Interaction between Galactomyces geotrichum KL20B, Lactobacillus plantarum LAT3 and Enterococcus faecalis KE06 during Milk Fermentation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microorganisms and Culture Conditions
2.2. Milk Preparation and Fermentation Conditions
2.3. Microorganisms Enumeration
2.4. pH Measurement
2.5. Determination of Organic Acids Content
2.6. Determination of Ethanol Content
2.7. Determination of Amino Acids Content
2.8. Statistical Analysis
3. Results
3.1. Growth Performance of the Co-Cultures
3.2. Organic Acid Accumulation
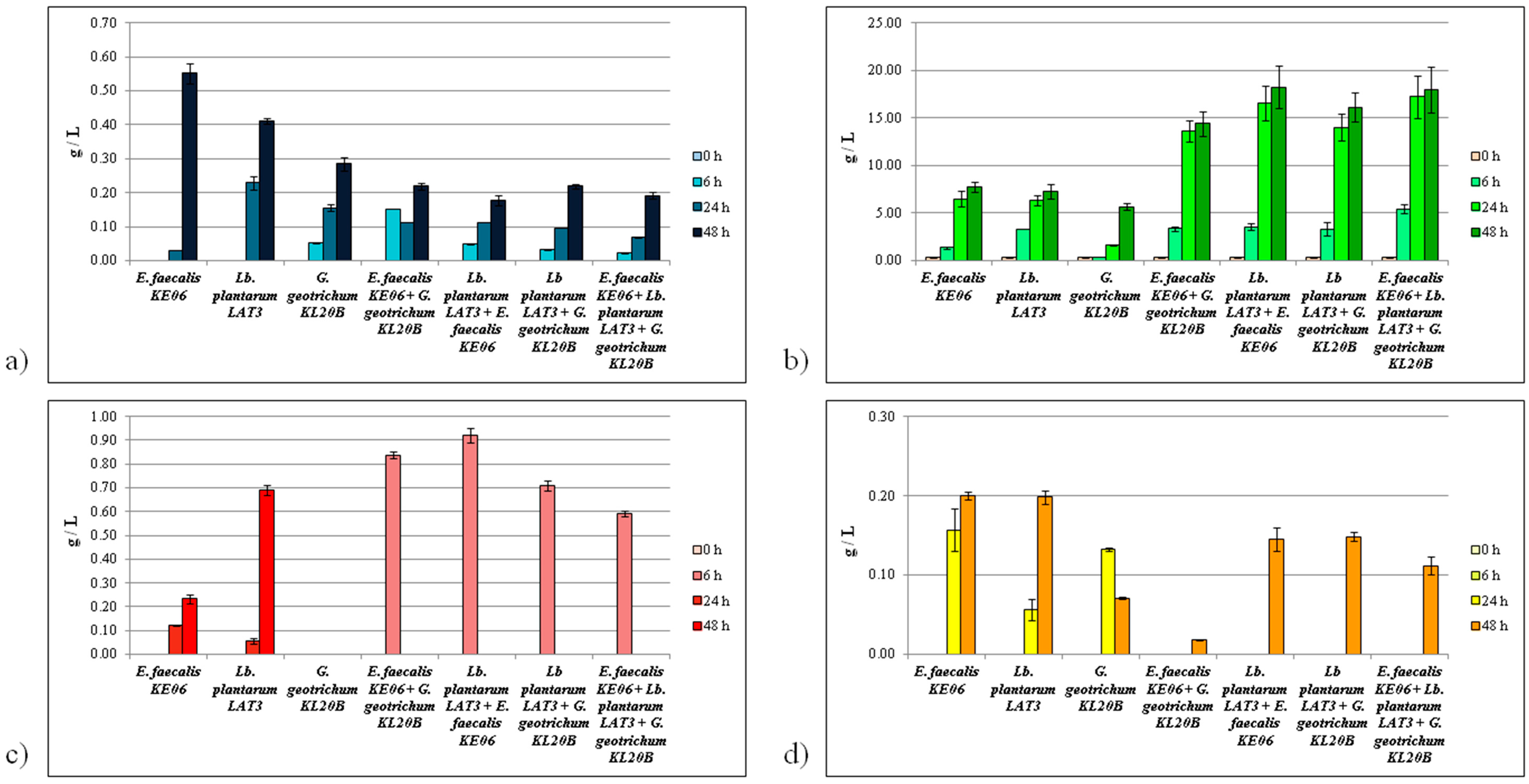
3.2.1. Citric Acid Consumption
3.2.2. Lactic Acid Production
3.2.3. Formic Acid Production
3.2.4. Acetate Production
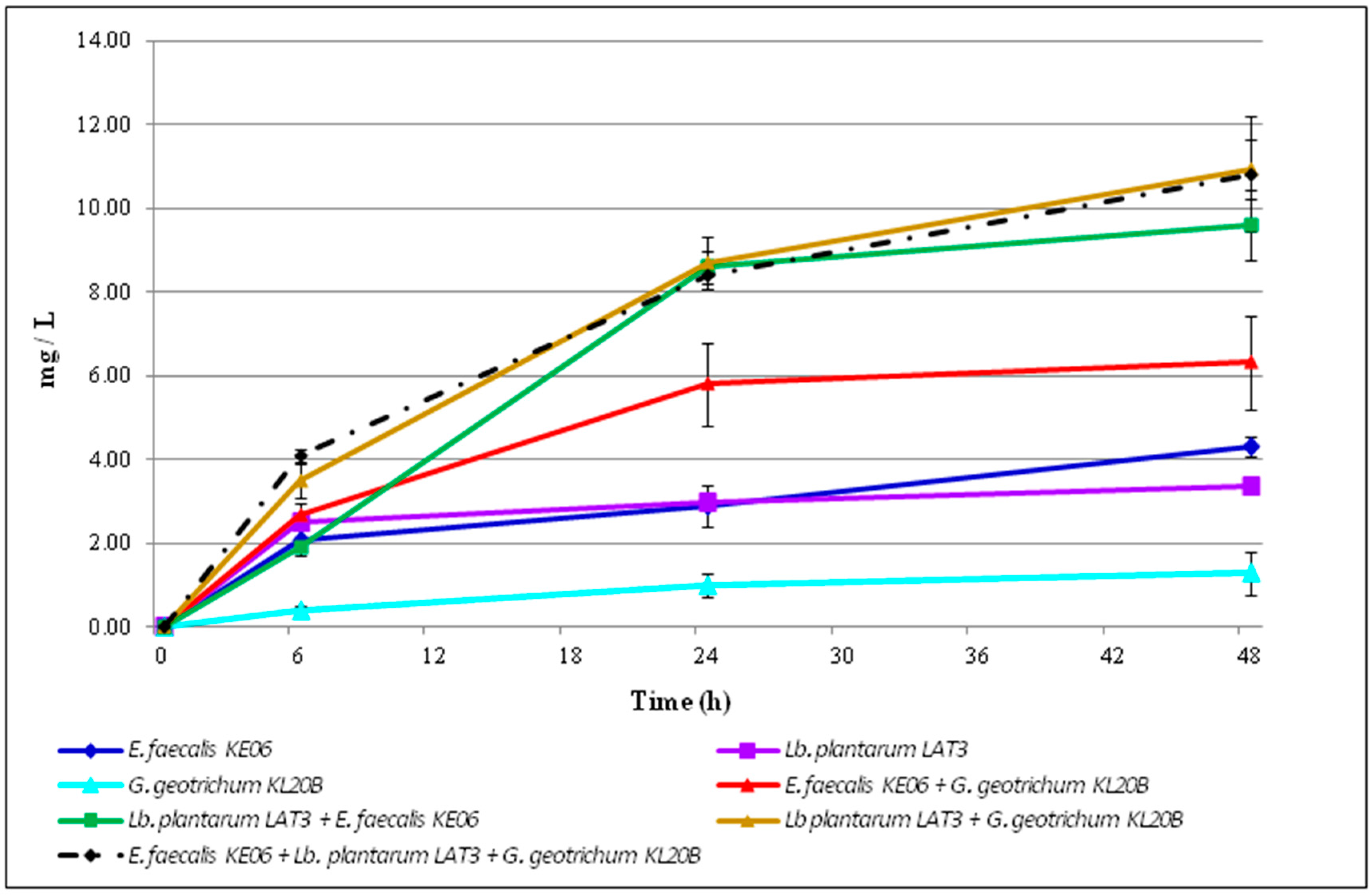
3.3. Ethanol Accumulation
3.4. Total Free Amino Acids Content (FAA)
Author Contributions
Conflicts of Interest
References
- Chaves-López, C.; Tofalo, R.; Serio, A.; Paparella, A.; Sacchetti, G.; Suzzi, G. Yeasts from Colombian Kumis as source of peptides with Angiotensin I converting enzyme (ACE) inhibitory activity in milk. Int. J. Food Microbiol. 2012, 159, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Chaves-López, C.; Serio, A.; Martuscelli, M.; Paparella, A.; Osorio-Cadavid, E.; Suzzi, G. Microbiological characteristics of kumis, a traditional fermented Colombian milk, with particular emphasis on enterococci population. Food Microbiol. 2011, 28, 1041–1047. [Google Scholar] [CrossRef] [PubMed]
- Chaves-López, C.; Serio, A.; Osorio-Cadavid, E.; Paparella, A.; Suzzi, G. Volatile compounds produced in wine by Colombian wild Saccharomyces cerevisiae strains. Ann. Microbiol. 2009, 59, 733–740. [Google Scholar] [CrossRef]
- Akabanda, F.; Owusu-Kwarteng, J.; Tano-Debrah, K.; Glover, R.L.K.; Nielsen, D.S.; Jespersen, L. Taxonomic and molecular characterization of lactic acid bacteria and yeasts in nunu, a Ghanaian fermented milk product. Food Microbiol. 2013, 34, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Akhmetsadykova, S.; Baubekova, A.; Konuspayeva, G.; Akhmetsadykov, N.; Loiseau, G. Microflora identification of fresh and fermented camel milk from Kazakhstan. Emir. J. Food Agric. 2014, 26, 327–332. [Google Scholar] [CrossRef]
- Rossi, C. Produzione di un Latte Fermentato Funzionale con Attività ACE-Inibitoria “In Vitro”. Bachelor’s Thesis, Faculty of Bioscience and Technology for Food, Agriculture and Environment, University of Teramo, Teramo, Italy, 25 October 2010. [Google Scholar]
- Chaves-López, C.; Serio, A.; Paparella, A.; Martuscelli, M.; Corsetti, A.; Tofalo, R.; Suzzi, G. Impact of microbial cultures on proteolysis and release of bioactive peptides in fermented milk. Food Microbiol. 2014, 42, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Jakobsen, M.; Narvhus, J. Yeasts and their possible beneficial and negative effects on the quality of dairy products. Int. Dairy J. 1996, 6, 755–768. [Google Scholar] [CrossRef]
- Viljoen, B. Yeast ecological interactions. Yeast-Yeast, Yeast-Bacteria, Yeast-Fungi interactions and yeasts as biocontrol agents. In Yeasts in Food and Beverages; Querol, A., Fleet, G.H., Eds.; Springer: Berlin, Germany, 2006; pp. 83–110. [Google Scholar]
- Folkertsma, B.; Fox, P.F. Use of the Cd-ninhydrin reagent to assess proteolysis in cheese during ripening. J. Dairy Res. 1992, 59, 217–224. [Google Scholar] [CrossRef]
- Sudun; Wulijideligen; Arakawa, K.; Miyamoto, M.; Miyamoto, T. Interaction between lactic acid bacteria and yeasts in airag, an alcoholic fermented milk. Anim. Sci. J. 2013, 84, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Martín, P.; Flórez, A.B.; Hernández-Barranco, A.; Mayo, B. Interaction between dairy yeasts and lactic acid bacteria strains during milk fermentation. Food Control 2008, 19, 62–70. [Google Scholar] [CrossRef]
- Gadaga, T.H.; Mutukumira, A.N.; Narvhus, J.A. The growth and interaction of yeasts and lactic acid bacteria isolated from Zimbabwean naturally fermented milk in UHT milk. Int. J. Food Microbiol. 2001, 68, 21–32. [Google Scholar] [CrossRef]
- Liu, S.Q.; Tsao, M. Enhancement of survival of probiotic and non-probiotic lactic acid bacteria by yeasts in fermented milk under non-refrigerated conditions. Int. J. Food Microbiol. 2009, 135, 34–38. [Google Scholar] [CrossRef] [PubMed]
- Roostita, R.; Fleet, G.H. Growth of yeasts in milk and associated changes to milk composition. Int. J. Food Microbiol. 1996, 31, 205–219. [Google Scholar] [CrossRef]
- Stadie, J.; Gulitz, A.; Ehrmann, M.A.; Vogel, R.F. Metabolic activity and symbiotic interactions of lactic acid bacteria and yeasts isolated from water kefir. Food Microbiol. 2013, 35, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Leroi, F.; Pidoux, M. Detection of interactions between yeasts and lactic acid bacteria isolated from sugary kefir grains. J. Appl. Microbiol. 1993, 74, 48–53. [Google Scholar] [CrossRef]
- Lonvaud-Funel, A.; Joyeux, A.; Desens, C. Inhibition of malolactic fermentation of wines by products of yeast metabolism. J. Sci. Food Agric. 1988, 44, 183–191. [Google Scholar] [CrossRef]
- Kantachote, D.; Prachyakij, P.; Charernjiratrakul, W.; Ongsakul, M.; Duangjitcharoen, Y.; Chaiyasut, C.; Nitoda, T.; Kanzaki, H. Characterization of the antiyeast compound and probiotic properties of a starter Lactobacillus plantarum DW3 for possible use in fermented plant beverages. Electron. J. Biotechnol. 2010, 13. [Google Scholar] [CrossRef]
- Durlu-Özkaya, F.; Karabicak, N.; Kayali, R.; Esen, B. Inhibition of yeasts isolated from traditional Turkish cheeses by Lactobacillus spp. Int. J. Dairy Technol. 2005, 58, 111–114. [Google Scholar] [CrossRef]
- Nayyeri, N.; Dovom, M.R.E.; Najafi, M.B.H.; Bahreini, M. A Preliminary study on antifungal activity of lactic acid bacteria isolated from different production stages of Lighvan cheese on Penicillium expansum and Rhodotorula mucilaginosa. J. Food Meas. Charact. 2017, 1–11. [Google Scholar] [CrossRef]
- Guo, Z.P.; Olsson, L. Physiological responses to acid stress by Saccharomyces cerevisiae when applying high initial cell density. FEMS Yeast Res. 2016, 16, fow072. [Google Scholar] [CrossRef] [PubMed]
- Rea, M.C.; Cogan, T.M. Catabolite repression in Enterococcus faecalis. Syst. Appl. Microbiol. 2003, 26, 159–164. [Google Scholar] [CrossRef] [PubMed]
- Pottier, I.; Gente, S.; Vernoux, J.P.; Guéguen, M. Safety assessment of dairy microorganisms: Geotrichum candidum. Int. J. Food Microbiol. 2008, 126, 327–332. [Google Scholar] [CrossRef] [PubMed]
- De Vuyst, L.; Vaningelgem, F.; Ghijsels, V.; Tsakalidou, E.; Leroy, F. New insights into the citrate metabolism of Enterococcus faecium FAIR-E 198 and its possible impact on the production of fermented dairy products. Int. Dairy J. 2011, 21, 580–585. [Google Scholar] [CrossRef]
- Sarantinopoulos, P.; Kalantzopoulos, G.; Tsakalidou, E. Citrate metabolism by Enterococcus faecalis FAIR-E 229. Appl. Environ. Microbiol. 2001, 67, 5482–5487. [Google Scholar] [CrossRef] [PubMed]
- Cabral, M.E.; Mukdsi, M.C.A.; Medina de Figueroa, R.B.; Gonzalez, S.N. Citrate metabolism by Enterococcus faecium and Enterococcus durans isolated from goat’s and ewe’s milk: Influence of glucose and lactose. Can. J. Microbiol. 2007, 53, 607–615. [Google Scholar] [CrossRef] [PubMed]
- Kebede, A.; Viljoen, B.C.; Gadaga, T.H.; Narvhus, J.A.; Lourens-Hattingh, A. The effect of container type on the growth of yeast and lactic acid bacteria during production of Sethemi, South African spontaneously fermented milk. Food Res. Int. 2007, 40, 33–38. [Google Scholar] [CrossRef]
- Fleet, G.H. Yeasts in dairy products. J. Appl. Bacteriol. 1990, 68, 199–211. [Google Scholar] [CrossRef] [PubMed]
- Sarais, I.; Piussi, D.; Aquili, V.; Stecchini, M.L. The behavior of yeast populations in Stracchino cheese packaged under various conditions. J. Food Prot. 1996, 59, 541–544. [Google Scholar] [CrossRef]
- Starke, I.C.; Zentek, J.; Vahjen, W. Effects of the probiotic Enterococcus faecium NCIMB 10415 on selected lactic acid bacteria and enterobacteria in co-culture. Benef. Microbes 2015, 6, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Sieuwerts, S. Analysis of Molecular Interactions between Yoghurt Bacteria by an Integrated Genomics Approach. Ph.D. Thesis, Wageningen University, Wageningen, The Netherlands, 27 October 2009. [Google Scholar]
- Hugenholtz, J. Citrate metabolism in lactic acid bacteria. FEMS Microbiol. Rev. 1993, 12, 165–178. [Google Scholar] [CrossRef]
- Oude Elferink, S.J.W.H.; Krooneman, J.; Gottschal, J.C.; Spoelstra, S.F.; Faber, F.; Driehuis, F. Anaerobic conversion of lactic acid to acetic acid and 1,2-propanediol by Lactobacillus buchneri. Appl. Environ. Microbiol. 2001, 67, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Zalán, Z.; Hudáček, J.; Štětina, J.; Chumchalová, J.; Halász, A. Production of organic acids by Lactobacillus strains in three different media. Eur. Food Res. Technol. 2010, 230, 395. [Google Scholar] [CrossRef]
- Salazar, N.; Prieto, A.; Leal, J.A.; Mayo, B.; Bada-Gancedo, J.C.; de Los Reyes-Gavilán, C.G.; Ruas-Madiedo, P. Production of exopolysaccharides by Lactobacillus and Bifidobacterium strains of human origin, and metabolic activity of the producing bacteria in milk. J. Dairy Sci. 2009, 92, 4158–4168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Vos, W.M.; Hugenholtz, J. Engineering metabolic highways in Lactococci and other lactic acid bacteria. Trends Biotechnol. 2004, 22, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Heath, E.C.; Hurwitz, J.; Horecker, B.L.; Ginsburg, A. Pentose fermentation by Lactobacillus plantarum. I. The cleavage of xylulose 5-phosphate by phosphoketolase. J. Biol. Chem. 1958, 231, 1009–1029. [Google Scholar] [PubMed]
- Chaves-López, C.; Serio, A.; Grande-Tovar, C.D.; Cuervo-Mulet, R.; Delgado-Ospina, J.; Paparella, A. Traditional fermented foods and beverages from a microbiological and nutritional perspective: The Colombian heritage. Compr. Rev. Food Sci. Food Saf. 2014, 13, 1031–1048. [Google Scholar] [CrossRef]
- Irigoyen, A.; Ortigosa, M.; Garcia, S.; Ibanez, F.C.; Torre, P. Comparison of free amino acids and volatile components in three fermented milks. Int. J. Dairy Technol. 2012, 65, 578–584. [Google Scholar] [CrossRef]
- Serio, A.; Chaves-López, C.; Paparella, A.; Suzzi, G. Evaluation of metabolic activities of enterococci isolated from Pecorino Abruzzese cheese. Int. Dairy J. 2010, 20, 459–464. [Google Scholar] [CrossRef]
- Serio, A.; Paparella, A.; Chaves-López, C.; Corsetti, A.; Suzzi, G. Enterococcus populations in Pecorino Abruzzese cheese: Biodiversity and safety aspects. J. Food Prot. 2007, 70, 1561–1568. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Bayjanov, J.R.; Renckens, B.; Nauta, A.; Siezen, R.J. The proteolytic system of lactic acid bacteria revisited: A genomic comparison. BMC Genom. 2010, 11, 36. [Google Scholar] [CrossRef] [PubMed]
- Grygier, A.; Myszka, K.; Rudzińska, M. Galactomyces geotrichum—Moulds from dairy products with high biotechnological potential. Acta Sci. Pol. Technol. Aliment. 2017, 16, 5–16. [Google Scholar] [CrossRef] [PubMed]

| Total Free Amino Acids (mM Leucine) | 0 h | 6 h | 24 h | 48 h |
|---|---|---|---|---|
| E. faecalis KE06 | 0.047 ± 0.003 a | 0.074 ± 0.012 b | 0.119 ± 0.031 c | 0.202 ± 0.042 d |
| Lb. plantarum LAT3 | 0.047 ± 0.003 a | 0.022 ± 0.010 b | 0.029 ± 0.007 b | 0.060 ± 0.010 c |
| G. geotrichum KL20B | 0.047 ± 0.003 a | 0.058 ± 0.004 b | 0.063 ± 0.016 b | 0.196 ± 0.022 c |
| E. faecalis KE06 + G. geotrichum KL20B | 0.047 ± 0.003 a | 0.111 ± 0.033 b | 0.068 ± 0.003 c | 0.061 ± 0.008 c |
| Lb. plantarum LAT3 + E. faecalis KE06 | 0.047 ± 0.003 a | 0.053 ± 0.005 ab | 0.057 ± 0.006 b | 0.057 ± 0.003 b |
| Lb plantarum LAT3 + G. geotrichum KL20B | 0.047 ± 0.003 a | 0.037 ± 0.004 b | 0.048 ± 0.004 a | 0.059 ± 0.002 b |
| E. faecalis KE06 + Lb. plantarum LAT3 + G. geotrichum KL20B | 0.047 ± 0.003 a | 0.037 ± 0.002 b | 0.064 ± 0.003 c | 0.053 ± 0.002 d |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chaves-López, C.; Serio, A.; Rossi, C.; Pepe, A.; Compagnone, E.; Paparella, A. Interaction between Galactomyces geotrichum KL20B, Lactobacillus plantarum LAT3 and Enterococcus faecalis KE06 during Milk Fermentation. Fermentation 2017, 3, 52. https://doi.org/10.3390/fermentation3040052
Chaves-López C, Serio A, Rossi C, Pepe A, Compagnone E, Paparella A. Interaction between Galactomyces geotrichum KL20B, Lactobacillus plantarum LAT3 and Enterococcus faecalis KE06 during Milk Fermentation. Fermentation. 2017; 3(4):52. https://doi.org/10.3390/fermentation3040052
Chicago/Turabian StyleChaves-López, Clemencia, Annalisa Serio, Chiara Rossi, Alessia Pepe, Elisabetta Compagnone, and Antonello Paparella. 2017. "Interaction between Galactomyces geotrichum KL20B, Lactobacillus plantarum LAT3 and Enterococcus faecalis KE06 during Milk Fermentation" Fermentation 3, no. 4: 52. https://doi.org/10.3390/fermentation3040052