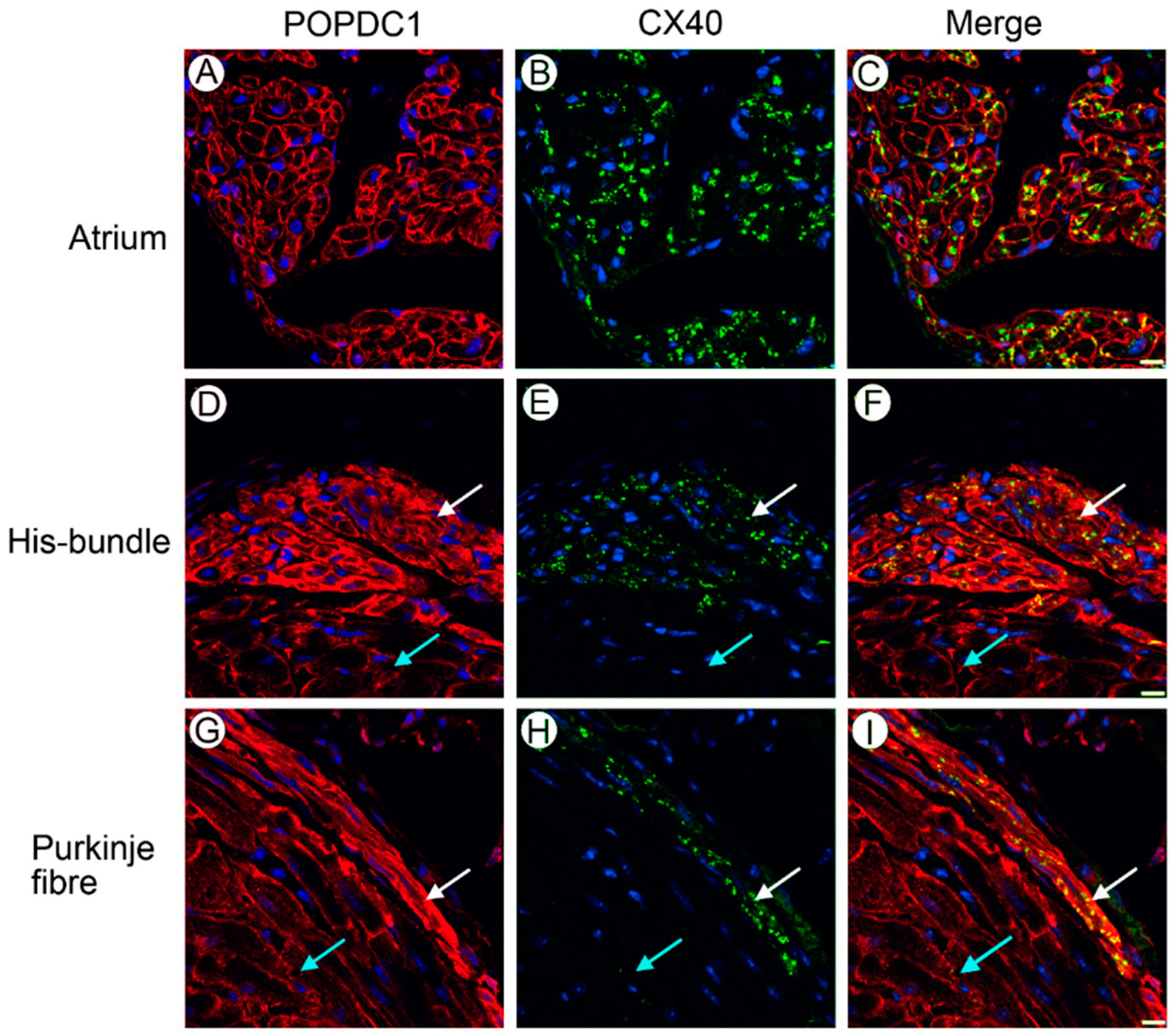
6. Mouse Popdc1 and Popdc2 Mutants Develop a Stress-Induced Sinus Node Bradycardia
7. Popdc2 Depletion in Zebrafish Is Associated with Atrioventricular Block and Muscular Dystrophy
8. A POPDC1S201F Mutation Causes Limb-Girdle Muscular Dystrophy and Cardiac Arrhythmia
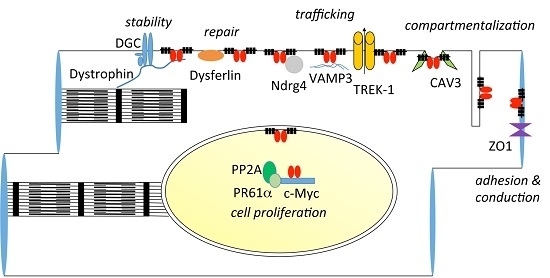
9. Interaction Partners of POPDC Proteins
9.1. Zonula Occludens-1
9.2. VAMP2/3, GEFT and NDRG4
9.3. TREK-1
9.4. Caveolin-3, Dystrophin and Dysferlin
9.5. c-Myc
10. Outlook
Acknowledgments
Conflicts of Interest
Abbreviations
| AKAP | A-kinase anchoring protein |
| ANO5 | anoctamin 5 |
| AV-block | atrioventricular block |
| AVN | atrioventricular node |
| BVES | blood vessel epicardial substance |
| cAMP | cyclic adenosine monophosphate |
| CAP | catabolite activator protein |
| CAV3 | caveolin 3 |
| Cdc42 | cell division control protein 42 homolog |
| CFP | cyan fluorescent protein |
| cGMP | cyclic guanosine monophosphate |
| cNGC | cyclic nucleotide-gated channel |
| cNMP | cyclic nucleotide monophosphate |
| CRIS | cyclic nucleotide receptor involved in sperm function |
| CRP | cAMP receptor protein |
| Cx40/43 | connexin 40/43 |
| DCM | dilated cardiomyopathy |
| DGC | dystrophin glycoprotein complex |
| DMD | Duchenne muscular dystrophy |
| dpf | days post-fertilization |
| DYS | dystrophin |
| DYSF | dysferlin |
| ECG | electrocardiography |
| EDMD | Emery-Dreifuss muscular dystrophy |
| EPAC | exchange protein directly activated by cAMP |
| FRET | Förster resonance energy transfer |
| GLUT4 | glucose transporter type 4 |
| GST | glutathione-S-transferase |
| HF | heart failure |
| IC50 | half maximal inhibitor concentration |
| kDa | kilo Dalton |
| K2P | two-pore domain potassium channel |
| LacZ | ß-galactosidase gene |
| LGMD | limb-girdle muscular dystrophy |
| MTJ | myotendinous junction |
| MyoD | myogenic differentiation gene |
| NCBI | national centre for biotechnology information |
| NDRG4 | N-myc downregulated gene 4 |
| PBC | phosphate binding cassette |
| PFAM | protein families |
| PLA | proximity ligation assay |
| POPDC | Popeye domain containing |
| PKA | protein kinase A |
| PP2A | protein phosphatase 2 |
| Rac1 | Ras-related C3 botulinum toxin substrate 1 |
| RhoA | Ras homolog family member A |
| SAN | sinoatrial node |
| SNARE | soluble N-ethylmaleimide-sensitive- factor attachment receptor |
| TREK-1 | TWIK related K+ channel 1 |
| VAMP2/3 | vesicle associated membrane protein 2/3 |
| Y2H | yeast-two-hybrid |
| YFP | yellow fluorescent protein |
| ZO1 | zona occludens protein 1 |
References
- Andrée, B.; Hillemann, T.; Kessler-Icekson, G.; Schmitt-John, T.; Jockusch, H.; Arnold, H.H.; Brand, T. Isolation and characterization of the novel popeye gene family expressed in skeletal muscle and heart. Dev. Biol. 2000, 223, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.C.; Maurice, D.H.; Baillie, G.S. Targeting protein-protein interactions within the cyclic amp signaling system as a therapeutic strategy for cardiovascular disease. Future Med. Chem. 2013, 5, 451–464. [Google Scholar] [CrossRef] [PubMed]
- Boularan, C.; Gales, C. Cardiac camp: Production, hydrolysis, modulation and detection. Front. Pharmacol. 2015, 6, 203. [Google Scholar] [CrossRef] [PubMed]
- Lefkimmiatis, K.; Zaccolo, M. Camp signaling in subcellular compartments. Pharmacol. Ther. 2014, 143, 295–304. [Google Scholar] [CrossRef] [PubMed]
- McCormick, K.; Baillie, G.S. Compartmentalisation of second messenger signalling pathways. Curr. Opin. Genet. Dev. 2014, 27, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Rehmann, H.; Wittinghofer, A.; Bos, J.L. Capturing cyclic nucleotides in action: Snapshots from crystallographic studies. Nat. Rev. Mol. Cell Biol. 2007, 8, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Krahling, A.M.; Alvarez, L.; Debowski, K.; Van, Q.; Gunkel, M.; Irsen, S.; Al-Amoudi, A.; Strunker, T.; Kremmer, E.; Krause, E.; et al. Cris-a novel camp-binding protein controlling spermiogenesis and the development of flagellar bending. PLoS Genet. 2013, 9, e1003960. [Google Scholar] [CrossRef] [PubMed]
- Schindler, R.F.; Brand, T. The popeye domain containing protein family—A novel class of camp effectors with important functions in multiple tissues. Prog. Biophys. Mol. Biol. 2016, 120, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Kirchmaier, B.C.; Poon, K.L.; Schwerte, T.; Huisken, J.; Winkler, C.; Jungblut, B.; Stainier, D.Y.; Brand, T. The popeye domain containing 2 (popdc2) gene in zebrafish is required for heart and skeletal muscle development. Dev. Biol. 2012, 363, 438–450. [Google Scholar] [CrossRef] [PubMed]
- Froese, A.; Breher, S.S.; Waldeyer, C.; Schindler, R.F.; Nikolaev, V.O.; Rinne, S.; Wischmeyer, E.; Schlueter, J.; Becher, J.; Simrick, S.; et al. Popeye domain containing proteins are essential for stress-mediated modulation of cardiac pacemaking in mice. J. Clin. Investig. 2012, 122, 1119–1130. [Google Scholar] [CrossRef] [PubMed]
- Andrée, B.; Fleige, A.; Arnold, H.H.; Brand, T. Mouse pop1 is required for muscle regeneration in adult skeletal muscle. Mol. Cell. Biol. 2002, 22, 1504–1512. [Google Scholar] [CrossRef] [PubMed]
- Schindler, R.F.; Scotton, C.; Zhang, J.; Passarelli, C.; Ortiz-Bonnin, B.; Simrick, S.; Schwerte, T.; Poon, K.L.; Fang, M.; Rinne, S.; et al. Popdc1s201f causes muscular dystrophy and arrhythmia by affecting protein trafficking. J. Clin. Investig. 2016, 126, 239–253. [Google Scholar] [CrossRef] [PubMed]
- Reese, D.E.; Zavaljevski, M.; Streiff, N.L.; Bader, D. Bves: A novel gene expressed during coronary blood vessel development. Dev. Biol. 1999, 209, 159–171. [Google Scholar] [CrossRef] [PubMed]
- Vasavada, T.K.; DiAngelo, J.R.; Duncan, M.K. Developmental expression of pop1/bves. J. Histochem. Cytochem. 2004, 52, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Knight, R.F.; Bader, D.M.; Backstrom, J.R. Membrane topology of bves/pop1a, a cell adhesion molecule that displays dynamic changes in cellular distribution during development. J. Biol. Chem. 2003, 278, 32872–32879. [Google Scholar] [CrossRef] [PubMed]
- Lundby, A.; Andersen, M.N.; Steffensen, A.B.; Horn, H.; Kelstrup, C.D.; Francavilla, C.; Jensen, L.J.; Schmitt, N.; Thomsen, M.B.; Olsen, J.V. In vivo phosphoproteomics analysis reveals the cardiac targets of beta-adrenergic receptor signaling. Sci. Signal 2013, 6, rs11. [Google Scholar] [CrossRef] [PubMed]
- Kannan, N.; Wu, J.; Anand, G.S.; Yooseph, S.; Neuwald, A.F.; Venter, J.C.; Taylor, S.S. Evolution of allostery in the cyclic nucleotide binding module. Genome Biol. 2007, 8, R264. [Google Scholar] [CrossRef] [PubMed]
- First glance in jmol. Available online: http://bioinformatics.org/firstglance/fgij/ (accessed on 18 February 2016).
- Pfam 29.0. Available online: http://pfam.xfam.org (accessed on 23 February 2016).
- Prosite database of protein domains, proteins, and functional sites. Available online: http://prosite.expasy.org (accessed on 23 February 2016).
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. Weblogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36, W465–W469. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Zhao, D.; Bownes, M. Blood vessel/epicardial substance (bves) expression, essential for embryonic development, is down regulated by grk/efgr signalling. Int. J. Dev. Biol. 2007, 51, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Brand, T. The popeye domain-containing gene family. Cell Biochem. Biophys. 2005, 43, 95–104. [Google Scholar] [CrossRef]
- Tree of the month: Zebrafish popeye-domain-containing proteins and heartbeat regulation. Available online: http://phylomedb.org/?q=node/659 (accessed on 31 March 2016).
- Smith, T.K.; Bader, D.M. Characterization of bves expression during mouse development using newly generated immunoreagents. Dev. Dyn. 2006, 235, 1701–1708. [Google Scholar] [CrossRef] [PubMed]
- Froese, A.; Brand, T. Expression pattern of popdc2 during mouse embryogenesis and in the adult. Dev. Dyn. 2008, 237, 780–787. [Google Scholar] [CrossRef] [PubMed]
- Torlopp, A.; Breher, S.S.; Schluter, J.; Brand, T. Comparative analysis of mrna and protein expression of popdc1 (bves) during early development in the chick embryo. Dev. Dyn. 2006, 235, 691–700. [Google Scholar] [CrossRef] [PubMed]
- Ripley, A.N.; Chang, M.S.; Bader, D.M. Bves is expressed in the epithelial components of the retina, lens, and cornea. Investig. Ophthalmol. Vis. Sci. 2004, 45, 2475–2483. [Google Scholar] [CrossRef] [PubMed]
- Wilkie, G.S.; Korfali, N.; Swanson, S.K.; Malik, P.; Srsen, V.; Batrakou, D.G.; de las Heras, J.; Zuleger, N.; Kerr, A.R.; Florens, L.; et al. Several novel nuclear envelope transmembrane proteins identified in skeletal muscle have cytoskeletal associations. Mol. Cell. Proteom. 2011, 10, M110.003129. [Google Scholar] [CrossRef] [PubMed]
- Korfali, N.; Wilkie, G.S.; Swanson, S.K.; Srsen, V.; de Las Heras, J.; Batrakou, D.G.; Malik, P.; Zuleger, N.; Kerr, A.R.; Florens, L.; et al. The nuclear envelope proteome differs notably between tissues. Nucleus 2012, 3, 552–564. [Google Scholar] [CrossRef] [PubMed]
- Schindler, R.; Simrick, S.; Brand, T. Nuclear localization of members of popeye domain containing (popdc) protein family. Cardiovasc. Res. 2012, 93, S98. [Google Scholar]
- Alcalay, Y.; Hochhauser, E.; Kliminski, V.; Dick, J.; Zahalka, M.A.; Parnes, D.; Schlesinger, H.; Abassi, Z.; Shainberg, A.; Schindler, R.F.; et al. Popeye domain containing 1 (popdc1/bves) is a caveolae-associated protein involved in ischemia tolerance. PLoS ONE 2013, 8, e71100. [Google Scholar] [CrossRef] [PubMed]
- #616812 muscular dystrophy, limb-girdle, type 2x; lgmd2x. Available online: http://www.omim.org (accessed on 20 October 2014).
- Thompson, R.; Straub, V. Limb-girdle muscular dystrophies—International collaborations for translational research. Nat. Rev. Neurol. 2016, 12, 294–309. [Google Scholar] [CrossRef] [PubMed]
- Petri, H.; Sveen, M.L.; Thune, J.J.; Vissing, C.; Dahlqvist, J.R.; Witting, N.; Bundgaard, H.; Kober, L.; Vissing, J. Progression of cardiac involvement in patients with limb-girdle type 2 and becker muscular dystrophies: A 9-year follow-up study. Int. J. Cardiol. 2015, 182, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Margeta, M.; Connolly, A.M.; Winder, T.L.; Pestronk, A.; Moore, S.A. Cardiac pathology exceeds skeletal muscle pathology in two cases of limb-girdle muscular dystrophy type 2i. Muscle Nerve 2009, 40, 883–889. [Google Scholar] [CrossRef]
- Tan, N.; Chung, M.K.; Smith, J.D.; Hsu, J.; Serre, D.; Newton, D.W.; Castel, L.; Soltesz, E.; Pettersson, G.; Gillinov, A.M.; et al. Weighted gene coexpression network analysis of human left atrial tissue identifies gene modules associated with atrial fibrillation. Circ. Cardiovasc. Genet. 2013, 6, 362–371. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Tucker, N.R.; Rizki, G.; Mills, R.; Krijger, P.H.; de Wit, E.; Subramanian, V.; Bartell, E.; Nguyen, X.X.; Ye, J.; et al. Discovery and validation of sub-threshold genome-wide association study loci using epigenomic signatures. eLife 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Gingold-Belfer, R.; Bergman, M.; Alcalay, Y.; Schlesinger, H.; Aravot, D.; Berman, M.; Salman, H.; Brand, T.; Kessler-Icekson, G. Popeye domain-containing 1 is down-regulated in failing human hearts. Int. J. Mol. Med. 2011, 27, 25–31. [Google Scholar] [PubMed]
- Poon, K.L.; Brand, T. The zebrafish model system in cardiovascular research: A tiny fish with mighty prospects. Glob. Cardiol. Sci. Pract. 2013, 2013, 9–28. [Google Scholar] [CrossRef] [PubMed]
- Osler, M.E.; Chang, M.S.; Bader, D.M. Bves modulates epithelial integrity through an interaction at the tight junction. J. Cell Sci. 2005, 118, 4667–4678. [Google Scholar] [CrossRef] [PubMed]
- Zihni, C.; Balda, M.S.; Matter, K. Signalling at tight junctions during epithelial differentiation and microbial pathogenesis. J. Cell Sci. 2014, 127, 3401–3413. [Google Scholar] [CrossRef] [PubMed]
- Rhett, J.M.; Jourdan, J.; Gourdie, R.G. Connexin 43 connexon to gap junction transition is regulated by zonula occludens-1. Mol. Biol. Cell 2011, 22, 1516–1528. [Google Scholar] [CrossRef] [PubMed]
- Soni, S.; Raaijmakers, A.J.A.; Raaijmakers, L.M.; Damen, J.M.A.; van Stuijvenberg, L.; Vos, M.A.; Heck, A.J.R.; van Veen, T.A.B.; Scholten, A. A proteomics approach to identify new putative cardiac intercalated disk proteins. PLoS ONE 2016, 11, e0152231. [Google Scholar] [CrossRef] [PubMed]
- Lindskog, C.; Linne, J.; Fagerberg, L.; Hallstrom, B.M.; Sundberg, C.J.; Lindholm, M.; Huss, M.; Kampf, C.; Choi, H.; Liem, D.A.; et al. The human cardiac and skeletal muscle proteomes defined by transcriptomics and antibody-based profiling. BMC Genom. 2015, 16, 475. [Google Scholar] [CrossRef] [PubMed]
- Somekawa, S.; Fukuhara, S.; Nakaoka, Y.; Fujita, H.; Saito, Y.; Mochizuki, N. Enhanced functional gap junction neoformation by protein kinase a-dependent and epac-dependent signals downstream of camp in cardiac myocytes. Circ. Res. 2005, 97, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.M.; Lin, S.Z.; Chang, N.C. Both pka and epac pathways mediate N-acetylcysteine-induced connexin43 preservation in rats with myocardial infarction. PLoS ONE 2013, 8, e71878. [Google Scholar] [CrossRef] [PubMed]
- Hager, H.A.; Roberts, R.J.; Cross, E.E.; Proux-Gillardeaux, V.; Bader, D.M. Identification of a novel bves function: Regulation of vesicular transport. EMBO J. 2010, 29, 532–545. [Google Scholar] [CrossRef] [PubMed]
- Benesh, E.C.; Miller, P.M.; Pfaltzgraff, E.R.; Grega-Larson, N.E.; Hager, H.A.; Sung, B.H.; Qu, X.; Baldwin, H.S.; Weaver, A.M.; Bader, D.M. Bves and ndrg4 regulate directional epicardial cell migration through autocrine extracellular matrix deposition. Mol. Biol. Cell 2013, 24, 3496–3510. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.K.; Hager, H.A.; Francis, R.; Kilkenny, D.M.; Lo, C.W.; Bader, D.M. Bves directly interacts with geft, and controls cell shape and movement through regulation of rac1/cdc42 activity. Proc. Natl. Acad. Sci. USA 2008, 105, 8298–8303. [Google Scholar] [CrossRef] [PubMed]
- Sudhof, T.C. Neurotransmitter release: The last millisecond in the life of a synaptic vesicle. Neuron 2013, 80, 675–690. [Google Scholar] [CrossRef] [PubMed]
- Schwenk, R.W.; Angin, Y.; Steinbusch, L.K.; Dirkx, E.; Hoebers, N.; Coumans, W.A.; Bonen, A.; Broers, J.L.; van Eys, G.J.; Glatz, J.F.; et al. Overexpression of vesicle-associated membrane protein (vamp) 3, but not vamp2, protects glucose transporter (glut) 4 protein translocation in an in vitro model of cardiac insulin resistance. J. Biol. Chem. 2012, 287, 37530–37539. [Google Scholar] [CrossRef] [PubMed]
- Braiman, L.; Alt, A.; Kuroki, T.; Ohba, M.; Bak, A.; Tennenbaum, T.; Sampson, S.R. Activation of protein kinase c zeta induces serine phosphorylation of vamp2 in the glut4 compartment and increases glucose transport in skeletal muscle. Mol. Cell. Biol. 2001, 21, 7852–7861. [Google Scholar] [CrossRef] [PubMed]
- Tajika, Y.; Sato, M.; Murakami, T.; Takata, K.; Yorifuji, H. Vamp2 is expressed in muscle satellite cells and up-regulated during muscle regeneration. Cell Tissue Res. 2007, 328, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Luftman, K.; Hasan, N.; Day, P.; Hardee, D.; Hu, C. Silencing of vamp3 inhibits cell migration and integrin-mediated adhesion. Biochem. Biophys. Res. Commun. 2009, 380, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Hasan, N.; Hu, C. Vesicle-associated membrane protein 2 mediates trafficking of alpha5beta1 integrin to the plasma membrane. Exp. Cell Res. 2010, 316, 12–23. [Google Scholar] [CrossRef] [PubMed]
- Sztal, T.E.; Sonntag, C.; Hall, T.E.; Currie, P.D. Epistatic dissection of laminin-receptor interactions in dystrophic zebrafish muscle. Hum. Mol. Genet. 2012, 21, 4718–4731. [Google Scholar] [CrossRef] [PubMed]
- Goody, M.F.; Sher, R.B.; Henry, C.A. Hanging on for the ride: Adhesion to the extracellular matrix mediates cellular responses in skeletal muscle morphogenesis and disease. Dev. Biol. 2015, 401, 75–91. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.E.; Bryson-Richardson, R.J.; Berger, S.; Jacoby, A.S.; Cole, N.J.; Hollway, G.E.; Berger, J.; Currie, P.D. The zebrafish candyfloss mutant implicates extracellular matrix adhesion failure in laminin alpha2-deficient congenital muscular dystrophy. Proc. Natl. Acad. Sci. USA 2007, 104, 7092–7097. [Google Scholar] [CrossRef] [PubMed]
- Riggs, K.A.; Hasan, N.; Humphrey, D.; Raleigh, C.; Nevitt, C.; Corbin, D.; Hu, C. Regulation of integrin endocytic recycling and chemotactic cell migration by syntaxin 6 and vamp3 interaction. J. Cell Sci. 2012, 125, 3827–3839. [Google Scholar] [CrossRef] [PubMed]
- Hager, H.A.; Bader, D.M. Bves: Ten years after. Histol. Histopathol. 2009, 24, 777–787. [Google Scholar] [PubMed]
- Bryan, B.A.; Mitchell, D.C.; Zhao, L.; Ma, W.; Stafford, L.J.; Teng, B.B.; Liu, M. Modulation of muscle regeneration, myogenesis, and adipogenesis by the rho family guanine nucleotide exchange factor geft. Mol. Cell. Biol. 2005, 25, 11089–11101. [Google Scholar] [CrossRef] [PubMed]
- Bryan, B.A.; Cai, Y.; Liu, M. The rho-family guanine nucleotide exchange factor geft enhances retinoic acid- and camp-induced neurite outgrowth. J. Neurosci. Res. 2006, 83, 1151–1159. [Google Scholar] [CrossRef] [PubMed]
- Honore, E. The neuronal background k2p channels: Focus on trek1. Nat. Rev. Neurosci. 2007, 8, 251–261. [Google Scholar] [CrossRef] [PubMed]
- Hund, T.J.; Snyder, J.S.; Wu, X.; Glynn, P.; Koval, O.M.; Onal, B.; Leymaster, N.D.; Unudurthi, S.D.; Curran, J.; Camardo, C.; et al. Betaiv-spectrin regulates trek-1 membrane targeting in the heart. Cardiovasc. Res. 2014, 102, 166–175. [Google Scholar] [CrossRef] [PubMed]
- Cunha, S.R.; Mohler, P.J. Ankyrin protein networks in membrane formation and stabilization. J. Cell. Mol. Med. 2009, 13, 4364–4376. [Google Scholar] [CrossRef] [PubMed]
- Hund, T.J.; Unudurthi, S.D.; Wu, X.; Qian, L.; Amari, F.; Onal, B.; Li, N.; Makara, M.; Smith, S.; Snyder, J.; et al. The two-pore K+ channel TREK-1 regulates sinoatrial node membrane excitability. J. Am. Heart Assoc. 2016, 20, e002865. [Google Scholar]
- Gazzerro, E.; Sotgia, F.; Bruno, C.; Lisanti, M.P.; Minetti, C. Caveolinopathies: From the biology of caveolin-3 to human diseases. Eur. J. Hum. Genet. 2010, 18, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Lo, H.P.; Nixon, S.J.; Hall, T.E.; Cowling, B.S.; Ferguson, C.; Morgan, G.P.; Schieber, N.L.; Fernandez-Rojo, M.A.; Bastiani, M.; Floetenmeyer, M.; et al. The caveolin-cavin system plays a conserved and critical role in mechanoprotection of skeletal muscle. J. Cell Biol. 2015, 210, 833–849. [Google Scholar] [CrossRef] [PubMed]
- Parton, R.G.; Way, M.; Zorzi, N.; Stang, E. Caveolin-3 associates with developing t-tubules during muscle differentiation. J. Cell Biol. 1997, 136, 137–154. [Google Scholar] [CrossRef] [PubMed]
- Galbiati, F.; Engelman, J.A.; Volonte, D.; Zhang, X.L.; Minetti, C.; Li, M.; Hou, H., Jr.; Kneitz, B.; Edelmann, W.; Lisanti, M.P. Caveolin-3 null mice show a loss of caveolae, changes in the microdomain distribution of the dystrophin-glycoprotein complex, and t-tubule abnormalities. J. Biol. Chem. 2001, 276, 21425–21433. [Google Scholar] [CrossRef] [PubMed]
- Woodman, S.E.; Park, D.S.; Cohen, A.W.; Cheung, M.W.; Chandra, M.; Shirani, J.; Tang, B.; Jelicks, L.A.; Kitsis, R.N.; Christ, G.J.; et al. Caveolin-3 knock-out mice develop a progressive cardiomyopathy and show hyperactivation of the p42/44 mapk cascade. J. Biol. Chem. 2002, 277, 38988–38997. [Google Scholar] [CrossRef] [PubMed]
- Roostalu, U.; Strahle, U. In vivo imaging of molecular interactions at damaged sarcolemma. Dev. Cell 2012, 22, 515–529. [Google Scholar] [CrossRef] [PubMed]
- Magri, F.; del Bo, R.; D’Angelo, M.G.; Sciacco, M.; Gandossini, S.; Govoni, A.; Napoli, L.; Ciscato, P.; Fortunato, F.; Brighina, E.; et al. Frequency and characterisation of anoctamin 5 mutations in a cohort of italian limb-girdle muscular dystrophy patients. Neuromuscul. Disord. 2012, 22, 934–943. [Google Scholar] [CrossRef] [PubMed]
- Kubisch, C.; Schoser, B.G.; von During, M.; Betz, R.C.; Goebel, H.H.; Zahn, S.; Ehrbrecht, A.; Aasly, J.; Schroers, A.; Popovic, N.; et al. Homozygous mutations in caveolin-3 cause a severe form of rippling muscle disease. Ann. Neurol. 2003, 53, 512–520. [Google Scholar] [CrossRef] [PubMed]
- Bansal, D.; Miyake, K.; Vogel, S.S.; Groh, S.; Chen, C.C.; Williamson, R.; McNeil, P.L.; Campbell, K.P. Defective membrane repair in dysferlin-deficient muscular dystrophy. Nature 2003, 423, 168–172. [Google Scholar] [CrossRef] [PubMed]
- Weiler, T.; Bashir, R.; Anderson, L.V.; Davison, K.; Moss, J.A.; Britton, S.; Nylen, E.; Keers, S.; Vafiadaki, E.; Greenberg, C.R.; et al. Identical mutation in patients with limb girdle muscular dystrophy type 2b or miyoshi myopathy suggests a role for modifier gene(s). Hum. Mol. Genet. 1999, 8, 871–877. [Google Scholar] [CrossRef] [PubMed]
- Han, R.; Bansal, D.; Miyake, K.; Muniz, V.P.; Weiss, R.M.; McNeil, P.L.; Campbell, K.P. Dysferlin-mediated membrane repair protects the heart from stress-induced left ventricular injury. J. Clin. Investig. 2007, 117, 1805–1813. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.Q.; McNally, E.M. The dystrophin complex: Structure, function, and implications for therapy. Compr. Physiol. 2015, 5, 1223–1239. [Google Scholar] [PubMed]
- Falzarano, M.S.; Scotton, C.; Passarelli, C.; Ferlini, A. Duchenne muscular dystrophy: From diagnosis to therapy. Molecules 2015, 20, 18168–18184. [Google Scholar] [CrossRef] [PubMed]
- Strehle, E.M.; Straub, V. Recent advances in the management of duchenne muscular dystrophy. Arch. Dis. Child. 2015, 100, 1173–1177. [Google Scholar] [CrossRef] [PubMed]
- Parang, B.; Kaz, A.; Barett, C.; Short, S.; Ning, W.; Keating, C.; Mittal, M.; Naik, R.; Washington, M.; Revetta, F.; et al. Bves regulates c-myc stability via pp2a and suppresses 1 colitis-induced tumorigenesis. Gut 2016. [Google Scholar] [CrossRef]
- Miner, J.H.; Wold, B.J. C-myc inhibition of myod and myogenin-initiated myogenic differentiation. Mol. Cell. Biol. 1991, 11, 2842–2851. [Google Scholar] [CrossRef] [PubMed]
- Jackson, T.; Allard, M.F.; Sreenan, C.M.; Doss, L.K.; Bishop, S.P.; Swain, J.L. The c-myc proto-oncogene regulates cardiac development in transgenic mice. Mol. Cell. Biol. 1990, 10, 3709–3716. [Google Scholar] [CrossRef] [PubMed]
- Komuro, I.; Kurabayashi, M.; Takaku, F.; Yazaki, Y. Expression of cellular oncogenes in the myocardium during the developmental stages and pressure-overloaded hypertrophy of the heart. Circ. Res. 1988, 62, 1075–1079. [Google Scholar] [CrossRef] [PubMed]
- Zhong, W.; Mao, S.; Tobis, S.; Angelis, E.; Jordan, M.C.; Roos, K.P.; Fishbein, M.C.; de Alboran, I.M.; MacLellan, W.R. Hypertrophic growth in cardiac myocytes is mediated by myc through a cyclin d2-dependent pathway. EMBO J. 2006, 25, 3869–3879. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.G.; Chen, Q.; Wolfram, J.A.; Richardson, S.L.; Liner, A.; Siedlak, S.L.; Zhu, X.; Ziats, N.P.; Fujioka, H.; Felsher, D.W.; et al. Cell cycle re-entry and mitochondrial defects in myc-mediated hypertrophic cardiomyopathy and heart failure. PLoS ONE 2009, 4, e7172. [Google Scholar] [CrossRef] [PubMed]
- McConnachie, G.; Langeberg, L.K.; Scott, J.D. Akap signaling complexes: Getting to the heart of the matter. Trends Mol. Med. 2006, 12, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Dodge-Kafka, K.L.; Soughayer, J.; Pare, G.C.; Carlisle Michel, J.J.; Langeberg, L.K.; Kapiloff, M.S.; Scott, J.D. The protein kinase a anchoring protein makap coordinates two integrated camp effector pathways. Nature 2005, 437, 574–578. [Google Scholar] [CrossRef] [PubMed]
- Parnell, E.; Smith, B.O.; Yarwood, S.J. The camp sensors, epac1 and epac2, display distinct subcellular distributions despite sharing a common nuclear pore localisation signal. Cell Signal. 2015, 27, 989–996. [Google Scholar] [CrossRef] [PubMed]
- Kritzer, M.D.; Li, J.; Dodge-Kafka, K.; Kapiloff, M.S. Akaps: The architectural underpinnings of local camp signaling. J. Mol. Cell. Cardiol. 2012, 52, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Kritzer, M.D.; Li, J.; Passariello, C.L.; Gayanilo, M.; Thakur, H.; Dayan, J.; Dodge-Kafka, K.; Kapiloff, M.S. The scaffold protein muscle a-kinase anchoring protein beta orchestrates cardiac myocyte hypertrophic signaling required for the development of heart failure. Circ. Heart Fail. 2014, 7, 663–672. [Google Scholar] [CrossRef] [PubMed]



| Function | Reference |
|---|---|
| POPDC genes are predominantly expressed in striated muscle | [1] |
| Popdc1−/− mice display a reduction in the number and an increase in the size of caveolae | [33] |
| Popdc1−/− mice display an increased vulnerability to ischemia-reperfusion injury | [33] |
| Popdc1 controls membrane trafficking of TREK-1 | [10] |
| Popdc1 or Popdc2 null mutant mice display an age-dependent stress-induced bradycardia phenotype | [10] |
| Depletion of either Popdc1 or Popdc2 in mice results in structural remodelling of SAN tissue | [10] |
| Depletion of popdc1 or popdc2 in zebrafish causes AV-block and muscular dystrophy | [9,12] |
| Popdc1 in zebrafish controls the formation of the myotendinous junction | [12] |
| The missense mutation POPDC1S191F causes cardiac arrhythmia and muscular dystrophy in patients | [12] |
| POPDC1 and POPDC3 are downregulated in human heart failure | [40] |
| POPDC1 has been associated with atrial fibrillation | [38] |
| POPDC1 is a novel genetic determinant of the QT interval and the QRS time | [39] |
| Protein | Evidence | Reference |
|---|---|---|
| TREK-1 | GST-PD, Co-IP, Co-IF, FRET, TEVC | [10] |
| Caveolin-3 | Co-IP, Co-IF | [33] |
| Dystrophin | Co-IP, Co-IF | [12] |
| Dysferlin | Co-IP, Co-IF | [12] |
| VAMP2, VAMP3 | Y2H, GST-PD, Co-IF | [49] |
| GEFT | Y2H, GST-PD, Co-IF | [51] |
| NDRG4 | Y2H, GST-PD, Co-IP, Co-IF | [50] |
| ZO1 | GST-PD, Co-IF, IG-EM | [42] |
| PR61α | Y2H, Co-IP | [83] |
| c-MYC | Co-IP, PLA | [83] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schindler, R.F.R.; Scotton, C.; French, V.; Ferlini, A.; Brand, T. The Popeye Domain Containing Genes and Their Function in Striated Muscle. J. Cardiovasc. Dev. Dis. 2016, 3, 22. https://doi.org/10.3390/jcdd3020022
Schindler RFR, Scotton C, French V, Ferlini A, Brand T. The Popeye Domain Containing Genes and Their Function in Striated Muscle. Journal of Cardiovascular Development and Disease. 2016; 3(2):22. https://doi.org/10.3390/jcdd3020022
Chicago/Turabian StyleSchindler, Roland F. R., Chiara Scotton, Vanessa French, Alessandra Ferlini, and Thomas Brand. 2016. "The Popeye Domain Containing Genes and Their Function in Striated Muscle" Journal of Cardiovascular Development and Disease 3, no. 2: 22. https://doi.org/10.3390/jcdd3020022