Enhanced Phycocyanobilin Production in Escherichia coli by Fusion-Expression of Apo-Proteins with Signal Peptides
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cyanobacterial Cultivation and DNA Extraction
2.2. Obtaining of the Gene Encoding Phycocyanin
2.3. Bacterial Strains and Culture Conditions
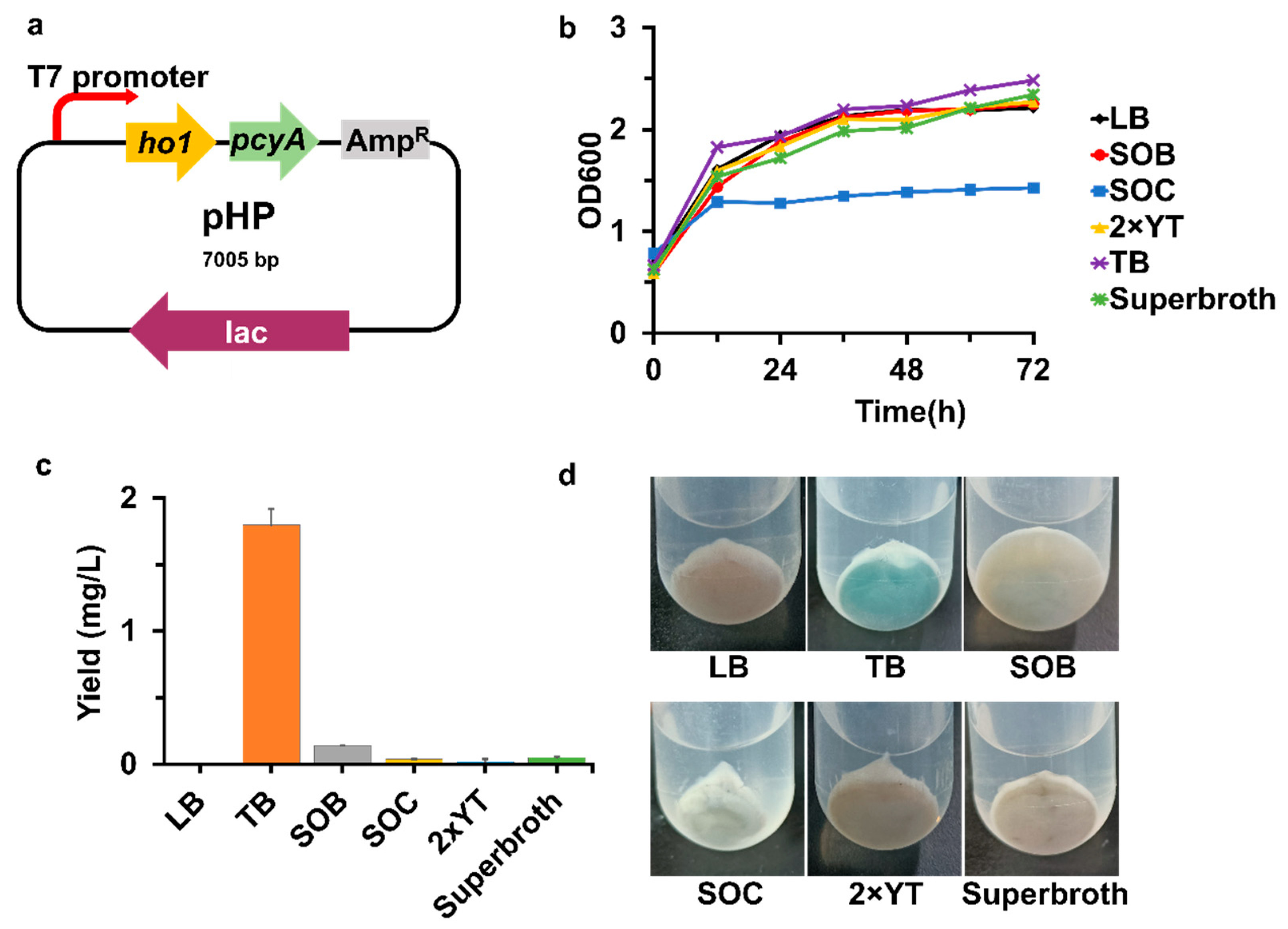
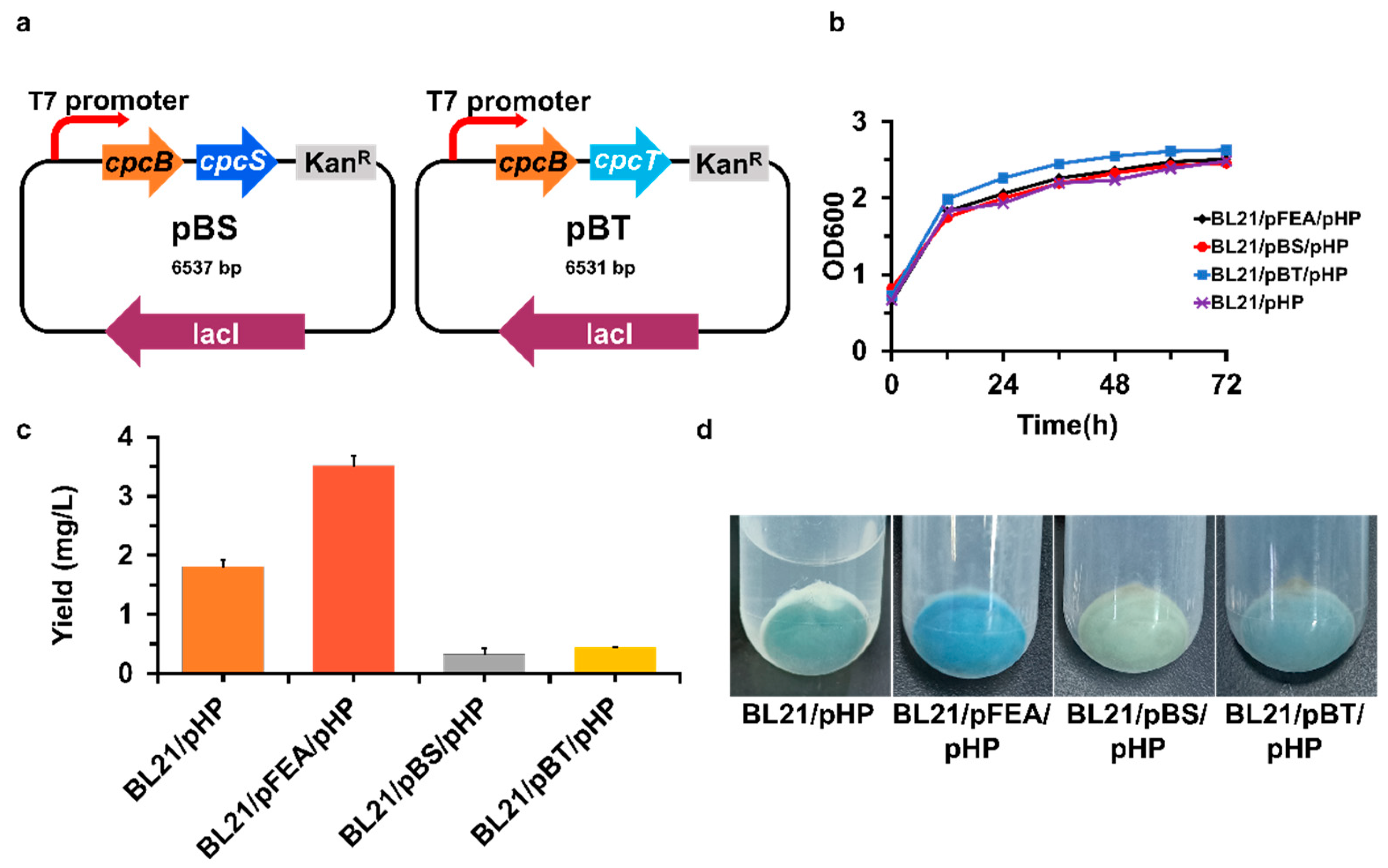
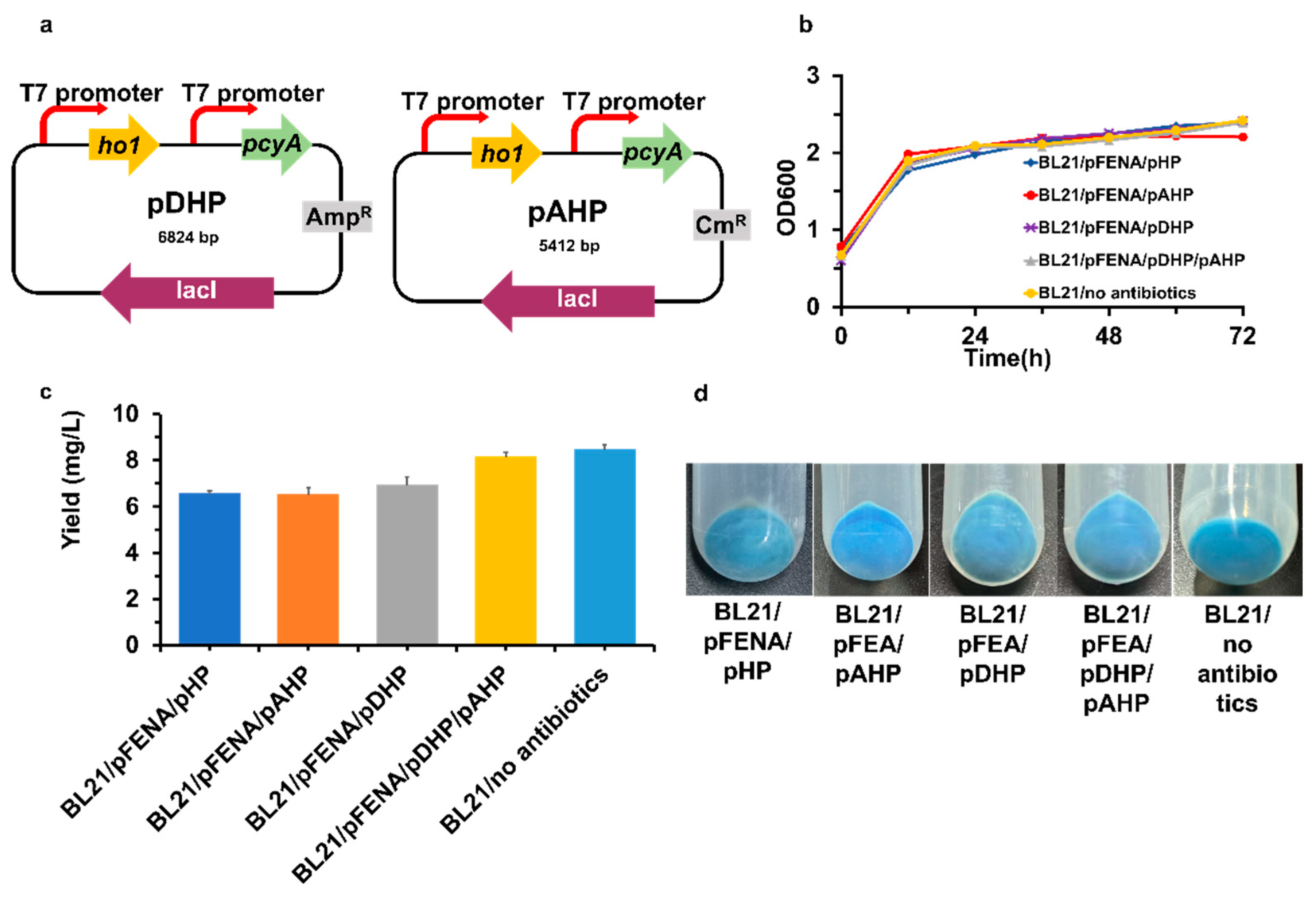
2.4. Plasmid Construction
2.5. PCB Standard Curve
2.6. SDS–PAGE Analysis
2.7. Alcoholysis
2.8. Abbreviations
3. Results
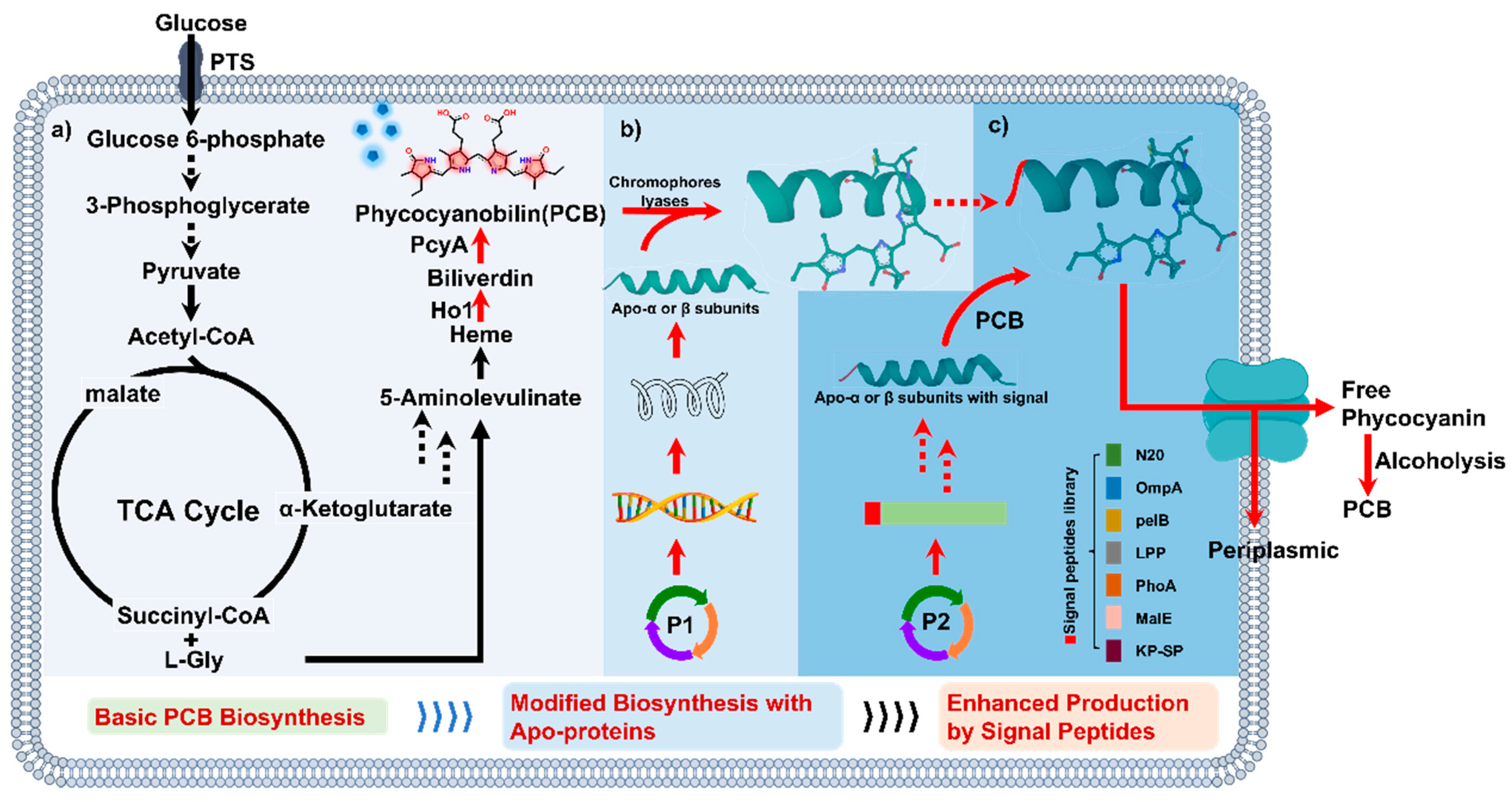
3.1. Heterologous Production of PCB in E. coli through the Heme Catabolic Pathway
3.2. Enhancing the PCB Yield by Introducing Apo-α or -β Protein
3.3. Enhancing PCB Production by Fusion-Expression of Apo-α Protein with Signal Peptide N20
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Biswas, A.; Vasquez, Y.M.; Dragomani, T.M.; Kronfel, M.L.; Williams, S.R.; Alvey, R.M.; Bryant, D.A.; Schluchter, W.M. Biosynthesis of cyanobacterial phycobiliproteins in Escherichia coli: Chromophorylation efficiency and specificity of all bilin lyases from Synechococcus sp. strain PCC 7002. Appl. Environ. Microbiol. 2010, 76, 2729–2739. [Google Scholar] [CrossRef] [PubMed]
- Malwade, C.R.; Roda-Serrat, M.C.; Christensen, K.V.; Fretté, X.; Christensen, L.P. Kinetics of Phycocyanobilin Cleavage from C-Phycocyanin by Methanolysis. Comput. Aided Chem. Eng. 2016, 38, 61–66. [Google Scholar] [CrossRef]
- Koníčková, R.; Vaňková, K.; Vaníková, J.; Váňová, K.; Muchová, L.; Subhanová, I.; Zadinová, M.; Zelenka, J.; Dvořák, A.; Kolář, M. Anti-cancer effects of blue-green alga Spirulina platensis, a natural source of bilirubin-like tetrapyrrolic compounds. Ann. Hepatol. 2014, 13, 273–283. [Google Scholar] [CrossRef]
- Marín-Prida, J.; Pavón-Fuentes, N.; Llópiz-Arzuaga, A.; Fernández-Massó, J.R.; Delgado-Roche, L.; Mendoza-Marí, Y.; Santana, S.P.; Cruz-Ramírez, A.; Valenzuela-Silva, C.; Nazábal-Gálvez, M. Phycocyanobilin promotes PC12 cell survival and modulates immune and inflammatory genes and oxidative stress markers in acute cerebral hypoperfusion in rats. Toxicol. Appl. Pharmacol. 2013, 272, 49–60. [Google Scholar] [CrossRef]
- Strasky, Z.; Zemankova, L.; Nemeckova, I.; Rathouska, J.; Nachtigal, P. Spirulina platensis and phycocyanobilin activate atheroprotective heme oxygenase-1: A possible implication for atherogenesis. Food Funct. 2013, 4, 1586–1594. [Google Scholar] [CrossRef]
- Zheng, J.; Inoguchi, T.; Sasaki, S.; Maeda, Y.; McCarty, M.F.; Fujii, M.; Ikeda, N.; Kobayashi, K.; Sonoda, N.; Takayanagi, R. Phycocyanin and phycocyanobilin from Spirulina platensis protect against diabetic nephropathy by inhibiting oxidative stress. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2013, 304, R110–R120. [Google Scholar] [CrossRef]
- Garcia-Pliego, E.; Franco-Colin, M.; Rojas-Franco, P.; Blas-Valdivia, V.; Serrano-Contreras, J.I.; Pentón-Rol, G.; Cano-Europa, E. Phycocyanobilin is the molecule responsible for the nephroprotective action of phycocyanin in acute kidney injury caused by mercury. Food Funct. 2021, 12, 2985–2994. [Google Scholar] [CrossRef]
- Encarnação, T.; Pais, A.A.C.C.; Campos, M.G.; Burrows, H.D. Cyanobacteria and Microalgae: A Renewable Source of Bioactive Compounds and Other Chemicals. Sci. Prog. 2015, 98, 145–168. [Google Scholar] [CrossRef]
- Beale, S.I. Biosynthesis of phycobilins. Chem. Rev. 1993, 93, 785–802. [Google Scholar] [CrossRef]
- Dammeyer, T.; Michaelsen, K.; Frankenberg-Dinkel, N. Biosynthesis of open-chain tetrapyrroles in Prochlorococcus marinus. FEMS Microbiol. Lett. 2007, 271, 251–257. [Google Scholar] [CrossRef]
- Ge, B.; Li, Y.; Sun, H.; Zhang, S.; Hu, P.; Qin, S.; Huang, F. Combinational biosynthesis of phycocyanobilin using genetically-engineered Escherichia coli. Biotechnol. Lett. 2013, 35, 689–693. [Google Scholar] [CrossRef]
- Craig, I.W.; Carr, N.G. C-phycocyanin and allophycocyanin in two species of blue-green algae. Biochem. J. 1968, 106, 361–366. [Google Scholar] [CrossRef] [PubMed]
- Pagels, F.; Guedes, A.C.; Amaro, H.M.; Kijjoa, A.; Vasconcelos, V. Phycobiliproteins from cyanobacteria: Chemistry and biotechnological applications. Biotechnol. Adv. 2019, 37, 422–443. [Google Scholar] [CrossRef]
- Wu, X.J.; Yang, H.; Chen, Y.T.; Li, P.P. Biosynthesis of Fluorescent beta Subunits of C-Phycocyanin from Spirulina subsalsa in Escherichia coli, and Their Antioxidant Properties. Molecules 2018, 23, 1369. [Google Scholar] [CrossRef]
- Guan, X.; Qin, S.; Su, Z.; Zhao, F.; Ge, B.; Li, F.; Tang, X. Combinational biosynthesis of a fluorescent cyanobacterial holo-alpha-phycocyanin in Escherichia coli by using one expression vector. Appl. Biochem. Biotechnol. 2007, 142, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Li, P.; Chen, X.; Chao, X. Combinatorial biosynthesis of Synechocystis PCC6803 phycocyanin holo-alpha-subunit (CpcA) in Escherichia coli and its activities. Appl. Microbiol. Biotechnol. 2016, 100, 5375–5388. [Google Scholar] [CrossRef]
- Zhao, K.H.; Su, P.; Tu, J.M.; Wang, X.; Liu, H.; Plöscher, M.; Eichacker, L.; Yang, B.; Zhou, M.; Scheer, H. Phycobilin: Cystein-84 biliprotein lyase, a near-universal lyase for cysteine-84-binding sites in cyanobacterial phycobiliproteins. Proc. Natl. Acad. Sci. USA 2007, 104, 14300–14305. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.; Xu, D.; Zang, X.; Yuan, D.; Zhao, B.; Tang, L.; Tan, Y.; Zhang, X. Lyase activities of heterologous CpcS and CpcT for phycocyanin holo-β-subunit from Arthrospira platensis in Escherichia coli. J. Ocean. Univ. China 2014, 13, 497–502. [Google Scholar] [CrossRef]
- Tooley, A.J.; Glazer, A.N. Biosynthesis of the Cyanobacterial Light-Harvesting Polypeptide Phycoerythrocyanin Holo-α Subunit in a Heterologous Host. J. Bacteriol. 2002, 184, 4666–4671. [Google Scholar] [CrossRef]
- Tooley, A.J.; Cai, Y.A.; Glazer, A.N. Biosynthesis of a fluorescent cyanobacterial C-phycocyanin holo-alpha subunit in a heterologous host. Proc. Natl. Acad. Sci. USA 2001, 98, 10560–10565. [Google Scholar] [CrossRef] [PubMed]
- Ge, B.; Lin, X.; Chen, Y.; Wang, X.; Chen, H.; Jiang, P.; Huang, F. Combinational biosynthesis of dual-functional streptavidin-phycobiliproteins for high-throughput-compatible immunoassay. Process Biochem. 2017, 58, 306–312. [Google Scholar] [CrossRef]
- Ge, B.; Chen, Y.; Yu, Q.; Lin, X.; Li, J.; Qin, S. Regulation of the heme biosynthetic pathway for combinational biosynthesis of phycocyanobilin in Escherichia coli. Process Biochem. 2018, 71, 23–30. [Google Scholar] [CrossRef]
- Danese, P.N.; Silhavy, T.J. Targeting and assembly of periplasmic and outer-membrane proteins in Escherichia coli. Annu. Rev. Genet. 1998, 32, 59–94. [Google Scholar] [CrossRef] [PubMed]
- von Heijne, G. The signal peptide. J. Membr. Biol. 1990, 115, 195–201. [Google Scholar] [CrossRef]
- Pan, X.; Yu, Q.; Chu, J.; Jiang, T.; He, B. Fitting replacement of signal peptide for highly efficient expression of three penicillin G acylases in E. coli. Appl. Microbiol. Biotechnol. 2018, 102, 7455–7464. [Google Scholar] [CrossRef]
- Pang, C.; Liu, S.; Zhang, G.; Zhou, J.; Du, G.; Li, J. Enhancing extracellular production of lipoxygenase in Escherichia coli by signal peptides and autolysis system. Microb. Cell Fact. 2022, 21, 42–53. [Google Scholar] [CrossRef]
- Miksch, G.; Kleist, S.; Friehs, K.; Flaschel, E. Overexpression of the phytase from Escherichia coli and its extracellular production in bioreactors. Appl. Microbiol. Biotechnol. 2002, 59, 685–694. [Google Scholar] [CrossRef]
- Jeong, K.J.; Lee, S.Y. Secretory production of human leptin in Escherichia coli. Biotechnol. Bioeng. 2000, 67, 398–407. [Google Scholar] [CrossRef]
- Fidan, O.; Zhan, J. Discovery and engineering of an endophytic Pseudomonas strain from Taxus chinensis for efficient production of zeaxanthin diglucoside. J. Biol. Eng. 2019, 13, 66–83. [Google Scholar] [CrossRef]
- Cao, X.; Zang, X.; Liu, Z.; Jin, Y.; Sun, D.; Guo, Y.; Wang, Z.; Zhang, F.; Lin, J. Molecular cloning of the cpeT gene encoding a bilin lyase responsible for attachment of phycoerythrobilin to Cys-158 on the β-subunit of phycoerythrin in Gracilariopsis lemaneiformis. J. Appl. Phycol. 2019, 31, 3331–3340. [Google Scholar] [CrossRef]
- Voss, M.; Schröder, B.; Fluhrer, R. Mechanism, specificity, and physiology of signal peptide peptidase (SPP) and SPP-like proteases. Biochim. Biophys. Acta. 2013, 1828, 2828–2839. [Google Scholar] [CrossRef]
- Cui, Y.; Meng, Y.; Zhang, J.; Cheng, B.; Yin, H.; Gao, C.; Xu, P.; Yang, C. Efficient secretory expression of recombinant proteins in Escherichia coli with a novel actinomycete signal peptide. Protein Expr. Purif. 2017, 129, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Gao, D.; Luan, Y.; Liang, Q.; Qi, Q. Exploring the N-terminal role of a heterologous protein in secreting out of Escherichia coli. Biotechnol. Bioeng. 2016, 113, 2561–2567. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, B.A.; Admiraal, S.J.; Gramajo, H.; Cane, D.E.; Khosla, C. Biosynthesis of complex polyketides in a metabolically engineered strain of E. coli. Science 2001, 291, 1790–1792. [Google Scholar] [CrossRef]
- Liu, X.; Hua, K.; Liu, D.; Wu, Z.; Wang, Y.; Zhang, H.; Deng, Z.; Pfeifer, B.A.; Jiang, M. Heterologous Biosynthesis of Type II Polyketide Products Using E. coli. ACS Chem. Biol. 2020, 15, 1177–1183. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Chen, R. Extracellular recombinant protein production from Escherichia coli. Biotechnol. Lett. 2009, 31, 1661–1670. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, H.; Zhou, C.; Ma, Y.; Li, J.; Song, J. Cloning, expression and characterization of a pectate lyase from Paenibacillus sp. 0602 in recombinant Escherichia coli. BMC Biotechnol. 2014, 14, 18–29. [Google Scholar] [CrossRef]
- Teng, C.; Jia, H.; Yan, Q.; Zhou, P.; Jiang, Z. High-level expression of extracellular secretion of a β-xylosidase gene from Paecilomyces thermophila in Escherichia coli. Bioresour. Technol. 2011, 102, 1822–1830. [Google Scholar] [CrossRef] [PubMed]

| Signal Sequences | Amino Acid Sequences | Expression Level | |
|---|---|---|---|
| CpcA | CpcB | ||
| pelB (pectate lyase B) [25] | MKYLLPTAAAGLLLLAAQPAMA | + | − |
| Kp-SP [32] | MSRRAPLLRAGAATAVAVLYLTAVPQAASA | + | − |
| PhoA (alkaline phosphatase) [25] | MKQSTIALALLPLLFTPVTKA | + | + |
| OmpA (outer membrane protein A) [25] | MKKTAIAIAVALAGFATVAQA | − | − |
| MalE (maltose-binding protein) [25] | MKIKTGARILALSALTTMMFSASALA | + | ++ |
| Lpp (murein lipoprotein) [25] | MKATKLVLGAVILGSTLLAG | − | − |
| N20 [33] | MEGNTREDNFKHLLGNDNVKRPSEA | +++ | |
| Native apo-CpcA | ++ | ||
| Native apo-CpcB | ++ | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Yu, J.; Che, Q.; Zhu, T.; Li, D.; Zhang, G. Enhanced Phycocyanobilin Production in Escherichia coli by Fusion-Expression of Apo-Proteins with Signal Peptides. Fermentation 2023, 9, 851. https://doi.org/10.3390/fermentation9090851
Liu X, Yu J, Che Q, Zhu T, Li D, Zhang G. Enhanced Phycocyanobilin Production in Escherichia coli by Fusion-Expression of Apo-Proteins with Signal Peptides. Fermentation. 2023; 9(9):851. https://doi.org/10.3390/fermentation9090851
Chicago/Turabian StyleLiu, Xiaolin, Jing Yu, Qian Che, Tianjiao Zhu, Dehai Li, and Guojian Zhang. 2023. "Enhanced Phycocyanobilin Production in Escherichia coli by Fusion-Expression of Apo-Proteins with Signal Peptides" Fermentation 9, no. 9: 851. https://doi.org/10.3390/fermentation9090851





