Copper-Chelated Chitosan Microgels for the Selective Enrichment of Small Cationic Peptides
Abstract
:1. Introduction
2. Results and Discussion
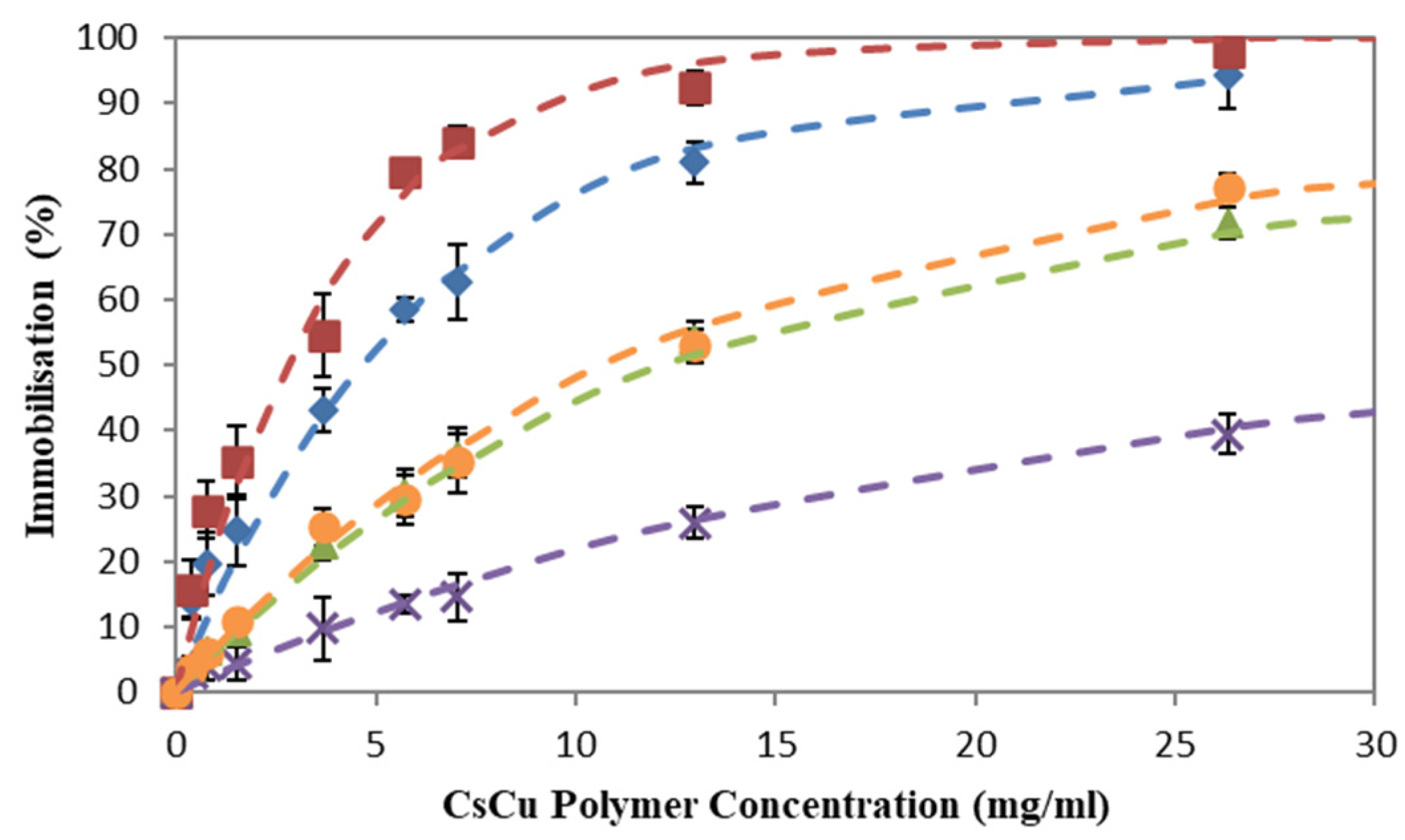
2.1. Amino Acids Immobilzsation
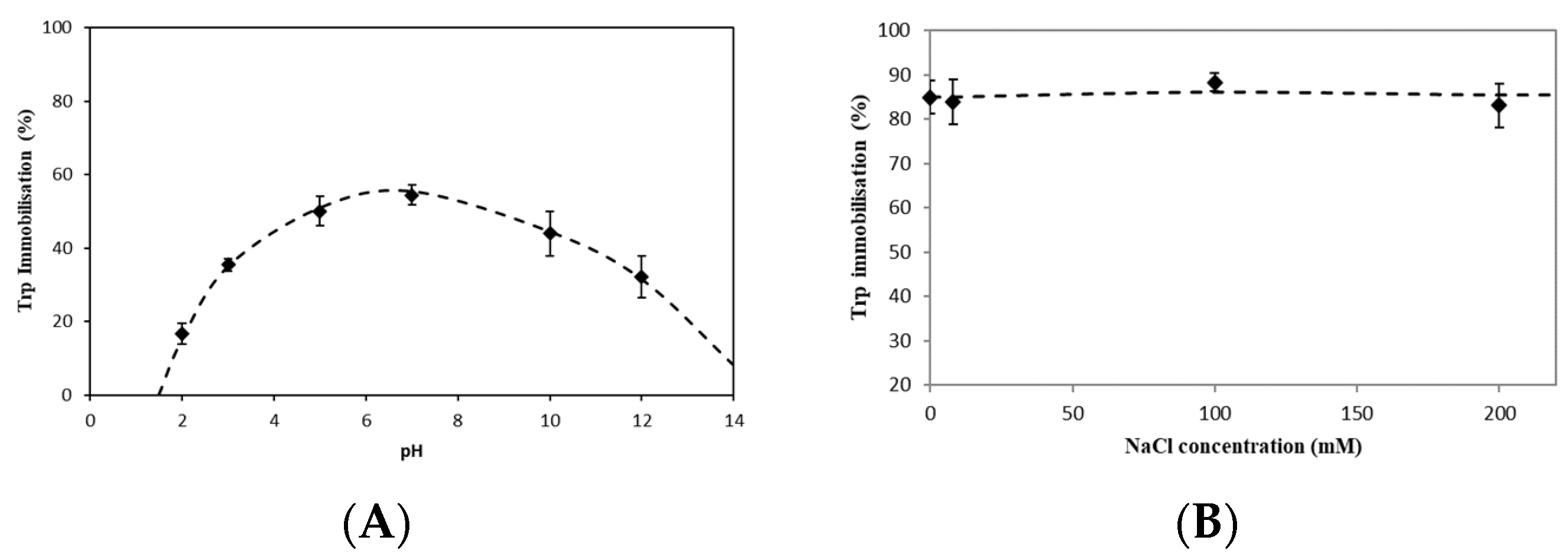
2.1.1. Effects of pH on Amino Acid Immobilization
2.1.2. Effects of Salt on Amino Acid Immobilization
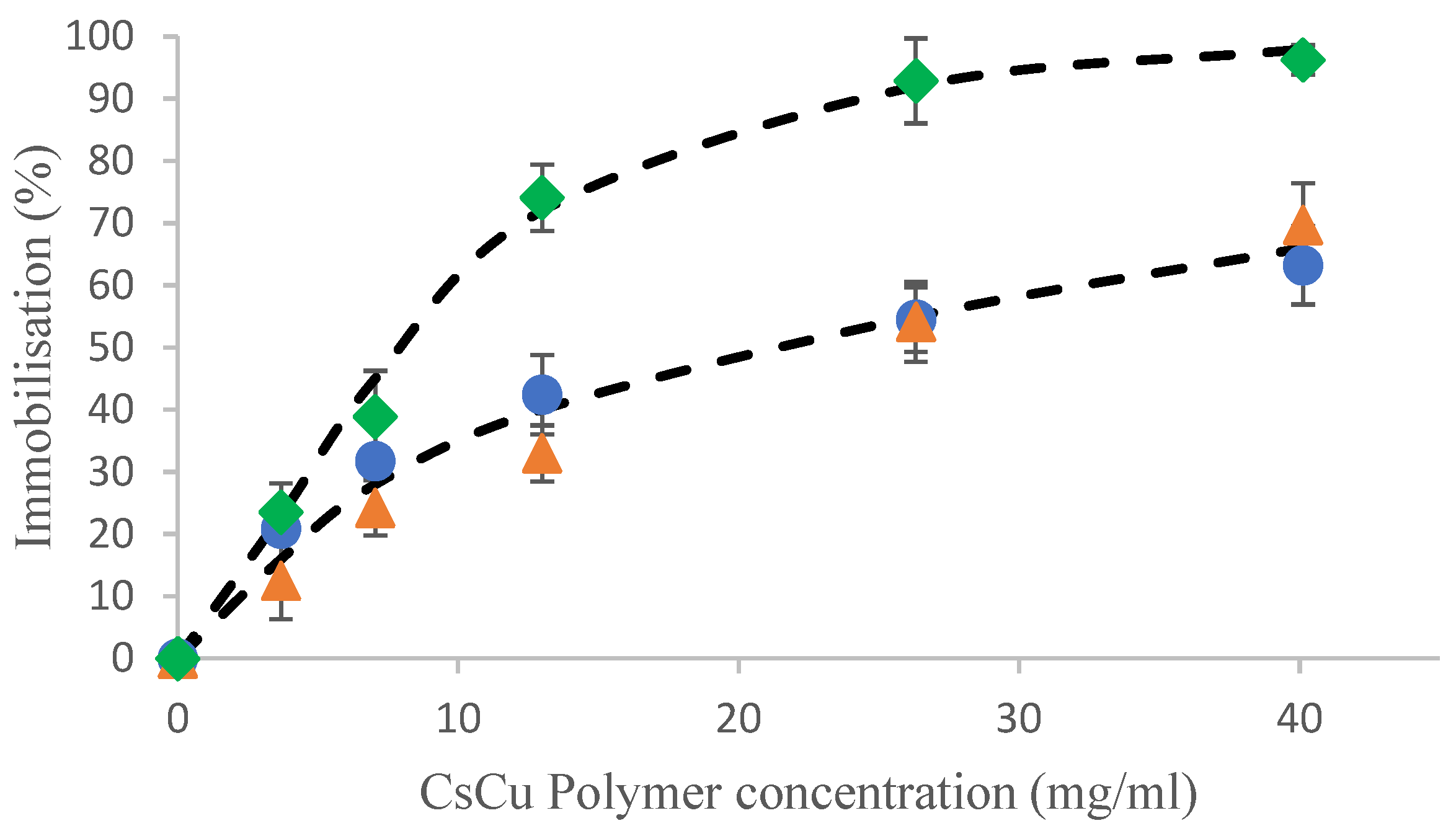
2.2. Hydrolysate Immobilzsation
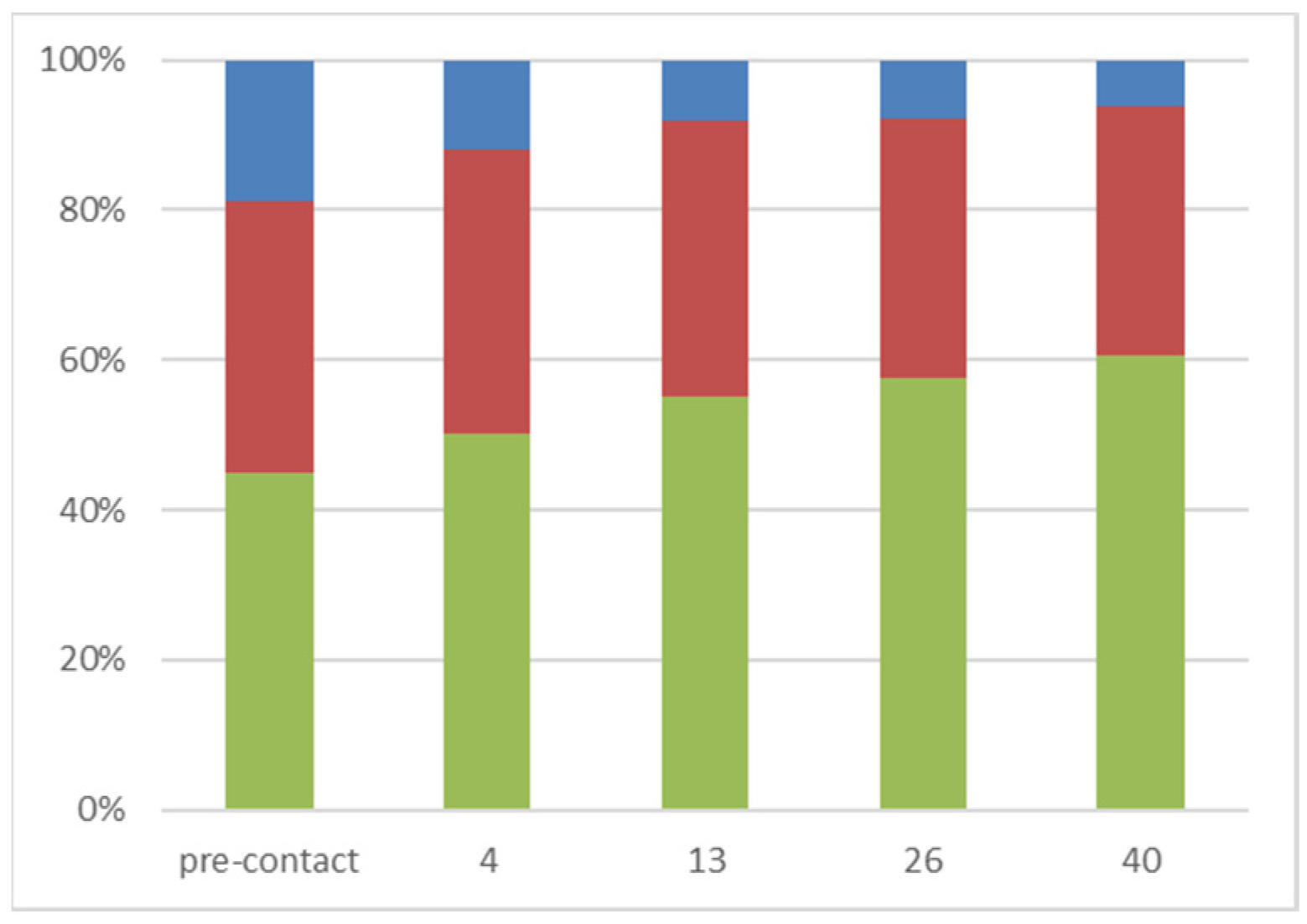
2.2.1. Characterization of the Immobilized Peptides Molecular Weights by SEC
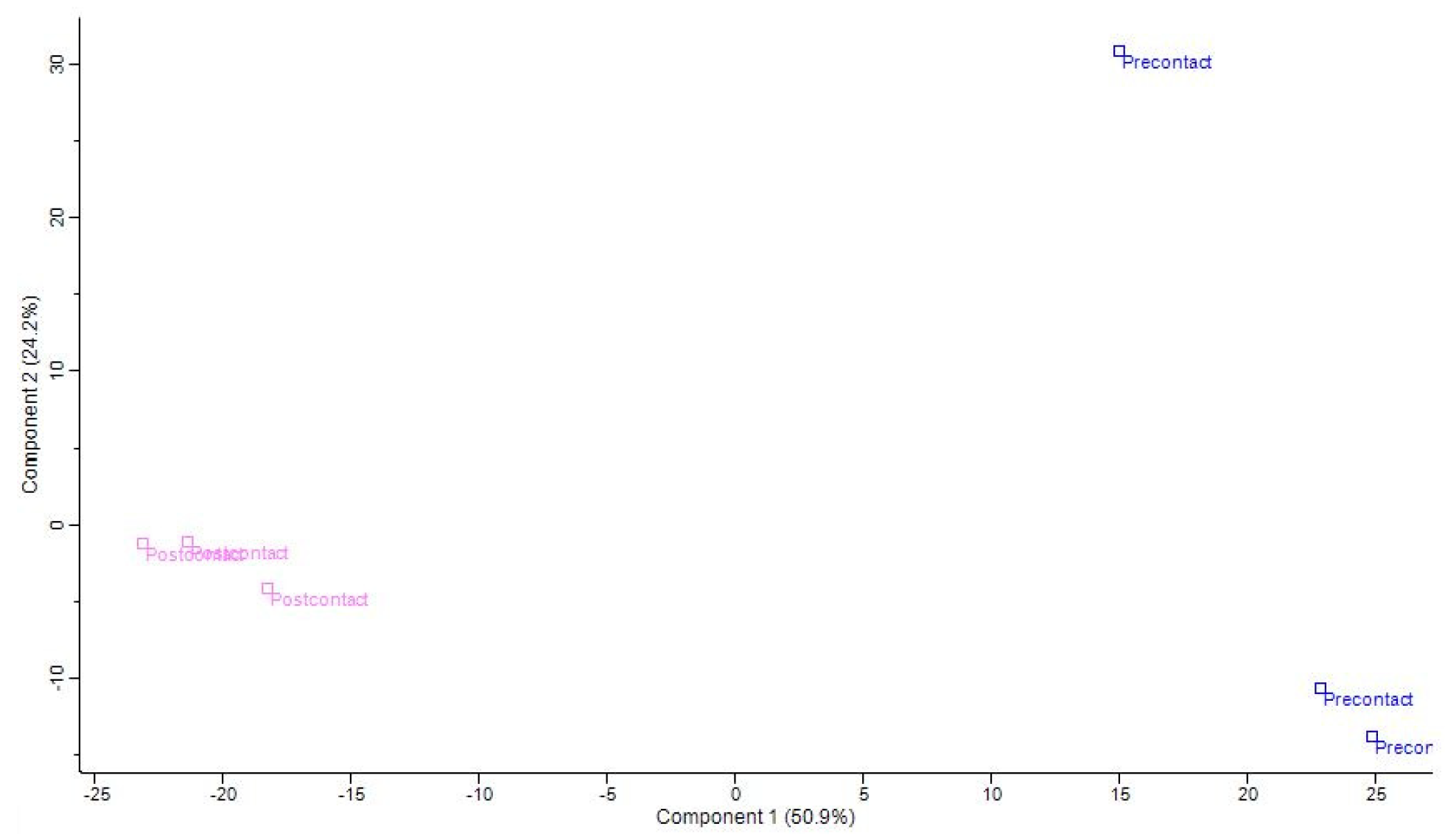
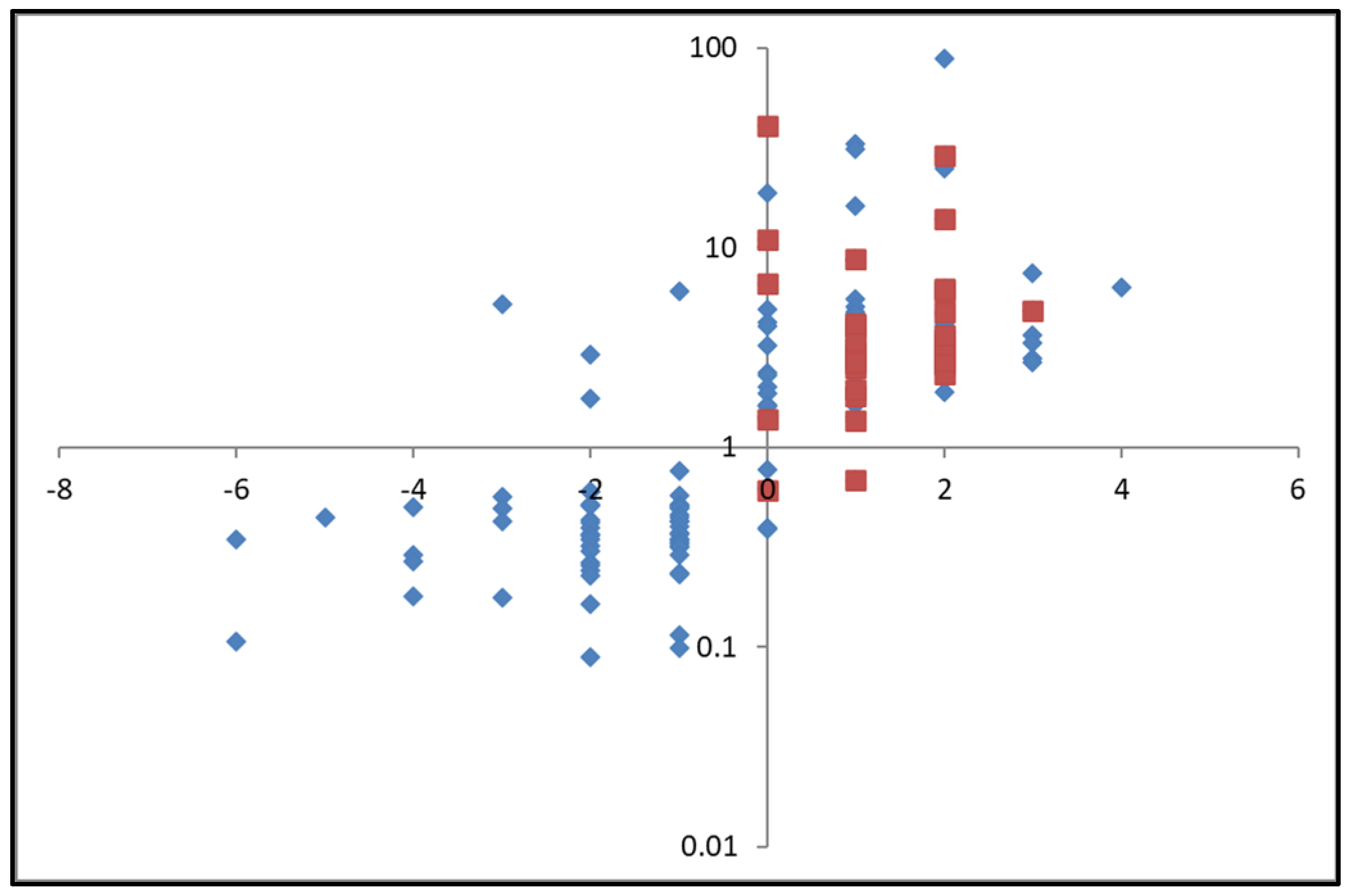
2.2.2. Characterization of the Enriched Peptides by LCMS
3. Conclusions
4. Materials and Methods
4.1. Materials
4.2. Preparation of Copper-Crosslinked Chitosan Gel Microbeads
4.3. Immobilisation of Amino Acids in Gel Beads
4.4. Immobilisation of Sodium Caseinate Proteins and Hydrolysate
LCMS Analysis of the Post-Contact Solution
4.5. Data Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Coates, R.J.; Young, M.T.; Scofield, S. Optimising expression and extraction of recombinant proteins in plants. Front. Plant Sci. 2022, 13, 1074531. [Google Scholar] [CrossRef]
- Du, M.; Hou, Z.; Liu, L.; Xuan, Y.; Chen, X.; Fan, L.; Li, Z.; Xu, B. 1Progress, applications, challenges and prospects of protein purification technology. Front. Bioeng. Biotechnol. 2022, 10, 1028691. [Google Scholar] [CrossRef]
- He, R.; Girgih, A.T.; Rozoy, E.; Bazinet, L.; Ju, X.-R.; Aluko, R.E. Selective separation and concentration of antihypertensive peptides from rapeseed protein hydrolysate by electrodialysis with ultrafiltration membranes. Food Chem. 2016, 197, 1008–1014. [Google Scholar] [CrossRef]
- Um, J.; Manguy, J.; Anes, J.; Jacquier, J.-C.; Hurley, D.; Dillon, E.T.; Wynne, K.; Fanning, S.; O’Sullivan, M.; Shields, D.C. Enriching antimicrobial peptides from milk hydrolysates using pectin/alginate food-gels. Food Chem. 2021, 352, 129220. [Google Scholar] [CrossRef]
- Cermeño, M.; Kleekayai, T.; Amigo-Benavent, M.; Harnedy-Rothwell, P.; FitzGerald, R.J. Current knowledge on the extraction, purification, identification, and validation of bioactive peptides from seaweed. Electrophoresis 2020, 41, 1694–1717. [Google Scholar] [CrossRef]
- Marson, G.V.; Belleville, M.-P.; Lacour, S.; Hubinger, M.D. Membrane Fractionation of Protein Hydrolysates from By-Products: Recovery of Valuable Compounds from Spent Yeasts. Membranes 2021, 11, 23. [Google Scholar] [CrossRef]
- He, J.; Xia, S.; Li, W.; Deng, J.; Lin, Q.; Zhang, L. Resource recovery and valorization of food wastewater for sustainable development: An overview of current approaches. J. Environ. Manag. 2023, 347, 119118. [Google Scholar] [CrossRef]
- Pinrattananon, S.; Courtes, F.; Chorhirankul, N.; Payongsri, P.; Pongtharangkul, T.; Janssen, A.E.M.; Niamsiri, N. The Effect of Different pH Conditions on Peptides’ Separation from the Skipjack Dark Meat Hydrolysate Using Ceramic Ultrafiltration. Foods 2023, 12, 3367. [Google Scholar] [CrossRef]
- Huang, Y.-P.; Robinson, R.C.; Dias, F.F.G.; de Moura Bell, J.M.L.N.; Barile, D. Solid-Phase Extraction Approaches for Improving Oligosaccharide and Small Peptide Identification with Liquid Chromatography-High-Resolution Mass Spectrometry: A Case Study on Proteolyzed Almond Extract. Foods 2022, 11, 340. [Google Scholar] [CrossRef]
- Porath, J. IMAC—Immobilized metal ion affinity based chromatography. TrAC Trends Anal. Chem. 1988, 7, 254–259. [Google Scholar] [CrossRef]
- Ueda, E.; Gout, P.; Morganti, L. Current and prospective applications of metal ion-protein binding. J. Chromatogr. A 2003, 988, 1–23. [Google Scholar] [CrossRef]
- Irankunda, R.; Echavarría, J.A.C.; Paris, C.; Stefan, L.; Desobry, S.; Selmeczi, K.; Muhr, L.; Canabady-Rochelle, L. Metal-Chelating Peptides Separation Using Immobilized Metal Ion Affinity Chromatography: Experimental Methodology and Simulation. Separations 2022, 9, 370. [Google Scholar] [CrossRef]
- Oshima, T.; Tachiyama, H.; Kanemaru, K.; Ohe, K.; Baba, Y. Adsorption and concentration of histidine-containing dipeptides using divalent transition metals immobilized on a chelating resin. Sep. Purif. Technol. 2009, 70, 79–86. [Google Scholar] [CrossRef]
- Wu, J.; Luan, M.; Zhao, J. Trypsin immobilization by direct adsorption on metal ion chelated macroporous chitosan-silica gel beads. Int. J. Biol. Macromol. 2006, 39, 185–191. [Google Scholar] [CrossRef]
- Muzzarelli RA, A.; Tanfani, F.; Muzzarelli, M.G.; Scarpini, G.; Rocchetti, R. Ligand-Exchange Chromatography of Amino Acids on Copper-Loaded Chitosan. Sep. Sci. Technol. 1978, 13, 869–879. [Google Scholar] [CrossRef]
- Hong, F.; Qiu, P.; Wang, Y.; Ren, P.; Liu, J.; Zhao, J.; Gou, D. Chitosan-based hydrogels: From preparation to applications, a review. Food Chem. X 2024, 21, 101095. [Google Scholar] [CrossRef]
- Guibal, E. Interactions of metal ions with chitosan-based sorbents: A review. Sep. Purif. Technol. 2004, 38, 43–74. [Google Scholar] [CrossRef]
- Hamacek, H.S.D.R.; Bresolin, I.T.L.; Fioravante, I.F.; Bueno, S.M.A. Synthesis and characterization of chitosan-polyacrylamide cryogels for the purification of human IgG by IMAC. Process Biochem. 2023, 131, 199–209. [Google Scholar] [CrossRef]
- Shi, Q.-H.; Tian, Y.; Dong, X.-Y.; Bai, S.; Sun, Y. Chitosan-coated silica beads as immobilized metal affinity support for protein adsorption. Biochem. Eng. J. 2003, 16, 317–322. [Google Scholar] [CrossRef]
- Bai, P.; Cao, F.; Lan, X.; Zhao, F.; Ma, Y.; Zhao, C. Chitosan gel beads immobilized Cu (II) for selective adsorption of amino acids. J. Biochem. Biophys. Methods 2008, 70, 903–908. [Google Scholar] [CrossRef]
- Duffy, C.; O’Riordan, D.; O’Sullivan, M.; Jacquier, J. In vitro evaluation of chitosan copper chelate gels as a multimicronutrient feed additive for cattle. J. Sci. Food Agric. 2018, 98, 4177–4183. [Google Scholar] [CrossRef]
- Egan, T.; O’riordan, D.; O’sullivan, M.; Jacquier, J.-C. Cold-set whey protein microgels as pH modulated immobilisation matrices for charged bioactives. Food Chem. 2014, 156, 197–203. [Google Scholar] [CrossRef]
- O’Neill, G.J.; Egan, T.; Jacquier, J.C.; O’sullivan, M.; O’riordan, E.D. Whey microbeads as a matrix for the encapsulation and immobilisation of riboflavin and peptides. Food Chem. 2014, 160, 46–52. [Google Scholar] [CrossRef]
- Kitts, D.D.; Weiler, K. Bioactive Proteins and Peptides from Food Sources. Applications of Bioprocesses used in Isolation and Recovery. Curr. Pharm. Des. 2003, 9, 1309–1323. [Google Scholar] [CrossRef]
- Brown, K.L.; Hancock, R.E. Cationic host defense (antimicrobial) peptides. Curr. Opin. Immunol. 2006, 18, 24–30. [Google Scholar] [CrossRef]
- Hancock, R.E.; Diamond, G. The role of cationic antimicrobial peptides in innate host defences. Trends Microbiol. 2000, 8, 402–410. [Google Scholar] [CrossRef]
- Megías, C.; Pedroche, J.; Yust, M.M.; Girón-Calle, J.; Alaiz, M.; Millán, F.; Vioque, J. Affinity Purification of Copper-Chelating Peptides from Sunflower Protein Hydrolysates. J. Agric. Food Chem. 2007, 55, 6509–6514. [Google Scholar] [CrossRef]
- Megías, C.; Pedroche, J.; Yust, M.M.; Girón-Calle, J.; Alaiz, M.; Millán, F.; Vioque, J. Affinity Purification of Copper Chelating Peptides from Chickpea Protein Hydrolysates. J. Agric. Food Chem. 2007, 55, 3949–3954. [Google Scholar] [CrossRef]
- Meisel, H.; FitzGerald, R.J. Biofunctional Peptides from Milk Proteins: Mineral Binding and Cytomodulatory Effects. Curr. Pharm. Des. 2003, 9, 1289–1295. [Google Scholar] [CrossRef]
- Veltri, D.; Kamath, U.; Shehu, A. Deep learning improves antimicrobial peptide recognition. Bioinformatics 2018, 34, 2740–2747. [Google Scholar] [CrossRef]
- Drummond, E.; Flynn, S.; Whelan, H.; Nongonierma, A.B.; Holton, T.A.; Robinson, A.; Egan, T.; Cagney, G.; Shields, D.C.; Gibney, E.R.; et al. Casein Hydrolysate with Glycemic Control Properties: Evidence from Cells, Animal Models, and Humans. J. Agric. Food Chem. 2018, 66, 4352–4363. [Google Scholar] [CrossRef]
- Yuan, D.; O’Riordan, E.D.; Jacquier, J.C. Development of a first order derivative spectrophotometry method to rapidly quantify protein in the presence of chitosan and its application in protein encapsulation systems. Food Chem. 2019, 289, 1–6. [Google Scholar] [CrossRef]
- Rappsilber, J.; Mann, M.; Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007, 2, 1896–1906. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. Computational principles of determining and improving mass precision and accuracy for proteome measurements in an Orbitrap. J. Am. Soc. Mass Spectrom. 2009, 20, 1477–1485. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate Proteome-wide Label-free Quantification by Delayed Normalization and Maximal Peptide Ratio Extraction, Termed MaxLFQ. Mol. Cell. Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jacquier, J.-C.; Duffy, C.; O’Sullivan, M.; Dillon, E. Copper-Chelated Chitosan Microgels for the Selective Enrichment of Small Cationic Peptides. Gels 2024, 10, 289. https://doi.org/10.3390/gels10050289
Jacquier J-C, Duffy C, O’Sullivan M, Dillon E. Copper-Chelated Chitosan Microgels for the Selective Enrichment of Small Cationic Peptides. Gels. 2024; 10(5):289. https://doi.org/10.3390/gels10050289
Chicago/Turabian StyleJacquier, Jean-Christophe, Ciara Duffy, Michael O’Sullivan, and Eugène Dillon. 2024. "Copper-Chelated Chitosan Microgels for the Selective Enrichment of Small Cationic Peptides" Gels 10, no. 5: 289. https://doi.org/10.3390/gels10050289




