In Vitro Antibacterial and Anti-Inflammatory Properties of Imidazolium Poly(ionic liquids) Microspheres Loaded in GelMA-PEG Hydrogels
Abstract
:1. Introduction
2. Results and Discussion
2.1. Preparation and Characterization of Linear Antibacterial Copolymers and Microspheres
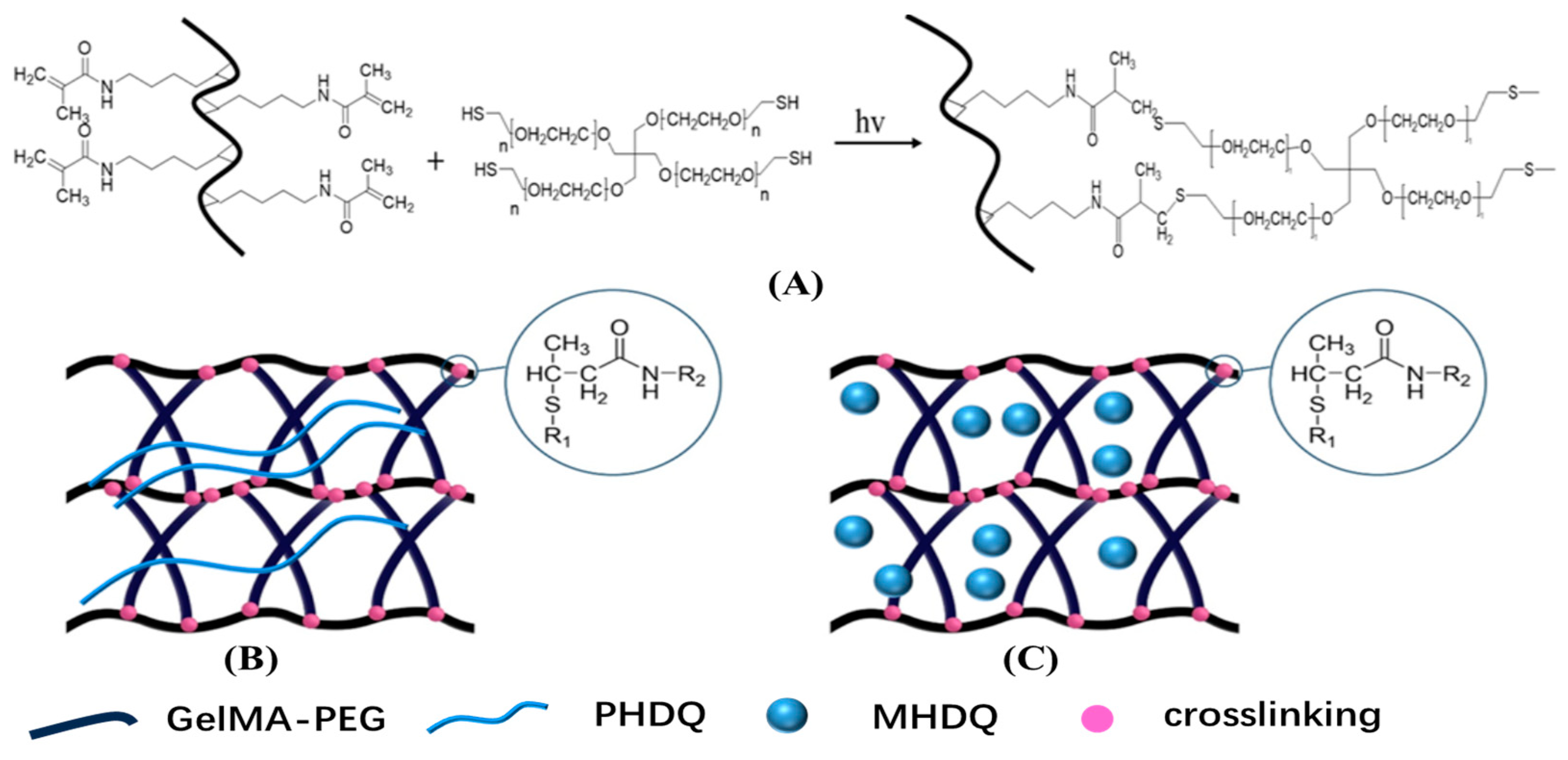
2.2. Structure and Physical Property of Antimicrobial Hydrogels
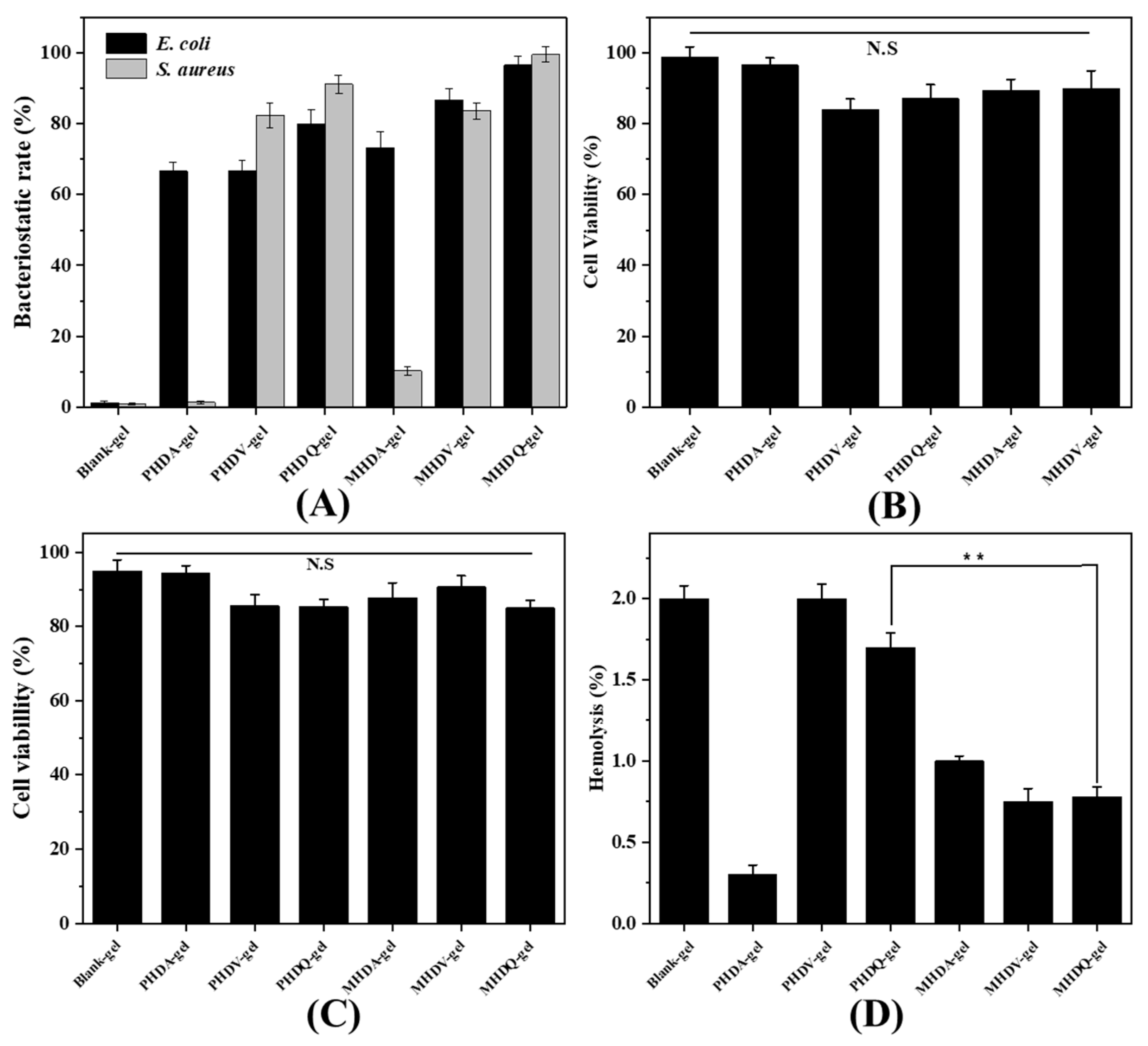
2.3. Antibacterial Activity and Biocompatible of Hydrogels
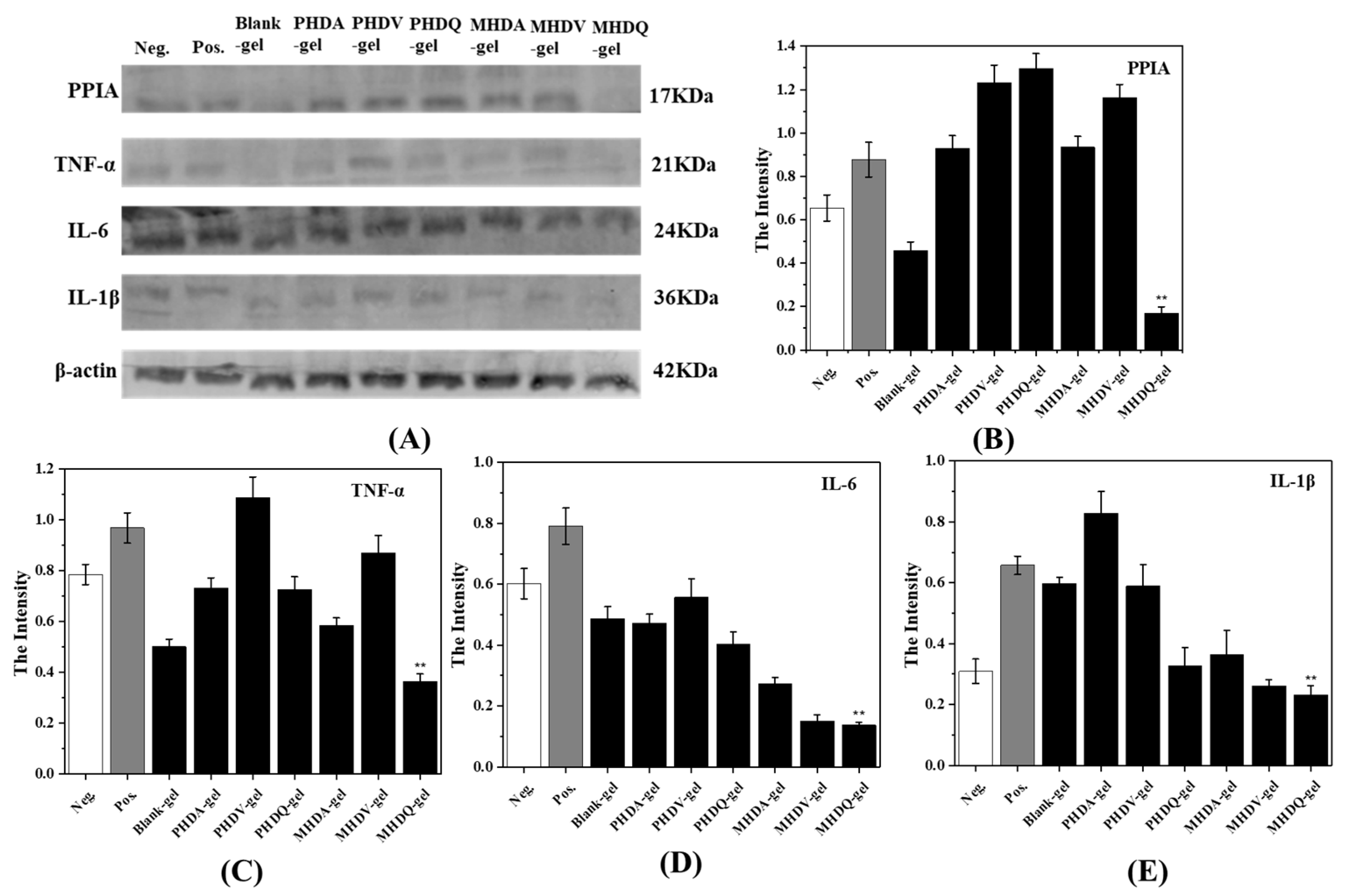
2.4. Anti-Inflammatory Activity of Hydrogels
2.5. In Vitro Skin Tissue Repairing by Hydrogel Treatment
2.6. In Vitro Bone Tissue Repairing by Hydrogel Treatment
3. Conclusions
4. Materials and Methods
4.1. Materials
4.2. Synthesis of Antibacterial Copolymers
4.3. Synthesis of Antibacterial Polymer Microspheres
4.4. Synthesis of Antibacterial Hydrogels
4.5. Chemical Structure Characterization
4.6. Zeta Potential and Particle Size Characterization
4.7. Rheological Performance and Surface Morphology of Antibacterial Hydrogel
4.8. Determination of In Vitro Biocompatibility of Hydrogels
4.9. Determination of In Vitro Antibacterial Activity of Copolymer, Microspheres, and Hydrogel
4.10. Determination of In Vitro Anti-Inflammatory Activity
4.10.1. Gene Expression by Quantitative PCR
4.10.2. Protein Expression by Western Blot
4.11. Determination of In Vitro Tissue Repairing
4.11.1. Cell Migration Assay
4.11.2. Immunofluorescence Staining Test
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhao, Y.; Song, S.L.; Ren, X.Z.; Zhang, J.M.; Lin, Q.; Zhao, Y.L. Supramolecular Adhesive Hydrogels for Tissue Engineering Applications. Chem. Rev. 2022, 122, 5604–5640. [Google Scholar] [CrossRef] [PubMed]
- Tu, Z.L.; Chen, M.; Wang, M.; Shao, Z.X.; Jiang, X.Q.; Wang, K.Y.; Yao, Z.; Yang, S.W.; Zhang, X.X.; Gao, W.Y.; et al. Engineering Bioactive M2 Macrophage-Polarized Anti-Inflammatory, Antioxidant, and Antibacterial Scaffolds for Rapid Angiogenesis and Diabetic Wound Repair. Adv. Funct. Mater. 2021, 31, 2100924. [Google Scholar] [CrossRef]
- Decambron, A.; Manassero, M.; Bensidhoum, M.; Lecuelle, B.; Logeart-Avramoglou, D.; Petite, H.; Viateau, V. A comparative study of tissue-engineered constructs from Acropora and Porites coral in a large animal bone defect model. Bone Jt. Res. 2017, 6, 208–215. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Tong, D.M.; Song, H.H.; Ruan, R.J.; Sun, Y.F.; Lin, Y.D.; Wang, J.; Hou, L.X.; Dai, J.Y.; Ding, J.X.; et al. Osteoimmunity-Regulating Biomimetically Hierarchical Scaffold for Augmented Bone Regeneration. Adv. Mater. 2022, 34, 2202044. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Duan, H.; Li, Y.Q.; She, Y.L.; Zhu, J.J.; Zhou, G.D.; Jiang, G.N.; Yang, Y. Nanofibrillar Decellularized Wharton’s Jelly Matrix for Segmental Tracheal Repair. Adv. Funct. Mater. 2020, 30, 1910067. [Google Scholar] [CrossRef]
- Gan, D.L.; Xu, T.; Xing, W.S.; Ge, X.; Fang, L.M.; Wang, K.F.; Ren, F.Z.; Lu, X. Mussel-Inspired Contact-Active Antibacterial Hydrogel with High Cell Affinity, Toughness, and Recoverability. Adv. Funct. Mater. 2019, 29, 1805964. [Google Scholar] [CrossRef]
- Godoy-Gallardo, M.; Eckhard, U.; Delgado, L.M.; Puente, Y.; Hoyos-Nogués, M.; Gil, F.J.; Perez, R.A. Antibacterial approaches in tissue engineering using metal ions and nanoparticles: From mechanisms to applications. Bioact. Mater. 2021, 6, 4470–4490. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, E.; Vigani, B.; Viseras, C.; Ferrari, F.; Rossi, S.; Sandri, G. Inorganic Nanomaterials in Tissue Engineering. Pharmaceutics 2022, 14, 1127. [Google Scholar] [CrossRef]
- Ye, Q.; Chen, S.H.; Zhang, Y.; Ruan, B.; Zhang, Y.J.; Zhang, X.K.; Jiang, T.; Wang, X.G.; Ma, N.; Tsai, F.C. Chitosan/Polyvinyl Alcohol/ Lauramidopropyl Betaine/2D-HOF Mixed Film with Abundant Hydrogen Bonds Acts as High Mechanical Strength Artificial Skin. Macromol. Biosci. 2021, 21, 2100317. [Google Scholar] [CrossRef]
- Liang, J.S.; Zeng, J.M.; Li, J.J.; She, J.Q.; Tan, R.X.; Liu, B. Cationic Antimicrobial Polymers. Prog. Chem. 2019, 31, 1263–1282. [Google Scholar]
- Zheng, S.J.; Li, W.Z.; Ren, Y.Y.; Liu, Z.Y.; Zou, X.Y.; Hu, Y.; Guo, J.N.; Sun, Z.; Yan, F. Moisture-Wicking, Breathable, and Intrinsically Antibacterial Electronic Skin Based on Dual-Gradient Poly(ionic liquid) Nanofiber Membranes. Adv. Mater. 2022, 34, 2106570. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.Y.; Wang, Y.; Ren, Y.Y.; Jin, G.Q.; Zhang, C.C.; Chen, W.; Yan, F. Poly(ionic liquid) hydrogel-based anti-freezing ionic skin for a soft robotic gripper. Mater. Horiz. 2020, 7, 919–927. [Google Scholar] [CrossRef]
- Qian, W.J.; Texter, J.; Yan, F. Frontiers in poly(ionic liquid)s: Syntheses and applications. Chem. Soc. Rev. 2017, 46, 1124–1159. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Zheng, Z.; Wang, B.; Mao, H.; Yan, F. Zinc ion coordinated poly(ionic liquid) antimicrobial membranes for wound healing. ACS Appl. Mater. Interfaces 2017, 9, 14656–14664. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Li, J.; Zhou, C.; Jia, W.; Song, H.; Zhang, L.; Zhao, F.; Lee, B.P.; Liu, B. In situ synthesis of biocompatible imidazolium salt hydrogels with antimicrobial activity. Acta Biomater. 2019, 99, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Xu, Q.; Guo, J.; Qin, J.; Mao, H.; Wang, B.; Yan, F. Structure-antibacterial activity relationships of imidazolium-type ionic liquid monomers, poly(ionic liquids) and poly(ionic liquid) membranes: Effect of alkyl chain length and cations. ACS Appl. Mater. Interfaces 2016, 8, 12684–12692. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Ding, X.J.; Guan, J.P.; Gu, Y.; Su, Z.K.; Zhao, Y.M.; Tu, Y.F.; Li, X.H.; Li, Y.J.; Li, J.Y. Ionic liquid-grafted polyamide 6 by radiation-induced grafting: New strategy to prepare covalently bonded ion-containing polymers and their application as functional fibers. ACS Appl. Mater. Interfaces 2019, 11, 5462–5475. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Nie, F.M.; Yang, J.X.; Pan, L.; Ma, Z.; Li, Y.S. Novel imidazolium-based poly(ionic liquid)s with different counterions for self-healing. J. Mater. Chem. A 2017, 5, 25220–25229. [Google Scholar] [CrossRef]
- Raucci, M.G.; Fasolino, I.; Pastore, S.G.; Soriente, A.; Capeletti, L.B.; Dessuy, M.B.; Giannini, C.; Schrekker, H.S.; Ambrosio, L. Antimicrobial imidazolium ionic liquids for the development of minimal invasive calcium phosphate-based bionanocomposites. Acs Appl. Mater. Interfaces 2018, 10, 42766–42776. [Google Scholar] [CrossRef]
- Rodrigues, M.; Calpena, A.C.; Amabilino, D.B.; Garduno-Ramirez, M.L.; Perez-Garcica, L. Supramolecular gels based on a gemini imidazolium amphiphile as molecular material for drug delivery. J. Mat. Chem. B 2014, 2, 5419–5429. [Google Scholar] [CrossRef]
- Zhou, C.; Sheng, C.J.; Chen, J.J.; Liang, Y.H.; Liu, Q.P.; Li, P.; Huang, X.J.; Liu, B. Gradual hydrogel degradation for programable repairing full-thickness skin defect wound. Chem. Eng. J. 2022, 450, 138200. [Google Scholar] [CrossRef]
- Zhou, C.; Sheng, C.J.; Gao, L.L.; Guo, J.; Li, P.; Liu, B. Engineering poly(ionic liquid) semi-IPN hydrogels with fast antibacterial and anti-inflammatory properties for wound healing. Chem. Eng. J. 2021, 413, 127429. [Google Scholar] [CrossRef]
- Chen, Y.; Dang, B.; Fu, J.; Wang, C.; Li, C.; Sun, Q.; Li, H. Cellulose-Based Hybrid Structural Material for Radiative Cooling. Nano Lett. 2021, 21, 397–404. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, A.M.; Hassan, K.T.; Hassan, O.M. Assessment of antimicrobial activity of chitosan/silver nanoparticles hydrogel and cryogel microspheres. Int. J. Biol. Macromol. 2023, 233, 123580. [Google Scholar] [CrossRef]
- Chen, M.M.; Tian, J.; Liu, Y.; Cao, H.; Li, R.Y.; Wang, J.H.; Wu, J.L.; Zhang, Q.Q. Dynamic covalent constructed self-healing hydrogel for sequential delivery of antibacterial agent and growth factor in wound healing. Chem. Eng. J. 2019, 373, 413–424. [Google Scholar] [CrossRef]
- Wang, F.; Zhang, Q.; Li, X.X.; Huang, K.X.; Shao, W.; Yao, D.W.; Huang, C.B. Redox-responsive blend hydrogel films based on carboxymethyl cellulose/chitosan microspheres as dual delivery carrier. Int. J. Biol. Macromol. 2019, 134, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.N.; Xing, X.D.; Tan, H.P.; Jia, Y.; Zhou, T.L.; Chen, Y.; Ling, Z.H.; Hu, X.H. Covalently antibacterial alginate-chitosan hydrogel dressing integrated gelatin microspheres containing tetracycline hydrochloride for wound healing. Mater. Sci. Eng. C-Mater. Biol. Appl. 2017, 70, 287–295. [Google Scholar] [CrossRef]
- Lacroix, C.; Sultan, E.; Fleury, E.; Charlot, A. Functional galactomannan platform from convenient esterification in imidazolium-based ionic liquids. Polym. Chem. 2012, 3, 538–546. [Google Scholar] [CrossRef]
- Wang, M.; Shi, J.; Mao, H.; Sun, Z.; Guo, S.; Guo, J.; Yan, F. Fluorescent Imidazolium-Type Poly(ionic liquid)s for Bacterial Imaging and Biofilm Inhibition. Biomacromolecules 2019, 20, 3161–3170. [Google Scholar] [CrossRef] [PubMed]
- Yue, K.; Trujillo-de Santiago, G.; Alvarez, M.M.; Tamayol, A.; Annabi, N.; Khademhosseini, A. Synthesis, properties, and biomedical applications of gelatin methacryloyl (GelMA) hydrogels. Biomaterials 2015, 73, 254–271. [Google Scholar] [CrossRef]
- Zhou, C.; Truong, V.X.; Qu, Y.; Lithgow, T.; Fu, G.D.; Forsythe, J.S. Antibacterial poly(ethylene glycol) hydrogels from combined epoxy-amine and thiol-ene click reaction. J. Polym. Sci. Part A-Polym. Chem. 2016, 54, 656–667. [Google Scholar] [CrossRef]
- Nishiguchi, A.; Taguchi, T. Designing an anti-inflammatory and tissue-adhesive colloidal dressing for wound treatment. Colloids Surf. B-Biointerfaces 2020, 188, 110737. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.T.; Bui, H.L.; Chakroborty, S.; Wang, Y.; Huang, C.J. Pegylated Metal-Phenolic Networks for Antimicrobial and Antifouling Properties. Langmuir 2019, 35, 8829–8839. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Zhou, J.T.; Ma, X.Q.; Pranantyo, D.; Li, J.J.; Xu, L.Q.; Truong, V.X. Robust anti-infective multilayer coatings with rapid self-healing property. Mater. Sci. Eng. C-Mater. Biol. Appl. 2021, 121, 111828. [Google Scholar] [CrossRef] [PubMed]




| Samples | GPC | Particle Size (nm) | Zeta Potential (mV) | Bacterial Inhibition (%) | Cell Viability (%) | |||
|---|---|---|---|---|---|---|---|---|
| Mn (g/mol) | Mw (g/mol) | PDI | E. coli | S. aureus | ||||
| PHDA | 3.7 × 104 | 9.1 × 104 | 2.4 | - | 0.2 | 36.1 | 1.0 | 88.9 |
| PHDV | 3.8 × 104 | 1.1 × 105 | 2.9 | - | 3.2 | 78.7 | 73.1 | 90.0 |
| PHDQ | 3.8 × 104 | 1.1 × 105 | 2.9 | - | 10.8 | 89.3 | 94.9 | 85.0 |
| MHDA | - | - | - | 28 | 5.2 | 54.7 | 2.9 | 90.0 |
| MHDV | - | - | - | 49 | 8.92 | 85.5 | 82.7 | 89.9 |
| MHDQ | - | - | - | 70 | 20.9 | 95.0 | 99.8 | 85.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, C.; Sun, M.; Wang, D.; Yang, M.; Loh, J.L.C.; Xu, Y.; Zhang, R. In Vitro Antibacterial and Anti-Inflammatory Properties of Imidazolium Poly(ionic liquids) Microspheres Loaded in GelMA-PEG Hydrogels. Gels 2024, 10, 278. https://doi.org/10.3390/gels10040278
Zhou C, Sun M, Wang D, Yang M, Loh JLC, Xu Y, Zhang R. In Vitro Antibacterial and Anti-Inflammatory Properties of Imidazolium Poly(ionic liquids) Microspheres Loaded in GelMA-PEG Hydrogels. Gels. 2024; 10(4):278. https://doi.org/10.3390/gels10040278
Chicago/Turabian StyleZhou, Chao, Mengdi Sun, Danni Wang, Mingmei Yang, Jia Ling Celestine Loh, Yawen Xu, and Ruzhi Zhang. 2024. "In Vitro Antibacterial and Anti-Inflammatory Properties of Imidazolium Poly(ionic liquids) Microspheres Loaded in GelMA-PEG Hydrogels" Gels 10, no. 4: 278. https://doi.org/10.3390/gels10040278





