Cryogels and Monoliths: Promising Tools for Chromatographic Purification of Nucleic Acids
Abstract
:1. Introduction
2. Monoliths
2.1. Monoliths for Nucleic Acid Purification
2.1.1. Monoliths for DNA Purification
Ion Exchange Chromatography Using Monoliths for DNA Purification
Hydrophobic Interaction Chromatography Using Monoliths for DNA Purification
Affinity Chromatography Using Monoliths for DNA Purification
Other Monolith Supports for DNA Purification
2.1.2. RNA Purification
Ion Exchange Chromatography Using Monoliths for RNA Purification
Other Monolith Supports for RNA Purification
3. Cryogels
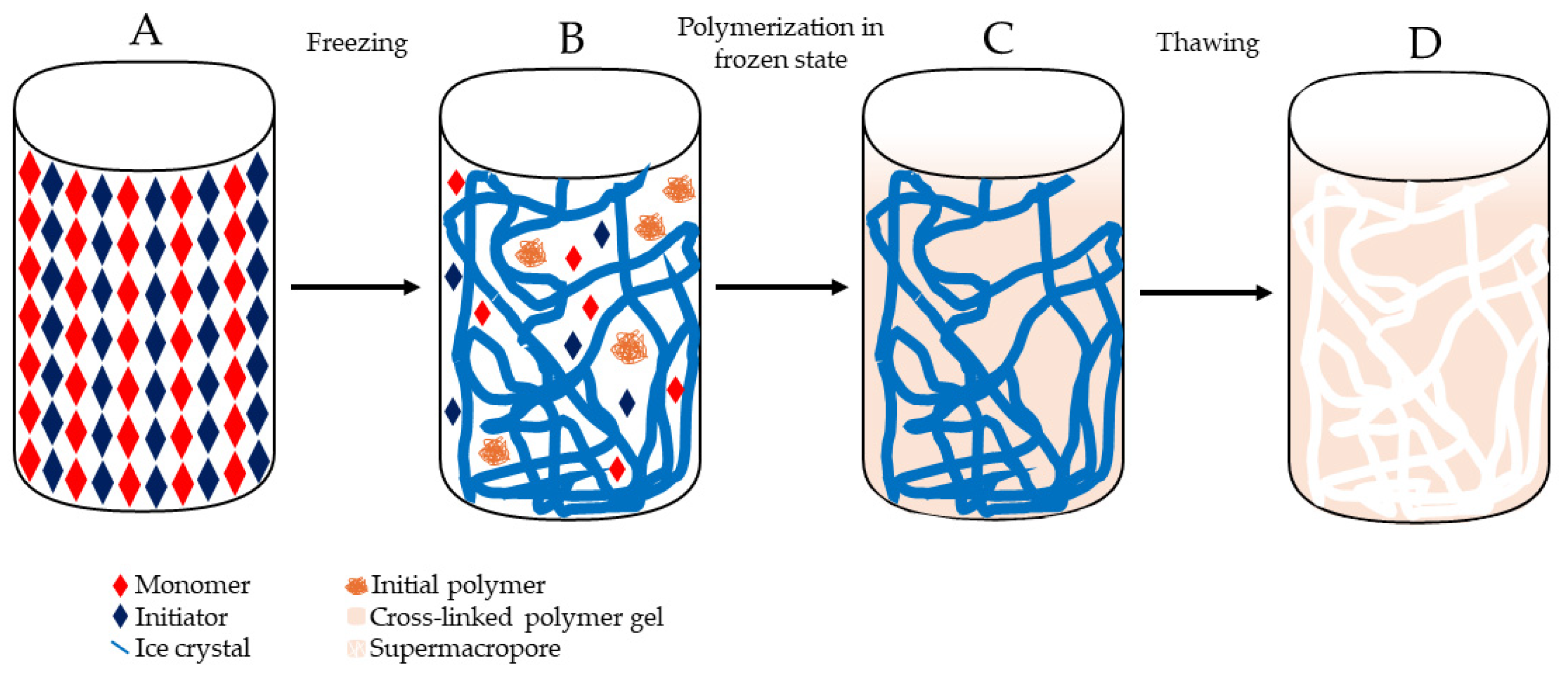
3.1. Preparation of Cryogels
3.2. Cryogels for Nucleic acid Purification
3.2.1. DNA Purification
3.2.2. RNA Purification
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| (NH4)2SO4 | Ammonium Sulfate |
| AC | Affinity chromatography |
| AEX | Anion exchange chromatography |
| APS | Ammonium Persulfate |
| C4 | Butyl |
| CDI | Carbonyldiimidazole |
| CIM® | Convective interaction media |
| DAPP | 3,8-Diamino-6-Phenylphenanthridine |
| DBC | Dynamic binding capacity |
| DEAE | Diethylaminoethyl |
| dsRNA | Double-stranded RNA |
| EDA | Ethylenediamine |
| EDMA | Polyethyleneglycol Dimethacrylate |
| FDA | Food and Drug Administration |
| G4s | Guanine Quadruplexes |
| gDNA | Genomic DNA |
| GMA | Glycidyl Methacrylate |
| HEMA | 2-Hydroxyethyl Methacrylate |
| IEX | Ion exchange chromatography |
| mcDNA | Minicircle DNA |
| mRNA | Messenger RNA |
| NaCl | Sodium Chloride |
| oc | Open circular |
| pDNA | Plasmid DNA |
| PEG | Polyethylene Glycol |
| pHEMA | Poly(2-Hydroxyethyl Methacrylate) |
| PHEMAH | Poly(Hydroxyethyl Methacrylate-N-Methacryloyl-(L)-Histidine Methyl Ester |
| pHEMA-MATrp | Poly(2-hydroxyethyl Methacrylate)-N-methacryloyl-L-tryptophan |
| PP | Parental plasmids |
| QA | Quaternary Ammonium |
| satRNA | Satellite RNA |
| sc | Supercoiled |
| SDC | Sample displacement chromatography |
| ssRNA | Single-stranded |
| TEMED | N,N,N,N-tetramethylethylenediamine |
| TMOS | Tetramethoxysilane |
| UFLMP | Unfrozen liquid microphase |
References
- Ita, K. Coronavirus Disease (COVID-19): Current Status and Prospects for Drug and Vaccine Development. Arch. Med. Res. 2021, 52, 15–24. [Google Scholar] [CrossRef]
- Alhakamy, N.A.; Curiel, D.T.; Berkland, C.J. The era of gene therapy: From preclinical development to clinical application. Drug Discov. Today 2021, 26, 1602–1619. [Google Scholar] [CrossRef]
- Baptista, B.; Carapito, R.; Laroui, N.; Pichon, C.; Sousa, F. mRNA, a Revolution in Biomedicine. Pharmaceutics 2021, 13, 2090. [Google Scholar] [CrossRef]
- Bicho, D.; Queiroz, J.A.; Tomaz, C.T. Influenza Plasmid DNA Vaccines: Progress and Prospects. Curr. Gene Ther. 2015, 15, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Caramelo-Nunes, C.; Gabriel, M.; Almeida, P.; Marcos, J.; Tomaz, C. Purification of plasmid DNA from clarified and non-clarified Escherichia coli lysates by berenil pseudo-affinity chromatography. J. Chromatogr. B 2012, 904, 81–87. [Google Scholar] [CrossRef]
- Bo, H.; Wang, J.; Chen, Q.; Shen, H.; Wu, F.; Shao, H.; Huang, S. Using a single hydrophobic-interaction chromatography to purify pharmaceutical-grade supercoiled plasmid DNA from other isoforms. Pharm. Biol. 2013, 51, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Almeida, A.; Eusébio, D.; Queiroz, J.; Sousa, F.; Sousa, A. The use of size-exclusion chromatography in the isolation of supercoiled minicircle DNA from Escherichia coli lysate. J. Chromatogr. A 2020, 1609, 460444. [Google Scholar] [CrossRef]
- Eon-Duval, A.; Burke, G. Purification of pharmaceutical-grade plasmid DNA by anion-exchange chromatography in an RNase-free process. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2004, 804, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Svec, F. Monolithic columns: A historical overview. Electrophoresis 2017, 38, 2810–2820. [Google Scholar] [CrossRef]
- Valente, J.; Queiroz, J.; Sousa, F. Dilemma on plasmid DNA purification: Binding capacity vs selectivity. J. Chromatogr. A 2021, 1637, 461848. [Google Scholar] [CrossRef]
- Saylan, Y.; Denizli, A. Supermacroporous Composite Cryogels in Biomedical Applications. Gels 2019, 5, 20. [Google Scholar] [CrossRef]
- Hahn, R.; Panzer, M.; Hansen, E.; Mollerup, J.; Jungbauer, A. Mass transfer properties of monoliths. Sep. Sci. Technol. 2002, 37, 1545–1565. [Google Scholar] [CrossRef]
- González-González, M.; Mayolo-Deloisa, K.; Rito-Palomares, M. Recent Advances in Antibody-Based Monolith Chromatography for Therapeutic Applications; Elsevier: Amsterdam, The Netherlands, 2020. [Google Scholar] [CrossRef]
- Hefnawy, M.; El-Gendy, M.; Al-Salem, H.; Marenga, H.; El-Azab, A.; Abdel-Aziz, A.; El Gamal, A.; Alanazi, M.; Obaidullah, A.; Al-Hossaini, A.; et al. Trends in monoliths: Packings, stationary phases and nanoparticles. J. Chromatogr. A 2023, 1691, 463819. [Google Scholar] [CrossRef]
- Feinle, A.; Elsaesser, M.S.; Hüsing, N. Sol–gel synthesis of monolithic materials with hierarchical porosity. Chem. Soc. Rev. 2016, 45, 3377–3399. [Google Scholar] [CrossRef]
- Gama, M.R.; Rocha, F.R.P.; Bottoli, C.B.G. Monoliths: Synthetic routes, functionalization and innovative analytical applications. TrAC Trends Anal. Chem. 2019, 115, 39–51. [Google Scholar] [CrossRef]
- Unger, K.K.; Skudas, R.; Schulte, M.M. Particle packed columns and monolithic columns in high-performance liquid chromatography-comparison and critical appraisal. J. Chromatogr. A 2008, 1184, 393–415. [Google Scholar] [CrossRef]
- Vergara-Barberán, M.; Carrasco-Correa, E.J.; Lerma-García, M.J.; Simó-Alfonso, E.F.; Herrero-Martínez, J.M. Current trends in affinity-based monoliths in microextraction approaches: A review. Anal. Chim. Acta 2019, 1084, 1–20. [Google Scholar] [CrossRef]
- Patel, N.; Gupta, K.R.; Umekar, M.J. An Overview of Monolithic Column: Types, Parameters and Applications. J. Drug Deliv. Ther. 2022, 12, 223–231. [Google Scholar] [CrossRef]
- Lima, I.d.P.; Valle, S.P.; de Oliveira, M.A.L.; Marques, F.F.d.C.; Vaz, F.A.S. Monolithic stationary phases preparation for use in chromatographic and electromigration techniques: The state-of-the-art. Microchem. J. 2023, 190, 108598. [Google Scholar] [CrossRef]
- Lynch, K.B.; Ren, J.; Beckner, M.A.; He, C.; Liu, S. Monolith columns for liquid chromatographic separations of intact proteins: A review of recent advances and applications. Anal. Chim. Acta 2019, 1046, 48–68. [Google Scholar] [CrossRef]
- Müllner, T.; Zankel, A.; Höltzel, A.; Svec, F.; Tallarek, U. Morphological Properties of Methacrylate-Based Polymer Monoliths: From Gel Porosity to Macroscopic Inhomogeneities. Langmuir 2017, 33, 2205–2214. [Google Scholar] [CrossRef]
- Hoegger, D.; Freitag, R. Acrylamide-based monoliths as robust stationary phases for capillary electrochromatography. J. Chromatogr. A 2001, 914, 211–222. [Google Scholar] [CrossRef]
- Zhang, Y.-P.; Li, W.; Wang, X.-J.; Qu, L.-B.; Zhao, G.-L.; Zhang, Y.-X. Fast preparation of polystyrene-based monolith using microwave irradiation for micro-column separation. Anal. Bioanal. Chem. 2009, 394, 617–623. [Google Scholar] [CrossRef]
- Nema, T.; Chan, E.C.; Ho, P.C. Applications of monolithic materials for sample preparation. J. Pharm. Biomed. Anal. 2014, 87, 130–141. [Google Scholar] [CrossRef]
- Sabarudin, A. Organic Polymer Monolith: Synthesis and Applications for bioanalytical. In Seminar Nasional Kimia—National Seminar on Chemistry (SNK 2018); Atlantis Press: Paris, France, 2018; pp. 1–7. [Google Scholar] [CrossRef]
- Cantó-Mirapeix, A.; Herrero-Martínez, J.M.; Mongay-Fernández, C.; Simó-Alfonso, E.F. Chemical initiation for butyl and lauryl acrylate monolithic columns for CEC. Electrophoresis 2009, 30, 599–606. [Google Scholar] [CrossRef]
- Acquah, C.; Moy, C.K.; Danquah, M.K.; Ongkudon, C.M. Development and characteristics of polymer monoliths for advanced LC bioscreening applications: A review. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2016, 1015–1016, 121–134. [Google Scholar] [CrossRef]
- Walsh, Z.; Paull, B.; Macka, M. Inorganic monoliths in separation science: A review. Anal. Chim. Acta 2012, 750, 28–47. [Google Scholar] [CrossRef]
- Randon, J.; Huguet, S.; Piram, A.; Puy, G.; Demesmay, C.; Rocca, J.-L. Synthesis of zirconia monoliths for chromatographic separations. J. Chromatogr. A 2006, 1109, 19–25. [Google Scholar] [CrossRef]
- Tokudome, Y.; Fujita, K.; Nakanishi, K.; Miura, K.; Hirao, K. Synthesis of monolithic Al2O3 with well-defined macropores and mesostructured skeletons via the sol-gel process accompanied by phase separation. Chem. Mater. 2007, 19, 3393–3398. [Google Scholar] [CrossRef]
- Zheng, J.Y.; Pang, J.B.; Qiu, K.Y.; Wei, Y. Synthesis of mesoporous titanium dioxide materials by using a mixture of organic compounds as a non-surfactant template. J. Mater. Chem. 2001, 11, 3367–3372. [Google Scholar] [CrossRef]
- Yu, S.; Geng, J.; Zhou, P.; Wang, J.; Chen, X.; Hu, J. New hydroxyapatite monolithic column for DNA extraction and its application in the purification of Bacillus subtilis crude lysate. J. Chromatogr. A 2008, 1183, 29–37. [Google Scholar] [CrossRef]
- Tanaka, N.; Kobayashi, H.; Ishizuka, N.; Minakuchi, H.; Nakanishi, K.; Hosoya, K.; Ikegami, T. Monolithic silica columns for high-efficiency chromatographic separations. J. Chromatogr. A 2002, 965, 35–49. [Google Scholar] [CrossRef]
- Zhu, T.; Row, K.H. Preparation and applications of hybrid organic-inorganic monoliths: A review. J. Sep. Sci. 2012, 35, 1294–1302. [Google Scholar] [CrossRef]
- Gunji, T.; Ozawa, M.; Abe, Y.; West, R. Preparation of C60-silica hybrid monolith by sol-gel process. J. Solgel. Sci. Technol. 2001, 22, 219–224. [Google Scholar] [CrossRef]
- Zhong, H.; Zhu, G.; Yang, J.; Wang, P.; Yang, Q. Periodic mesoporous hybrid monolith with hierarchical macro-mesopores. Microporous Mesoporous Mater. 2007, 100, 259–267. [Google Scholar] [CrossRef]
- Bai, L.; Liu, H.; Liu, Y.; Zhang, X.; Yang, G.; Ma, Z. Preparation of a novel hybrid organic-inorganic monolith for the separation of lysozyme by high performance liquid chromatography. J. Chromatogr. A 2011, 1218, 100–106. [Google Scholar] [CrossRef]
- Urthaler, J.; Schlegl, R.; Podgornik, A.; Strancar, A.; Jungbauer, A.; Necina, R. Application of monoliths for plasmid DNA purification development and transfer to production. J. Chromatogr. A 2005, 1065, 93–106. [Google Scholar] [CrossRef]
- Forcic, D.; Branovic-Cakanic, K.; Ivancic, J.; Jug, R.; Barut, M.; Strancar, A. Purification of genomic DNA by short monolithic columns. J. Chromatogr. A 2005, 1065, 115–120. [Google Scholar] [CrossRef]
- Ongkudon, C.M.; Danquah, M.K. Anion exchange chromatography of 4.2 kbp plasmid based vaccine (pcDNA3F) from alkaline lysed E. coli lysate using amino functionalised polymethacrylate conical monolith. Sep. Purif. Technol. 2011, 78, 303–310. [Google Scholar] [CrossRef]
- Černigoj, U.; Vidič, J.; Ferjančič, A.; Sinur, U.; Božič, K.; Mencin, N.; Celjar, A.M.; Gagnon, P.; Štrancar, A. Guanidine improves DEAE anion exchange-based analytical separation of plasmid DNA. Electrophoresis 2021, 42, 2619–2625. [Google Scholar] [CrossRef]
- Kazarian, A.A.; Barnhart, W.; Campuzano, I.D.; Cabrera, J.; Fitch, T.; Long, J.; Sham, K.; Wu, B.; Murray, J.K. Purification of guanine-quadruplex using monolithic stationary phase under ion-exchange conditions. J. Chromatogr. A 2020, 1634, 461633. [Google Scholar] [CrossRef]
- Sousa, A.; Tomaz, C.; Sousa, F.; Queiroz, J. Successful application of monolithic innovative technology using a carbonyldiimidazole disk to purify supercoiled plasmid DNA suitable for pharmaceutical applications. J. Chromatogr. A 2011, 1218, 8333–8343. [Google Scholar] [CrossRef]
- Černigoj, U.; Martinuč, U.; Cardoso, S.; Sekirnik, R.; Krajnc, N.L.; Štrancar, A. Sample displacement chromatography of plasmid DNA isoforms. J. Chromatogr. A 2015, 1414, 103–109. [Google Scholar] [CrossRef]
- Miladys, L.; Lourdes, Z.; Urska, V.; Nika, L.K. Comparison of CIM® C4-HLD monolithic column with Sartobind phenyl membrane column for pIDKE2 purification. Chin. J. Chromatogr. 2017, 35, 1028–1036. [Google Scholar] [CrossRef]
- Han, Y.; Forde, G.M. Single step purification of plasmid DNA using peptide ligand affinity chromatography. J. Chromatogr. B 2008, 874, 21–26. [Google Scholar] [CrossRef]
- Černigoj, U.; Vidic, U.; Barut, M.; Podgornik, A.; Peterka, M.; Štrancar, A. A multimodal histamine ligand for chromatographic purification of plasmid DNA. J. Chromatogr. A 2013, 1281, 87–93. [Google Scholar] [CrossRef]
- Sousa, A.; Almeida, A.; Černigoj, U.; Sousa, F.; Queiroz, J. Histamine monolith versatility to purify supercoiled plasmid deoxyribonucleic acid from Escherichia coli lysate. J. Chromatogr. A 2014, 1355, 125–133. [Google Scholar] [CrossRef]
- Cardoso, S.; Černigoj, U.; Krajnc, N.L.; Štrancar, A. Chromatographic purification of plasmid DNA on hydrophobic methacrylate monolithic supports. Sep. Purif. Technol. 2015, 147, 139–146. [Google Scholar] [CrossRef]
- Soares, A.; Queiroz, J.; Sousa, F.; Sousa, A. Purification of human papillomavirus 16 E6/E7 plasmid deoxyribonucleic acid-based vaccine using an arginine modified monolithic support. J. Chromatogr. A 2013, 1320, 72–79. [Google Scholar] [CrossRef]
- Almeida, A.; Queiroz, J.; Sousa, F.; Sousa, A. Optimization of supercoiled HPV-16 E6/E7 plasmid DNA purification with arginine monolith using design of experiments. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2015, 978–979, 145–150. [Google Scholar] [CrossRef]
- Amorim, L.F.A.; Sousa, F.; Queiroz, J.A.; Cruz, C.; Sousa, Â. Screening of L-histidine-based ligands to modify monolithic supports and selectively purify the supercoiled plasmid DNA isoform. J. Mol. Recognit. 2015, 28, 349–358. [Google Scholar] [CrossRef]
- Amorim, L.F.A.; Gaspar, R.; Pereira, P.; Černigoj, U.; Sousa, F.; Queiroz, J.A.; Sousa, Â. Chromatographic HPV-16 E6/E7 plasmid vaccine purification employing L-histidine and 1-benzyl-L-histidine affinity ligands. Electrophoresis 2017, 38, 2975–2980. [Google Scholar] [CrossRef]
- Mann, A.; Thakur, G.; Shukla, V.; Singh, A.K.; Khanduri, R.; Naik, R.; Jiang, Y.; Kalra, N.; Dwarakanath, B.S.; Langel, U.; et al. Differences in DNA Condensation and Release by Lysine and Arginine Homopeptides Govern Their DNA Delivery Efficiencies. Mol. Pharm. 2011, 8, 1729–1741. [Google Scholar] [CrossRef]
- Bicho, D.; Santos, B.; Caramelo-Nunes, C.; Sousa, A.; Sousa, F.; Queiroz, J.; Tomaz, C. Application of ethylenediamine monolith to purify a hemagglutinin influenza deoxyribonucleic acid-based vaccine. Sep. Purif. Technol. 2015, 154, 320–327. [Google Scholar] [CrossRef]
- Bicho, D.; Caramelo-Nunes, C.; Sousa, A.; Sousa, F.; Queiroz, J.; Tomaz, C. Purification of influenza deoxyribonucleic acid-based vaccine using agmatine monolith. J. Chromatogr. B 2016, 1012–1013, 153–161. [Google Scholar] [CrossRef]
- Almeida, A.; Černigoj, U.; Queiroz, J.; Sousa, F.; Sousa, A. Quality assessment of supercoiled minicircle DNA by cadaverine-modified analytical chromatographic monolith. J. Pharm. Biomed. Anal. 2020, 180, 113037. [Google Scholar] [CrossRef]
- Almeida, A.; Queiroz, J.; Sousa, F.; Sousa, A. Minicircle DNA purification: Performance of chromatographic monoliths bearing lysine and cadaverine ligands. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2019, 1118–1119, 7–16. [Google Scholar] [CrossRef]
- Almeida, A.M.; Costa, D.; Simões, A.R.; Queiroz, J.A.; Sousa, F.; Sousa, Â. Enhancement of a biotechnological platform for the purification and delivery of a human papillomavirus supercoiled plasmid DNA vaccine. New Biotechnol. 2020, 59, 1–9. [Google Scholar] [CrossRef]
- Wu, Q.; Bienvenue, J.M.; Hassan, B.J.; Kwok, Y.C.; Giordano, B.C.; Norris, P.M.; Landers, J.P.; Ferrance, J.P. Microchip-Based Macroporous Silica Sol–Gel Monolith for Efficient Isolation of DNA from Clinical Samples. Anal. Chem. 2006, 78, 5704–5710. [Google Scholar] [CrossRef]
- Holdšvendová, P.; Suchánková, J.; Bunček, M.; Bačkovská, V.; Coufal, P. Hydroxymethyl methacrylate-based monolithic columns designed for separation of oligonucleotides in hydrophilic-interaction capillary liquid chromatography. J. Biochem. Biophys. Methods 2007, 70, 23–29. [Google Scholar] [CrossRef]
- Smrekar, F.; Podgornik, A.; Ciringer, M.; Kontrec, S.; Raspor, P.; Štrancar, A.; Peterka, M. Preparation of pharmaceutical-grade plasmid DNA using methacrylate monolithic columns. Vaccine 2010, 28, 2039–2045. [Google Scholar] [CrossRef]
- Smrekar, V.; Smrekar, F.; Štrancar, A.; Podgornik, A. Single step plasmid DNA purification using methacrylate monolith bearing combination of ion-exchange and hydrophobic groups. J. Chromatogr. A 2013, 1276, 58–64. [Google Scholar] [CrossRef]
- Ghanem, A.; Healey, R.; Adly, F.G. Current trends in separation of plasmid DNA vaccines: A review. Anal. Chim. Acta 2013, 760, 1–15. [Google Scholar] [CrossRef]
- FDA. Guidance for Industry: Considerations for Plasmid DNA Vaccines for Infectious Disease Indications. 2007. Available online: http://www.fda.gov/cber/guidelines.htm (accessed on 6 May 2023).
- Tomaz, C.T. Hydrophobic interaction chromatography. In Liquid Chromatography; Elsevier: Amsterdam, The Netherlands, 2017; pp. 171–190. [Google Scholar] [CrossRef]
- Sousa, F.; Tomaz, C.; Prazeres, D.; Queiroz, J. Separation of supercoiled and open circular plasmid DNA isoforms by chromatography with a histidine–agarose support. Anal. Biochem. 2005, 343, 183–185. [Google Scholar] [CrossRef]
- Sousa, F.; Prazeres, D.M.F.; Queiroz, J.A. Dynamic binding capacity of plasmid DNA in histidine–agarose chromatography. Biomed. Chromatogr. 2007, 21, 993–998. [Google Scholar] [CrossRef]
- Bicho, D.; Sousa, Â.; Sousa, F.; Queiroz, J.; Tomaz, C. Effect of chromatographic conditions and plasmid DNA size on the dynamic binding capacity of a monolithic support. J. Sep. Sci. 2014, 37, 2284–2292. [Google Scholar] [CrossRef]
- Poddar, S.; Sharmeen, S.; Hage, D.S. Affinity monolith chromatography: A review of general principles and recent developments. Electrophoresis 2021, 42, 2577–2598. [Google Scholar] [CrossRef]
- Sousa, F.; Prazeres, D.M.; Queiroz, J.A. Affinity chromatography approaches to overcome the challenges of purifying plasmid DNA. Trends Biotechnol. 2008, 26, 518–525. [Google Scholar] [CrossRef]
- Caramelo-Nunes, C.; Almeida, P.; Marcos, J.; Tomaz, C. Aromatic ligands for plasmid deoxyribonucleic acid chromatographic analysis and purification: An overview. J. Chromatogr. A 2014, 1327, 1–13. [Google Scholar] [CrossRef]
- Caramelo-Nunes, C.; Tente, T.; Almeida, P.; Marcos, J.; Tomaz, C. Specific berenil–DNA interactions: An approach for separation of plasmid isoforms by pseudo-affinity chromatography. Anal. Biochem. 2011, 412, 153–158. [Google Scholar] [CrossRef]
- Caramelo-Nunes, C.; Gabriel, M.; Almeida, P.; Marcos, J.; Tomaz, C. Negative pseudo-affinity chromatography for plasmid DNA purification using berenil as ligand. J. Chromatogr. B 2014, 944, 39–42. [Google Scholar] [CrossRef]
- Caramelo-Nunes, C.; Bicho, D.; Almeida, P.; Marcos, J.; Tomaz, C. Dynamic binding capacity and specificity of 3,8-diamino-6-phenylphenanthridine-Sepharose support for purification of supercoiled plasmid deoxyribonucleic acid. J. Chromatogr. A 2013, 1307, 91–98. [Google Scholar] [CrossRef]
- Caramelo-Nunes, C.; Tomaz, C.T. Specific Recognition of Supercoiled Plasmid DNA by Affinity Chromatography Using a Synthetic Aromatic Ligand. Affin. Chromatogr. Methods Protoc. 2015, 2015, 47–54. [Google Scholar] [CrossRef]
- Caramelo-Nunes, C.; Almeida, P.; Marcos, J.; Tomaz, C. Specific recognition of supercoiled plasmid DNA by affinity chromatography using the intercalator DAPP as ligand. J. Chromatogr. B 2013, 928, 121–124. [Google Scholar] [CrossRef]
- Santos, T.; Proença, Z.; Queiroz, J.; Tomaz, C.; Cruz, C. Plasmid purification by using a new naphthalene tripodal support. Sep. Purif. Technol. 2017, 188, 81–89. [Google Scholar] [CrossRef]
- Cardoso, S.; Filho, P.d.A.P.; Sousa, F.; Azzoni, A.R. Arginine and di-arginine ligands for plasmid DNA purification using negative chromatography. Sep. Purif. Technol. 2018, 202, 281–289. [Google Scholar] [CrossRef]
- Cardoso, S.; Sousa, Â.; Queiroz, J.A.; Azzoni, A.R.; Sousa, F. Arginine homopeptides for plasmid DNA purification using monolithic supports. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2018, 1087–1088, 149–157. [Google Scholar] [CrossRef]
- Kotagale, N.; Bhondekar, S.; Bhad, M.; Pise, S.; Charpe, A.; Umekar, M.; Taksande, B. Agmatine prevents development of tolerance to anti-nociceptive effect of ethanol in mice. Alcohol 2022, 101, 1–8. [Google Scholar] [CrossRef]
- Krajačić, M.; Ivancic-Jelecki, J.; Forcic, D.; Vrdoljak, A.; Škorić, D. Purification of plant viral and satellite double-stranded RNAs on DEAE monoliths. J. Chromatogr. A 2007, 1144, 111–119. [Google Scholar] [CrossRef]
- Perica, M.; Šola, I.; Urbas, L.; Smrekar, F.; Krajačić, M. Separation of hypoviral double-stranded RNA on monolithic chromatographic supports. J. Chromatogr. A 2009, 1216, 2712–2716. [Google Scholar] [CrossRef]
- Romanovskaya, A.; Sarin, L.P.; Bamford, D.H.; Poranen, M.M. High-throughput purification of double-stranded RNA molecules using convective interaction media monolithic anion exchange columns. J. Chromatogr. A 2013, 1278, 54–60. [Google Scholar] [CrossRef]
- Thayer, J.R.; Flook, K.J.; Woodruff, A.; Rao, S.; Pohl, C.A. New monolith technology for automated anion-exchange purification of nucleic acids. J. Chromatogr. B 2010, 878, 933–941. [Google Scholar] [CrossRef]
- Satterfield, B.C.; Stern, S.; Caplan, M.R.; Hukari, K.W.; West, J.A.A. Microfluidic Purification and Preconcentration of mRNA by Flow-Through Polymeric Monolith. Anal. Chem. 2007, 79, 6230–6235. [Google Scholar] [CrossRef]
- Pereira, P.; Sousa, Â.; Queiroz, J.A.; Figueiras, A.; Sousa, F. Pharmaceutical-grade pre-miR-29 purification using an agmatine monolithic support. J. Chromatogr. A 2014, 1368, 173–182. [Google Scholar] [CrossRef]
- Çorman, M.E.; Bereli, N.; Özkara, S.; Uzun, L.; Denizli, A. Hydrophobic cryogels for DNA adsorption: Effect of embedding of monosize microbeads into cryogel network on their adsorptive performances. Biomed. Chromatogr. 2013, 27, 1524–1531. [Google Scholar] [CrossRef]
- Pereira, P.; Sousa, Â.; Queiroz, J.; Correia, I.; Figueiras, A.; Sousa, F. Purification of pre-miR-29 by arginine-affinity chromatography. J. Chromatogr. B 2014, 951–952, 16–23. [Google Scholar] [CrossRef]
- Ertürk, G.; Mattiasson, B. Cryogels-versatile tools in bioseparation. J. Chromatogr. A 2014, 1357, 24–35. [Google Scholar] [CrossRef]
- Lozinsky, V.I.; Galaev, I.Y.; Plieva, F.M.; Savina, I.N.; Jungvid, H.; Mattiasson, B. Polymeric cryogels as promising materials of biotechnological interest. Trends Biotechnol. 2003, 21, 445–451. [Google Scholar] [CrossRef]
- Shakya, A.K. Functionalized cryogel monoliths for fast and selective separation of nucleic acids directly from crude lysate. Biomed. Chromatogr. 2022, 36, e5333. [Google Scholar] [CrossRef]
- Memic, A.; Colombani, T.; Eggermont, L.J.; Rezaeeyazdi, M.; Steingold, J.; Rogers, Z.J.; Navare, K.J.; Mohammed, H.S.; Bencherif, S.A. Latest Advances in Cryogel Technology for Biomedical Applications. Adv. Ther. 2019, 2, 1800114. [Google Scholar] [CrossRef]
- Bakhshpour, M.; Idil, N.; Perçin, I.; Denizli, A. Biomedical Applications of Polymeric Cryogels. Appl. Sci. 2019, 9, 553. [Google Scholar] [CrossRef]
- Alkan, H.; Bereli, N.; Baysal, Z.; Denizli, A. Selective removal of the autoantibodies from rheumatoid arthritis patient plasma using protein A carrying affinity cryogels. Biochem. Eng. J. 2010, 51, 153–159. [Google Scholar] [CrossRef]
- Özgür, E.; Bereli, N.; Türkmen, D.; Ünal, S.; Denizli, A. PHEMA cryogel for in-vitro removal of anti-dsDNA antibodies from SLE plasma. Mater. Sci. Eng. C 2011, 31, 915–920. [Google Scholar] [CrossRef]
- Bereli, N.; Şener, G.; Yavuz, H.; Denizli, A. Oriented immobilized anti-LDL antibody carrying poly(hydroxyethyl methacrylate) cryogel for cholesterol removal from human plasma. Mater. Sci. Eng. C 2011, 31, 1078–1083. [Google Scholar] [CrossRef]
- La Spina, R.; Tripisciano, C.; Mecca, T.; Cunsolo, F.; Weber, V.; Mattiasson, B. Chemically modified poly(2-hydroxyethyl methacrylate) cryogel for the adsorption of heparin. J. Biomed. Mater. Res. B Appl. Biomater. 2014, 102, 1207–1216. [Google Scholar] [CrossRef]
- Baydemir, G.; Denizli, A. Heparin removal from human plasma using molecular imprinted cryogels. Artif. Cells Nanomed. Biotechnol. 2015, 43, 403–412. [Google Scholar] [CrossRef]
- Baydemir, G.; Bereli, N.; Andaç, M.; Say, R.; Galaev, I.Y.; Denizli, A. Bilirubin recognition via molecularly imprinted supermacroporous cryogels. Colloids Surf. B Biointerfaces 2009, 68, 33–38. [Google Scholar] [CrossRef]
- Baydemir, G.; Bereli, N.; Andaç, M.; Say, R.; Galaev, I.Y.; Denizli, A. Supermacroporous poly(hydroxyethyl methacrylate) based cryogel with embedded bilirubin imprinted particles. React. Funct. Polym. 2009, 69, 36–42. [Google Scholar] [CrossRef]
- Rabieizadeh, M.; Kashefimofrad, S.M.; Naeimpoor, F. Monolithic molecularly imprinted cryogel for lysozyme recognition. J. Sep. Sci. 2014, 37, 2983–2990. [Google Scholar] [CrossRef]
- Gao, F.-X.; Zhao, X.-L.; He, X.-W.; Li, W.-Y.; Zhang, Y.-K. A pH and temperature dual-responsive macroporous molecularly imprinted cryogel for enhanced recognition capability towards ovalbumin. Anal. Methods 2013, 5, 6700–6708. [Google Scholar] [CrossRef]
- Derazshamshir, A.; Baydemir, G.; Andac, M.; Say, R.; Galaev, I.Y.; Denizli, A. Molecularly Imprinted PHEMA-Based Cryogel for Depletion of Hemoglobin from Human Blood. Macromol. Chem. Phys. 2010, 211, 657–668. [Google Scholar] [CrossRef]
- Andac, M.; Galaev, I.Y.; Denizli, A. Molecularly imprinted poly(hydroxyethyl methacrylate) based cryogel for albumin depletion from human serum. Colloids Surf. B Biointerfaces 2013, 109, 259–265. [Google Scholar] [CrossRef]
- Andac, M.; Galaev, I.; Denizli, A. Dye attached poly(hydroxyethyl methacrylate) cryogel for albumin depletion from human serum. J. Sep. Sci. 2012, 35, 1173–1182. [Google Scholar] [CrossRef]
- Demiryas, N.; Tüzmen, N.; Galaev, I.Y.; Pişkin, E.; Denizli, A. Poly(acrylamide-allyl glycidyl ether) cryogel as a novel stationary phase in dye-affinity chromatography. J. Appl. Polym. Sci. 2007, 105, 1808–1816. [Google Scholar] [CrossRef]
- Bakhshpour, M.; Derazshamshir, A.; Bereli, N.; Elkak, A.; Denizli, A. [PHEMA/PEI]–Cu(II) based immobilized metal affinity chromatography cryogels: Application on the separation of IgG from human plasma. Mater. Sci. Eng. C 2016, 61, 824–831. [Google Scholar] [CrossRef]
- Hajizadeh, S. Application of composite cryogels in downstream processing—A review. React. Funct. Polym. 2023, 191, 105693. [Google Scholar] [CrossRef]
- Santos, T.; Brito, A.; Boto, R.; Sousa, P.; Almeida, P.; Cruz, C.; Tomaz, C. Influenza DNA vaccine purification using pHEMA cryogel support. Sep. Purif. Technol. 2018, 206, 192–198. [Google Scholar] [CrossRef]
- Plieva, F.M.; Savina, I.N.; Deraz, S.; Andersson, J.; Galaev, I.Y.; Mattiasson, B. Characterization of supermacroporous monolithic polyacrylamide based matrices designed for chromatography of bioparticles. J. Chromatogr. B 2004, 807, 129–137. [Google Scholar] [CrossRef]
- Tripathi, A.; Kumar, A. Multi-Featured Macroporous Agarose–Alginate Cryogel: Synthesis and Characterization for Bioengineering Applications. Macromol. Biosci. 2010, 11, 22–35. [Google Scholar] [CrossRef]
- Hwang, C.M.; Sant, S.; Masaeli, M.; Kachouie, N.N.; Zamanian, B.; Lee, S.-H.; Khademhosseini, A. Fabrication of three-dimensional porous cell-laden hydrogel for tissue engineering. Biofabrication 2010, 2, 035003. [Google Scholar] [CrossRef]
- Jain, E.; Karande, A.A.; Kumar, A. Supermacroporous polymer-based cryogel bioreactor for monoclonal antibody production in continuous culture using hybridoma cells. Biotechnol. Prog. 2010, 27, 170–180. [Google Scholar] [CrossRef]
- Andaç, M.; Galaev, I.Y.; Denizli, A. Affinity based and molecularly imprinted cryogels: Applications in biomacromolecule purification. J. Chromatogr. B 2016, 1021, 69–80. [Google Scholar] [CrossRef]
- Hanora, A.; Savina, I.; Plieva, F.M.; Izumrudov, V.A.; Mattiasson, B.; Galaev, I.Y. Direct capture of plasmid DNA from non-clarified bacterial lysate using polycation-grafted monoliths. J. Biotechnol. 2006, 123, 343–355. [Google Scholar] [CrossRef]
- Perçin, I.; Sağlar, E.; Yavuz, H.; Aksöz, E.; Denizli, A. Poly(hydroxyethyl methacrylate) based affinity cryogel for plasmid DNA purification. Int. J. Biol. Macromol. 2011, 48, 577–582. [Google Scholar] [CrossRef]
- Üzek, R.; Uzun, L.; Şenel, S.; Denizli, A. Nanospines incorporation into the structure of the hydrophobic cryogels via novel cryogelation method: An alternative sorbent for plasmid DNA purification. Colloids Surf. B Biointerfaces 2013, 102, 243–250. [Google Scholar] [CrossRef]
- Çimen, D.; Yılmaz, F.; Perçin, I.; Türkmen, D.; Denizli, A. Dye affinity cryogels for plasmid DNA purification. Mater. Sci. Eng. C 2015, 56, 318–324. [Google Scholar] [CrossRef]
- Önal, B.; Odabaşı, M. Design and application of a newly generated bio/synthetic cryogel column for DNA capturing. Polym. Bull. 2021, 78, 6011–6028. [Google Scholar] [CrossRef]
- Galaev, I.Y.; Dainiak, M.B.; Plieva, F.M.; Hatti-Kaul, R.; Mattiasson, B. High throughput processing of particulate-containing samples using supermacroporous elastic monoliths in microtiter (multiwell) plate format. J. Chromatogr. A 2005, 1065, 169–175. [Google Scholar] [CrossRef]
- Srivastava, A.; Shakya, A.K.; Kumar, A. Boronate affinity chromatography of cells and biomacromolecules using cryogel matrices. Enzym. Microb. Technol. 2012, 51, 373–381. [Google Scholar] [CrossRef]
- Köse, K.; Erol, K.; ÿzgür, E.; Uzun, L.; Denizli, A. PolyAdenine cryogels for fast and effective RNA purification. Colloids Surf. B Biointerfaces 2016, 146, 678–686. [Google Scholar] [CrossRef]
- Köse, K.; Uzun, L. PolyGuanine methacrylate cryogels for ribonucleic acid purification. J. Sep. Sci. 2016, 39, 1998–2005. [Google Scholar] [CrossRef] [PubMed]

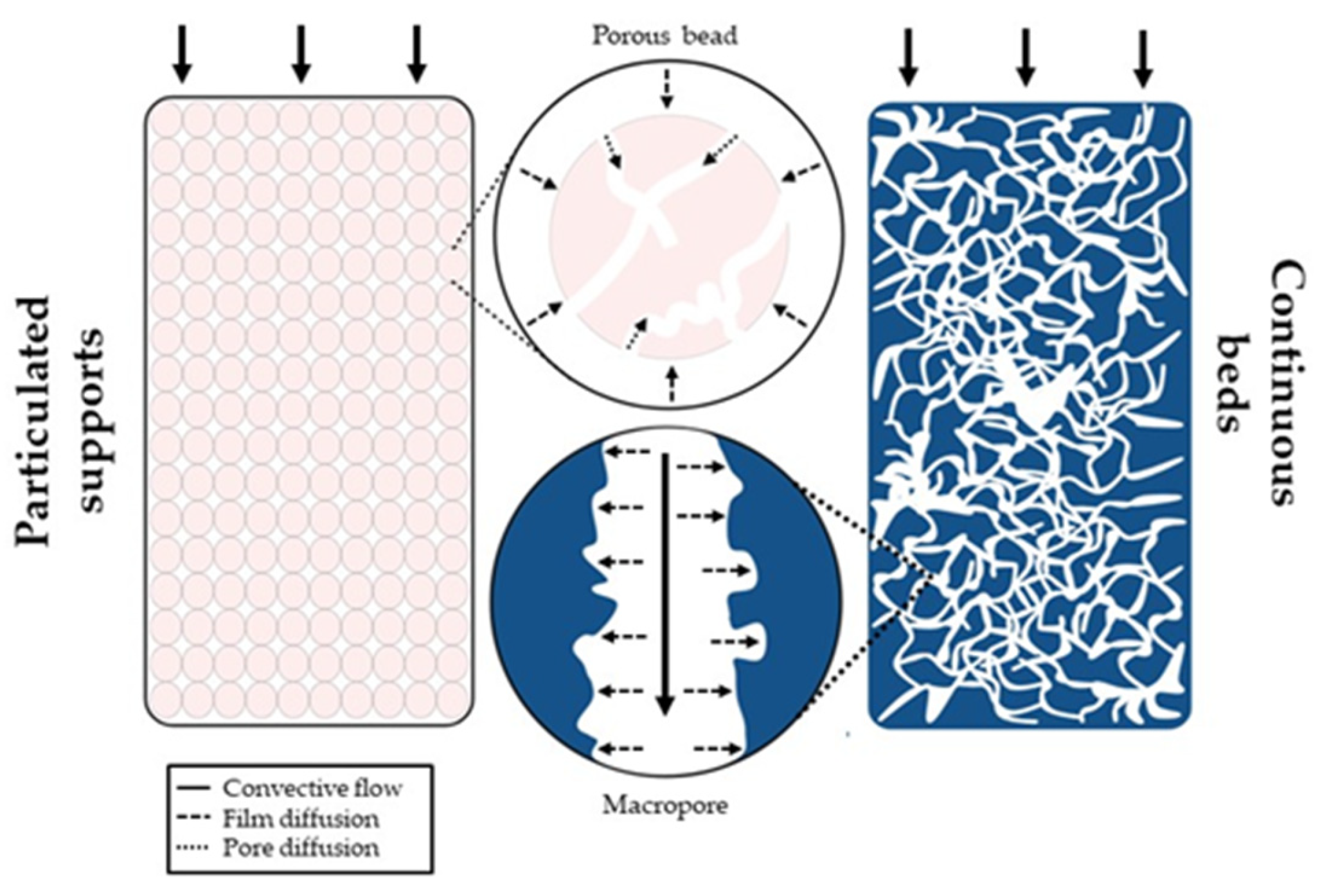
| Target | Ligand | Matrix | Binding Capacity (mg/mL) | Purity (%) | Recovery Yield (%) | Application | Reference |
|---|---|---|---|---|---|---|---|
| pDNA | DEAE | CIM® | 8.9 | 92.0 | 100.0 | - | [39] |
| gDNA | DEAE | CIM® | - | - | - | - | [40] |
| pcDNA3F | Triethylamine | Poly (GMA-EDMA) | 15.8 | - | >95.0 | - | [41] |
| Diethylamine | 21.5 | ||||||
| pDNA | DEAE and QA | CIM® | - | 82.0–100.0 | - | [42] | |
| G-quadruplex | QA | CIMmultusTM | 5.5 | 92.0 | - | - | [43] |
| pDNA | - | CDI | - | 100.0 | 74.0 | - | [44] |
| pDNA | Pyridine | CIM® | 0.5 | >95.0 | - | - | [45] |
| pDNA | C4 HLD | CIM® | 0.9–2.0 | 92.8–99.4 | 80.9–100.0 | Hepatitis C DNA vaccine | [46] |
| pDNA | 16mer (oligonucleotide) | poly (GMA-co-EDMA) | 0.02 | 92.0 | 81.0 | - | [47] |
| pDNA | Histamine | CDI | 4.0 | 98.5 | 97.0 | - | [48] |
| pDNA | Histamine | CDI | 2.7–4.0 | 78.9–96.7 | 91.6–99.3 | - | [49] |
| pDNA | Pyridine | CIM® | 1.3 | >90.0 | 98.0 | - | [50] |
| pDNA | Arginine | CIM® epoxy | 5.2 | >99.0 | 39.2 | HPV DNA vaccine | [51] |
| pDNA | Arginine | CIM® epoxy | - | 100.0 | 75.8–88.8 | HPV DNA vaccine | [52] |
| pDNA | L-Histidine | CIMacTM | 11.0 | - | - | Ligand screening | [53] |
| 1-benzyl-L-Histidine | - | - | - | ||||
| pDNA | L-Histidine | CIMacTM | 2.4 | >99.0 | 74.4 | - | [54] |
| 1-benzyl-L-Histidine | - | 31.6 | - | ||||
| pDNA | Arginine | CIM® epoxy | 1.5 | >97.0 | 88.0 | - | [55] |
| di-Arginine | 3.5 | 66.1 | - | ||||
| tri-Arginine | 3.6 | >99.0 | 52.7 | - | |||
| pDNA | Ethylenediamine | CIM® polymethacrylate | - | 97.1 | 47.0 | Influenza DNA vaccine | [56] |
| pDNA | Agmatine | CDI | 8.6 | 99.8 | 98.3–99.6 | Influenza DNA vaccine | [57] |
| mcDNA | Cadaverine | CIM® | - | - | - | Analytical approach | [58] |
| mcDNA | Lysine | CIM® epoxy | - | - | - | - | [59] |
| Cadaverine | 2.4 | 98.4 | 78.6 | ||||
| pDNA | Arginine + Space arm | Epoxy | 2.5 | 93.3 | 72.0 | HPV DNA vaccine | [60] |
| DNA | - | TMOS + PEG | - | - | - | Separation of several DNA types | [61] |
| Oligonucleotides | - | HMMAA-(EDMA) | - | - | - | - | [62] |
| pDNA | DEAE and C4 | CIM® OH (hydroxy) | - | 98.0 | 81.0 | - | [63] |
| pDNA | Octylamine (IEX conditions) | CIM® epoxy | 1.6 | - | >80.0 | - | [64] |
| Octylamine (HIC conditions) | 0.6 | - | |||||
| gBuMA + DEAE (IEX conditions) | 4.7 | - | |||||
| gBuMA + DEAE (HIC conditions) | 2.1 | - |
| Target | Ligand | Matrix | Binding Capacity | Purity (%) | Recovery Yield (%) | Application | Reference |
|---|---|---|---|---|---|---|---|
| satRNA and dsRNA | DEAE | CIM® | - | - | - | detection and identification | [83] |
| dsRNA | DEAE | CIM® | - | - | - | - | [84] |
| dsRNA and siRNA | DEAE | CIM® | 8.0 mg dsRNA/mL resin | - | 39.0 | - | [85] |
| Quaternary amine | 5.5 mg dsRNA/mL resin | - | 52.0 | ||||
| Oligonucleotides | Nanobeads w/Quartenary amine | ProSwift SCX-1S | - | >90.0 | 75.0 | - | [86] |
| mRNA | Oligo dT | EGDMA-(GMA) | - | - | - | - | [87] |
| Pre-miR-29 | Agmatine | CDI (NaCl gradient) | 8.1 mg RNA/mL support | 75.2 | 97.3 | - | [88] |
| Agmatine | CDI (Arginine gradient) | 90.1 | 94.9 | - | |||
| RNA | OH (hydroxy) | CIM® | - | - | 80.0 | - | [89] |
| Target | Ligand | Matrix | Binding Capacity (mg/g) | Purity (%) | Recovery Yield (%) | Application | Reference |
|---|---|---|---|---|---|---|---|
| DNA | p(HEMA-MATrp) monosize particles | pHEMA | 38.0 | - | - | - | [89] |
| pDNA | - | pHEMA | - | 98.1 | 69.2 | Influenza DNA vaccine | [111] |
| pDNA | Polycations | Polyacrylamide | - | - | - | - | [117] |
| pDNA | MAH | pHEMA | - | 90.0 | - | - | [118] |
| DNA | MATrp | pHEMA-MATrp | 15.0 | - | - | - | [119] |
| pDNA | Cibacron Blue F3GA | pHEMA | 32.5 | - | - | - | [120] |
| pDNA | Fe3+ and EPS | pHEMA | 39.7 | - | - | - | [121] |
| Target | Ligand | Matrix | Binding Capacity | Application | Reference |
|---|---|---|---|---|---|
| RNA | Boronate | PHEMA-co-VPBA) | 1.1 mg/mL cryogel | - | [123] |
| RNA | Adenine | pHEMA | 11.9 mg/g cryogel | - | [124] |
| RNA | Guanine | pHEMA | 5.6 mg/g cryogel | - | [125] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ribeiro, J.; Luís, M.Â.; Rodrigues, B.; Santos, F.M.; Mesquita, J.; Boto, R.; Tomaz, C.T. Cryogels and Monoliths: Promising Tools for Chromatographic Purification of Nucleic Acids. Gels 2024, 10, 198. https://doi.org/10.3390/gels10030198
Ribeiro J, Luís MÂ, Rodrigues B, Santos FM, Mesquita J, Boto R, Tomaz CT. Cryogels and Monoliths: Promising Tools for Chromatographic Purification of Nucleic Acids. Gels. 2024; 10(3):198. https://doi.org/10.3390/gels10030198
Chicago/Turabian StyleRibeiro, João, Marco Â. Luís, Bruno Rodrigues, Fátima Milhano Santos, Joana Mesquita, Renato Boto, and Cândida Teixeira Tomaz. 2024. "Cryogels and Monoliths: Promising Tools for Chromatographic Purification of Nucleic Acids" Gels 10, no. 3: 198. https://doi.org/10.3390/gels10030198





