The Controlled Release of Abscisic Acid (ABA) Utilizing Alginate–Chitosan Gel Blends: A Synergistic Approach for an Enhanced Small-Molecule Delivery Controller
Abstract
1. Introduction
2. Results and Discussion
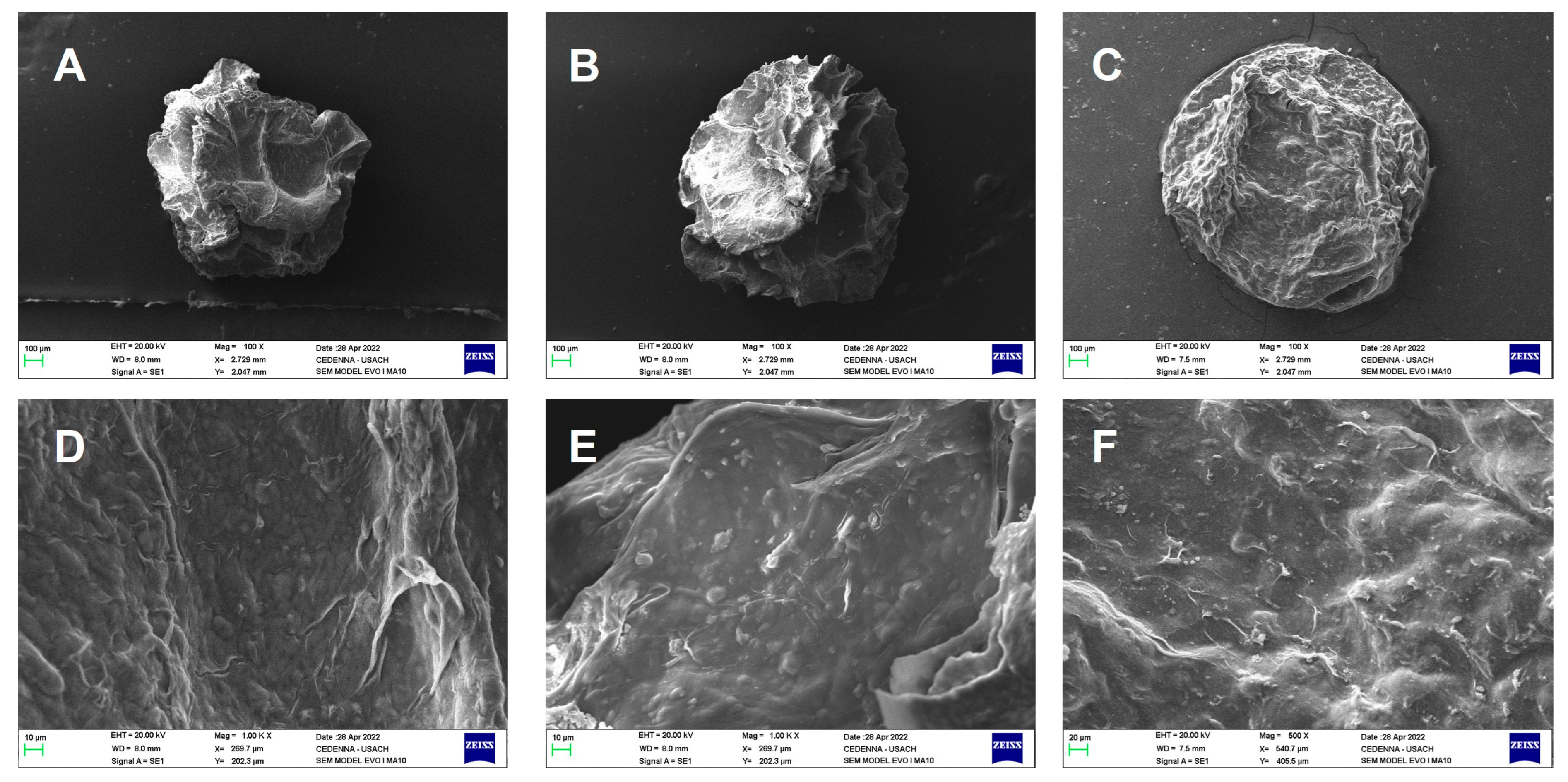
2.1. Morphological Analysis by Scanning Electron Microscopy (SEM)
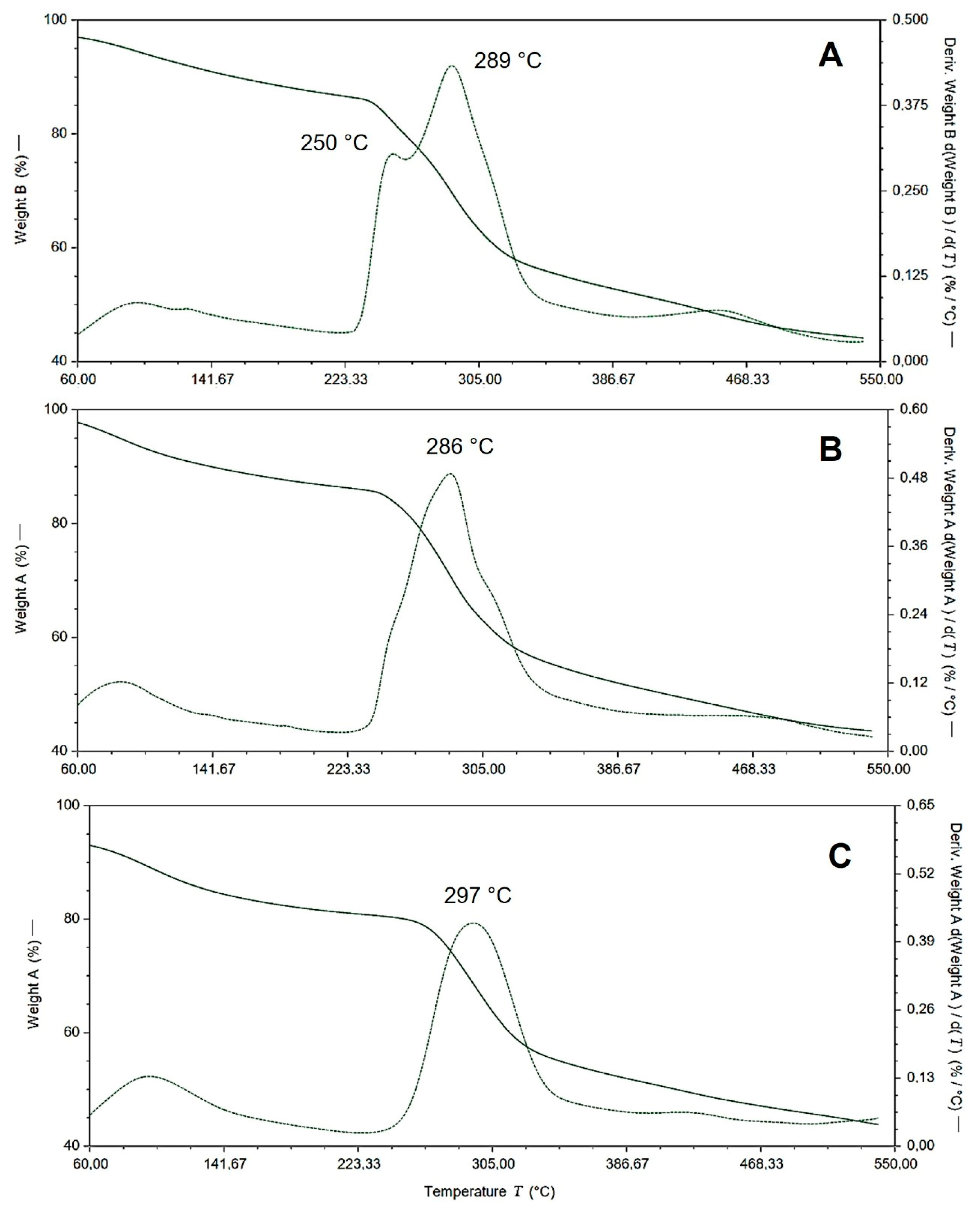
2.2. Thermogravimetric Analysis
2.3. Characterization of Complex by ATR-FTIR Spectroscopy
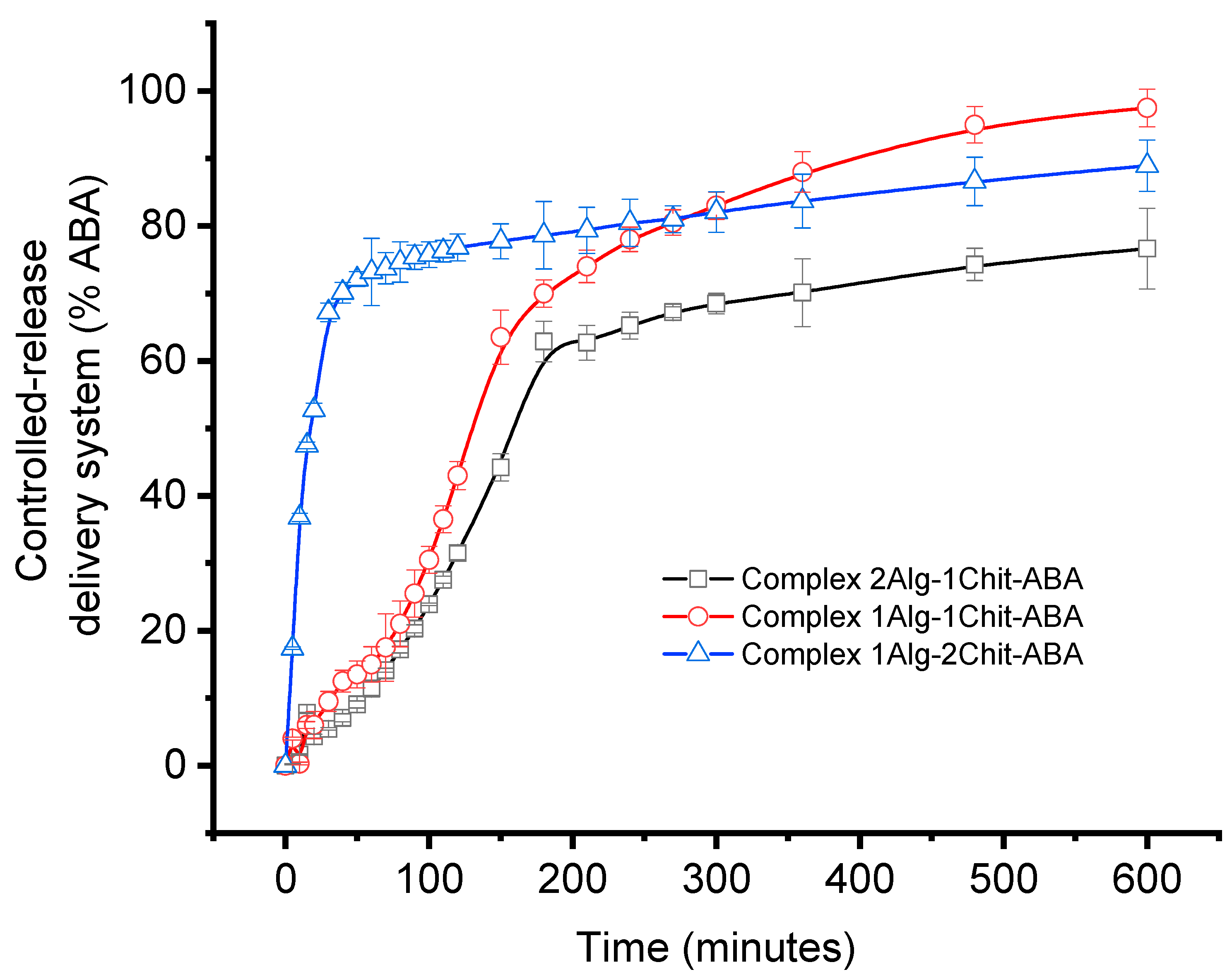
2.4. In Vitro Release Studies
2.5. Kinetic Swelling Study
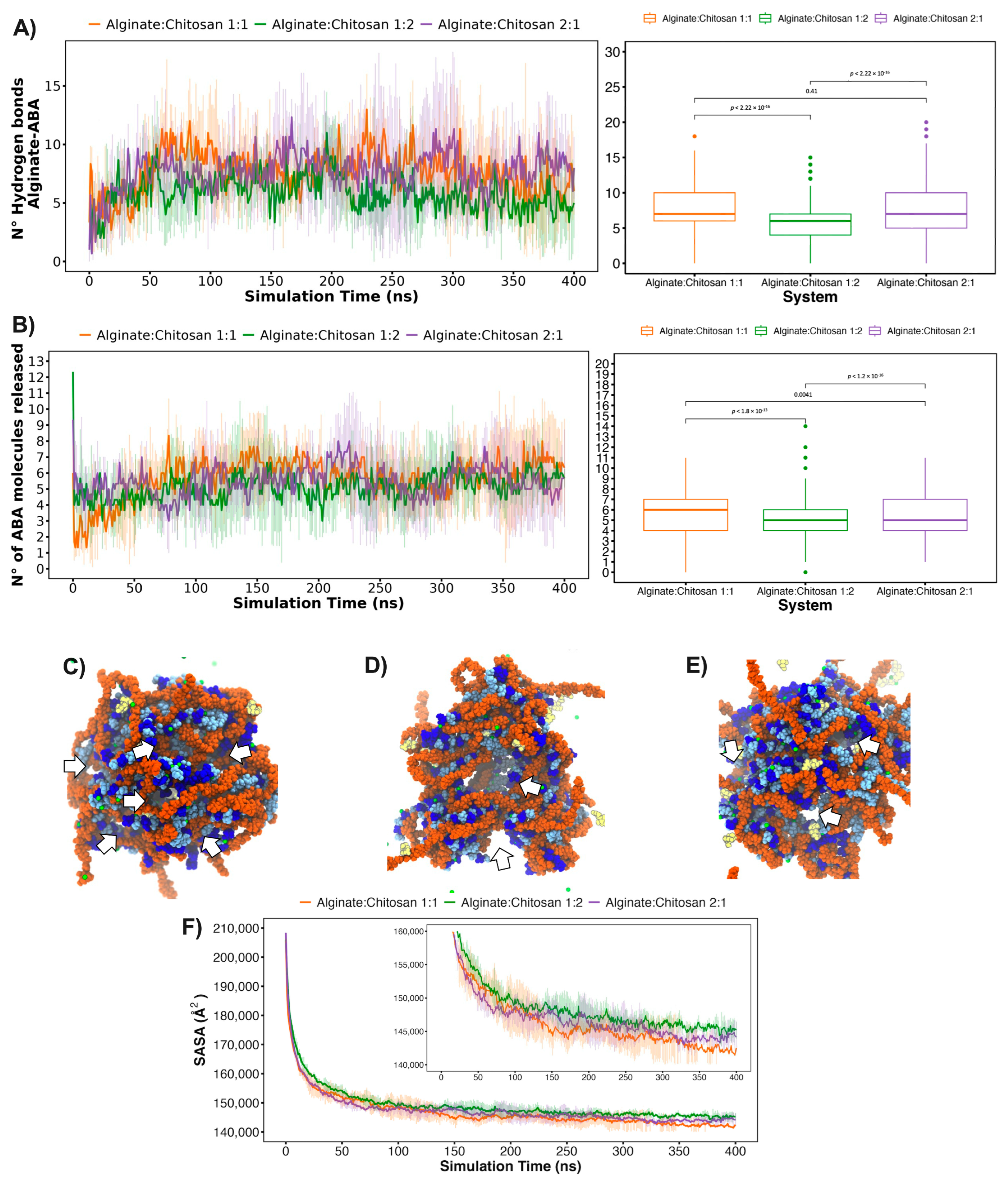
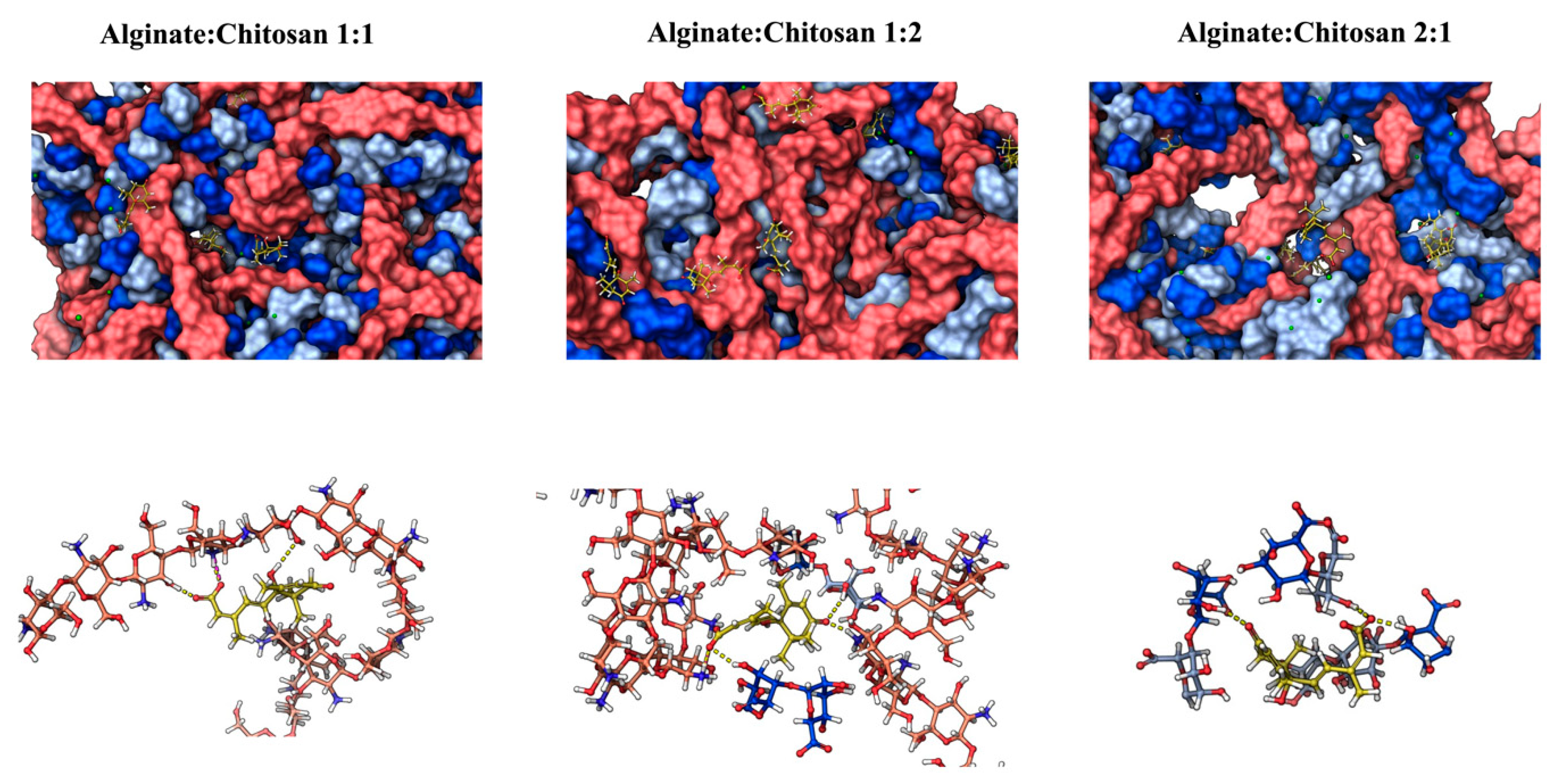
2.6. In Silico ABA Delivery Evaluations
3. Conclusions
4. Material and Methods
4.1. Materials to Form the Molecular Blend
4.2. Gel Blend Preparation
4.3. Scanning Electron Microscopy (SEM)
4.4. Thermogravimetric Analysis (TGA)
4.5. Attenuated Total Reflection–Fourier-Transform Infrared Spectroscopy (ATR-FTIR Spectroscopy)
4.6. Quantification of Abscisic Acid (ABA)
4.7. Sustained Release of Abscisic Acid (ABA) and Release Kinetics Study
4.8. Computational Assays
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bustos, D.; Guzmán, L.; Valdés, O.; Muñoz-Vera, M.; Morales-Quintana, L.; Castro, R.I. Development and Evaluation of Cross-Linked Alginate–Chitosan–Abscisic Acid Blend Gel. Polymers 2023, 15, 3217. [Google Scholar] [CrossRef]
- Wang, J.J.; Zeng, Z.W.; Xiao, R.Z.; Xie, T.; Zhou, G.L.; Zhan, X.R.; Wang, S.L. Recent Advances of Chitosan Nanoparticles as Drug Carriers. Int. J. Nanomedicine 2011, 6, 765–774. [Google Scholar]
- Yuliana, M.; Ismadji, S.; Lie, J.; Santoso, S.P.; Soetaredjo, F.E.; Waworuntu, G.; Putro, J.N.; Wijaya, C.J. Low-Cost Structured Alginate-Immobilized Bentonite Beads Designed for an Effective Removal of Persistent Antibiotics from Aqueous Solution. Environ. Res. 2022, 207, 112162. [Google Scholar] [CrossRef]
- Hesan, M.; Gholipour-Kanani, A.; Lotfi, M.; Shafiee, M. The Synthesis and Characterization of Core-Shell Nanogels Based on Alginate and Chitosan for the Controlled Delivery of Mupirocin. Biochem. Eng. J. 2023, 190, 108742. [Google Scholar] [CrossRef]
- Khalil, H.P.S.A.; Yahya, E.B.; Jummaat, F.; Adnan, A.S.; Olaiya, N.G.; Rizal, S.; Abdullah, C.K.; Pasquini, D.; Thomas, S. Biopolymers Based Aerogels: A Review on Revolutionary Solutions for Smart Therapeutics Delivery. Prog. Mater. Sci. 2023, 131, 101014. [Google Scholar] [CrossRef]
- Fazal, T.; Murtaza, B.N.; Shah, M.; Iqbal, S.; Rehman, M.; Jaber, F.; Dera, A.A.; Awwad, N.S.; Ibrahium, H.A. Recent Developments in Natural Biopolymer Based Drug Delivery Systems. RSC Adv. 2023, 13, 23087–23121. [Google Scholar] [CrossRef]
- Ilić-Stojanović, S.; Nikolić, L.; Cakić, S. A Review of Patents and Innovative Biopolymer-Based Hydrogels. Gels 2023, 9, 556. [Google Scholar] [CrossRef]
- Albrecht, M.T.; Schiller, N.L. Alginate Lyase (AlgL) Activity Is Required for Alginate Biosynthesis in Pseudomonas Aeruginosa. J. Bacteriol. 2005, 187, 3869–3872. [Google Scholar] [CrossRef]
- Chen, J.; Mao, L.; Lu, W.; Ying, T.; Luo, Z. Transcriptome Profiling of Postharvest Strawberry Fruit in Response to Exogenous Auxin and Abscisic Acid. Planta 2016, 243, 183–197. [Google Scholar] [CrossRef]
- Castro, R.I.; Morales-Quintana, L.; Alvarado, N.; Guzmán, L.; Forero-Doria, O.; Valenzuela-Riffo, F.; Laurie, V.F. Design and Optimization of a Self-Assembling Complex Based on Microencapsulated Calcium Alginate and Glutathione (CAG) Using Response Surface Methodology. Polymers 2021, 13, 2080. [Google Scholar] [CrossRef]
- Jia, H.-F.; Chai, Y.-M.; Li, C.-L.; Lu, D.; Luo, J.-J.; Qin, L.; Shen, Y.-Y. Abscisic Acid Plays an Important Role in the Regulation of Strawberry Fruit Ripening. Plant Physiol. 2011, 157, 188–199. [Google Scholar] [CrossRef]
- Symons, G.M.; Chua, Y.-J.; Ross, J.J.; Quittenden, L.J.; Davies, N.W.; Reid, J.B. Hormonal Changes during Non-Climacteric Ripening in Strawberry. J. Exp. Bot. 2012, 63, 4741–4750. [Google Scholar] [CrossRef]
- Cook, M.T.; Tzortzis, G.; Khutoryanskiy, V.V.; Charalampopoulos, D. Layer-by-Layer Coating of Alginate Matrices with Chitosan–Alginate for the Improved Survival and Targeted Delivery of Probiotic Bacteria after Oral Administration. J. Mater. Chem. B 2013, 1, 52–60. [Google Scholar] [CrossRef]
- Yáñez, O.; Alegría-Arcos, M.; Suardiaz, R.; Morales-Quintana, L.; Castro, R.I.; Palma-Olate, J.; Galarza, C.; Catagua-González, Á.; Rojas-Pérez, V.; Urra, G. Calcium-Alginate-Chitosan Nanoparticle as a Potential Solution for Pesticide Removal, a Computational Approach. Polymers 2023, 15, 3020. [Google Scholar] [CrossRef]
- Eltaweil, A.S.; Abd El-Monaem, E.M.; Elshishini, H.M.; El-Aqapa, H.G.; Hosny, M.; Abdelfatah, A.M.; Ahmed, M.S.; Hammad, E.N.; El-Subruiti, G.M.; Fawzy, M. Recent Developments in Alginate-Based Adsorbents for Removing Phosphate Ions from Wastewater: A Review. RSC Adv. 2022, 12, 8228–8248. [Google Scholar] [CrossRef]
- Ilyas, R.A.; Aisyah, H.A.; Nordin, A.H.; Ngadi, N.; Zuhri, M.Y.M.; Asyraf, M.R.M.; Sapuan, S.M.; Zainudin, E.S.; Sharma, S.; Abral, H. Natural-Fiber-Reinforced Chitosan, Chitosan Blends and Their Nanocomposites for Various Advanced Applications. Polymers 2022, 14, 874. [Google Scholar] [CrossRef]
- In Lee, S.; Choi, S.H.; Lee, E.Y.; Kim, H.S. Molecular Cloning, Purification, and Characterization of a Novel PolyMG-Specific Alginate Lyase Responsible for Alginate MG Block Degradation in Stenotrophomas Maltophilia KJ-2. Appl. Microbiol. Biotechnol. 2012, 95, 1643–1653. [Google Scholar] [CrossRef]
- Lillo, L.E.; Matsuhiro, B. Chemical Modifications of 1→ 4-2-Amino-2-Deoxy-α-d-Galactan. Carbohydr. Polym. 2003, 51, 317–325. [Google Scholar] [CrossRef]
- Wang, L.; Gang, X.; Xiao, Y.; Ren, Y.; Wang, J.; Niu, B.; Li, W. Sodium Alginate/Carboxymethyl Chitosan-CuO Hydrogel Beads as a PH-Sensitive Carrier for the Controlled Release of Curcumin. Eur. Polym. J. 2023, 192, 112069. [Google Scholar] [CrossRef]
- Tan, J.; Luo, Y.; Guo, Y.; Zhou, Y.; Liao, X.; Li, D.; Lai, X.; Liu, Y. Development of Alginate-Based Hydrogels: Crosslinking Strategies and Biomedical Applications. Int. J. Biol. Macromol. 2023, 239, 124275. [Google Scholar] [CrossRef]
- Lai, H.L.; Abu’Khalil, A.; Craig, D.Q.M. The Preparation and Characterisation of Drug-Loaded Alginate and Chitosan Sponges. Int. J. Pharm. 2003, 251, 175–181. [Google Scholar] [CrossRef]
- Takka, S.; Gürel, A. Evaluation of Chitosan/Alginate Beads Using Experimental Design: Formulation and in Vitro Characterization. Aaps Pharmscitech 2010, 11, 460–466. [Google Scholar] [CrossRef]
- Simsek-Ege, F.A.; Bond, G.M.; Stringer, J. Polyelectrolyte Complex Formation between Alginate and Chitosan as a Function of PH. J. Appl. Polym. Sci. 2003, 88, 346–351. [Google Scholar] [CrossRef]
- Teng, K.; An, Q.; Chen, Y.; Zhang, Y.; Zhao, Y. Recent Development of Alginate-Based Materials and Their Versatile Functions in Biomedicine, Flexible Electronics, and Environmental Uses. ACS Biomater. Sci. Eng. 2021, 7, 1302–1337. [Google Scholar] [CrossRef]
- Castro, R.I.; Laurie, V.F.; Padilla, C.; Carrasco-Sánchez, V. Removal of Ochratoxin A from Red Wine Using Alginate-PVA-L. Plantarum (APLP) Complexes: A Preliminary Study. Toxins 2022, 14, 230. [Google Scholar] [CrossRef]
- Kabir, S.F.; Rahman, A.; Yeasmin, F.; Sultana, S.; Masud, R.A.; Kanak, N.A.; Haque, P. Occurrence, Distribution, and Structure of Natural Polysaccharides. In Radiation-Processed Polysaccharides; Elsevier: Amsterdam, The Netherlands, 2022; pp. 1–27. [Google Scholar]
- Fan, L.; Du, Y.; Zhang, B.; Yang, J.; Zhou, J.; Kennedy, J.F. Preparation and Properties of Alginate/Carboxymethyl Chitosan Blend Fibers. Carbohydr. Polym. 2006, 65, 447–452. [Google Scholar] [CrossRef]
- Kulig, D.; Zimoch-Korzycka, A.; Jarmoluk, A.; Marycz, K. Study on Alginate–Chitosan Complex Formed with Different Polymers Ratio. Polymers 2016, 8, 167. [Google Scholar] [CrossRef]
- Donoso, W.; Castro, R.I.; Guzmán, L.; López-Cabaña, Z.; Nachtigall, F.M.; Santos, L.S. Fast Detection of Listeria Monocytogenes through a Nanohybrid Quantum Dot Complex. Anal. Bioanal. Chem. 2017, 409, 5359–5371. [Google Scholar] [CrossRef]
- Lawrie, G.; Keen, I.; Drew, B.; Chandler-Temple, A.; Rintoul, L.; Fredericks, P.; Grøndahl, L. Interactions between Alginate and Chitosan Biopolymers Characterized Using FTIR and XPS. Biomacromolecules 2007, 8, 2533–2541. [Google Scholar] [CrossRef]
- Wittaya-areekul, S.; Kruenate, J.; Prahsarn, C. Preparation and in Vitro Evaluation of Mucoadhesive Properties of Alginate/Chitosan Microparticles Containing Prednisolone. Int. J. Pharm. 2006, 312, 113–118. [Google Scholar] [CrossRef]
- Abbasnezhad, N.; Zirak, N.; Shirinbayan, M.; Kouidri, S.; Salahinejad, E.; Tcharkhtchi, A.; Bakir, F. Controlled Release from Polyurethane Films: Drug Release Mechanisms. J. Appl. Polym. Sci. 2021, 138, 50083. [Google Scholar] [CrossRef]
- Shah, J.C.; Deshpande, A. Kinetic Modeling and Comparison of Invitro Dissolution Profiles. World J. Pharm. Sci. 2014, 2, 302–309. [Google Scholar]
- Peppas, N.A.; Sahlin, J.J. A Simple Equation for the Description of Solute Release. III. Coupling of Diffusion and Relaxation. Int. J. Pharm. 1989, 57, 169–172. [Google Scholar] [CrossRef]
- Bruschi, M.L. Strategies to Modify the Drug Release from Pharmaceutical Systems; Woodhead Publishing: Cambridge, UK; Elsevier: Kidlington, UK, 2015; ISBN 0081001126. [Google Scholar]
- Serra, L.; Doménech, J.; Peppas, N.A. Drug Transport Mechanisms and Release Kinetics from Molecularly Designed Poly (Acrylic Acid-g-Ethylene Glycol) Hydrogels. Biomaterials 2006, 27, 5440–5451. [Google Scholar] [CrossRef]
- Maestro, S. Schrödinger Release 2021-1; LLC: New York, NY, USA, 2020. [Google Scholar]
- Jorgensen, W.L.; Tirado-Rives, J. The OPLS [Optimized Potentials for Liquid Simulations] Potential Functions for Proteins, Energy Minimizations for Crystals of Cyclic Peptides and Crambin. J. Am. Chem. Soc. 1988, 110, 1657–1666. [Google Scholar] [CrossRef]




| Sample | Complex + ABA | ||
|---|---|---|---|
| Alginate/Chitosan Ratio | 2:1 (System 1) | 1:1 (System 2) | 1:2 (System 3) |
| Concentration of ABA in the complex per 0.9 mg of complex | 0.035 ± 0.002 | 0.029 ± 0.003 | 0.012 ± 0.001 |
| Higuchi | |||||
| k | RSS | ||||
| System 1 | 2.6590 ± 0.2900 | 0.6701 | |||
| System 2 | 3.0688 ± 0.3061 | 0.7028 | |||
| System 3 | 11.7861 ± 0.4767 | 0.9699 | |||
| Ritger–Peppas | |||||
| n | k | RSS | |||
| System 1 | −61.8348 ± 8.9384 × 1042 | −22.4446 ± 1.9096 × 1041 | 1.0622 | ||
| System 2 | 1.5041 ± 0.0929 | 0.0321 ± 0.0144 | 0.9753 | ||
| System 3 | 0.6340 ± 0.0642 | 7.9518 ± 1.5493 | 0.9864 | ||
| Lindner–Lippold | |||||
| n | kD | b | RSS | ||
| System 1 | −5.6696 × 10−4 ± 0.3957 | −23,235.3678 ± 1.6186 × 107 | 23,202.3017 ± 1.6183 × 107 | 0.6628 | |
| System 2 | 1.8102 ± 0.1195 | 0.0067 ± 0.0040 | 3.5523 ± 0.9965 | 0.9863 | |
| System 3 | 0.6228 ± 0.0886 | 8.3559 ± 2.6245 | −0.8131 ± 3.6201 | 0.9867 | |
| Peppas–Sahlin | |||||
| n | kD | kR | RSS | ||
| System 1 | 1.0690 ± 0.8637 | 0.1037 ± 0.1977 | 5.2375 × 10−4 ± 0.0058 | 0.9912 | |
| System 2 | 1.0730 ± 1.3880 | 0.1323 ± 0.4052 | 7.0062 × 10−4 ± 0.0124 | 0.9933 | |
| System 3 | 0.9205 ± 0.1342 | 4.8247 ± 1.5454 | −0.0836 ± 0.0481 | 0.9936 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valdes, O.; Bustos, D.; Guzmán, L.; Muñoz-Vera, M.; Urra, G.; Castro, R.I.; Morales-Quintana, L. The Controlled Release of Abscisic Acid (ABA) Utilizing Alginate–Chitosan Gel Blends: A Synergistic Approach for an Enhanced Small-Molecule Delivery Controller. Gels 2024, 10, 185. https://doi.org/10.3390/gels10030185
Valdes O, Bustos D, Guzmán L, Muñoz-Vera M, Urra G, Castro RI, Morales-Quintana L. The Controlled Release of Abscisic Acid (ABA) Utilizing Alginate–Chitosan Gel Blends: A Synergistic Approach for an Enhanced Small-Molecule Delivery Controller. Gels. 2024; 10(3):185. https://doi.org/10.3390/gels10030185
Chicago/Turabian StyleValdes, Oscar, Daniel Bustos, Luis Guzmán, Marcelo Muñoz-Vera, Gabriela Urra, Ricardo I. Castro, and Luis Morales-Quintana. 2024. "The Controlled Release of Abscisic Acid (ABA) Utilizing Alginate–Chitosan Gel Blends: A Synergistic Approach for an Enhanced Small-Molecule Delivery Controller" Gels 10, no. 3: 185. https://doi.org/10.3390/gels10030185
APA StyleValdes, O., Bustos, D., Guzmán, L., Muñoz-Vera, M., Urra, G., Castro, R. I., & Morales-Quintana, L. (2024). The Controlled Release of Abscisic Acid (ABA) Utilizing Alginate–Chitosan Gel Blends: A Synergistic Approach for an Enhanced Small-Molecule Delivery Controller. Gels, 10(3), 185. https://doi.org/10.3390/gels10030185








