Tenacibaculum ovolyticum 16S rDNA Quantitative-PCR Assay Development and Field Testing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Assay Development
2.1.1. Isolates and DNA Used
2.1.2. Alignments and Oligonucleotide Generation
2.1.3. Temperature Gradient Test and Amplicon Sequencing
2.1.4. Primer and Probe Optimization
2.1.5. Standard Curves (Sensitivity and Amplification Efficiency Testing)
2.1.6. Outgroup Testing and Fluorescence in Mixed Cultures
2.2. Net-Pen Sample Screening
2.2.1. Sample Descriptions
2.2.2. DNA Extractions
2.2.3. qPCR Application
3. Results
3.1. Assay Development
3.1.1. Temperature Gradient Test and Amplicon Sequencing
3.1.2. Primer and Probe Optimization
3.1.3. Standard Curves (Sensitivity and Amplification Efficiency Testing)
3.1.4. Outgroup Testing and Fluorescence in Mixed Cultures
3.2. Net-Pen Sample Screening
4. Discussion
4.1. Assay Development
4.2. Net-Pen Sample Screening
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hikida, M.; Wakabayash, H.; Egusa, S.; Masumura, K. Flexibacter sp., a gliding bacterium pathogenic to some marine fishes in Japan. Bull. Jpn. Soc. Sci. Fish. 1979, 45, 421–428. [Google Scholar] [CrossRef]
- Avendaño-Herrera, R.; Toranzo, A.E.; Magariños, B. Tenacibaculosis infection in marine fish caused by Tenacibaculum maritimum: A review. Dis. Aquat. Org. 2006, 71, 255–266. [Google Scholar] [CrossRef]
- Nowlan, J.P.; Lumsden, J.S.; Russell, S. Advancements in characterizing Tenacibaculum infections in Canada. Pathogens 2020, 9, 1029. [Google Scholar] [CrossRef] [PubMed]
- Nowlan, J.P.; Britney, S.R.; Lumsden, J.S.; Russell, S. Application of quantitative-PCR to monitor netpen sites in British Columbia (Canada) for Tenacibaculum species. Pathogens 2021, 10, 414. [Google Scholar] [CrossRef] [PubMed]
- Frisch, K.; Småge, S.B.; Vallestad, C.; Duesund, H.; Brevik, J.; Klevan, A.; Olsen, R.H.; Sjaatil, S.T.; Gauthier, D.; Brudeseth, B. Experimental induction of mouthrot in Atlantic salmon smolts using Tenacibaculum maritimum from Western Canada. J. Fish. Dis. 2018, 41, 1247–1258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nowlan, J.P.; Britney, S.R.; Lumsden, J.S.; Russell, S. Experimental induction of tenacibaculosis in Atlantic salmon (Salmo salar L.) using Tenacibaculum maritimum, T. dicentrarchi, and T. finnmarkense. Pathogens 2021, 10, 1439. [Google Scholar] [CrossRef]
- Hewison, T. Yellow mouth—The Grieg Seafood experience in BC. In Tenacibaculum 2: Current Knowledge and Future Directions Workshop; Maritime Heritage Centre: Campbell River, BC, Canada, 2019; Available online: https://www.cahs-bc.ca/wp-content/uploads/2019/10/4Tim_Yellow_Mouth_The_Grieg_Seafood_BC_Experience.pdf (accessed on 24 September 2022).
- Irgang, R.; Mancilla, M.; Avendaño-Herrera, R. Florfenicol and oxytetracycline susceptibility patterns in Chilean isolates of Tenacibaculum dicentrarchi: An emerging pathogen for farmed salmonids. J. Fish Dis. 2021, 44, 1043–1046. [Google Scholar] [CrossRef]
- Míguez, B.; Combarro, M.P. Bacteria associated with sardine (Sardina pilchardus) eggs in a natural environment (Ría de Vigo, Galicia, northwestern Spain). FEMS Microbiol. Ecol. 2003, 44, 329–334. [Google Scholar] [CrossRef]
- Quinn, R.A.; Cawthorn, R.J.; Summerfield, R.L.; Smolowitz, R.; Chistoserdov, A.Y. Bacterial communities associated with lesions of two forms of shell disease in the American lobster (Homarus americanus, Milne Edwards) from Atlantic Canada. Can. J. Microbiol. 2013, 59, 380–390. [Google Scholar] [CrossRef]
- Olsen, A.B.; Gulla, S.; Steinum, T.; Colquhoun, D.J.; Nilsen, H.K.; Duchaud, E. Multilocus sequence analysis reveals extensive genetic variety within Tenacibaculum spp. associated with ulcers in sea-farmed fish in Norway. Vet. Microbiol. 2017, 205, 39–45. [Google Scholar] [CrossRef]
- Avendaño-Herrera, R.; Saldarriaga-Córdoba, M.; Irgang, R. Draft Genome Sequence of Tenacibaculum ovolyticum To-7Br, Recovered from a Farmed Atlantic Salmon (Salmo salar). Microbiol. Resour. Announc. 2022, 11, e00254-22. [Google Scholar] [CrossRef] [PubMed]
- Hansen, G.H.; Bergh, O.; Michaelsen, J.; Knappskog, D. Flexibacter ovolyticus sp. nov., a pathogen of eggs and larvae of Atlantic halibut, Hippoglossus hippoglossus L. Int. J. Syst. Bacteriol. 1992, 42, 451–458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Habib, C.; Houel, A.; Lunazzi, A.; Bernardet, J.F.; Olsen, A.B.; Nilsen, H.; Toranzo, A.E.; Castro, N.; Nicolas, P.; Duchaud, E. Multilocus sequence analysis of the marine bacterial genus Tenacibaculum suggests parallel evolution of fish pathogenicity and endemic colonization of aquaculture systems. Appl. Environ. Microbiol. 2014, 80, 5503–5514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teramoto, M.; Zhai, Z.; Komatsu, A.; Shibayama, K.; Suzuki, M. Genome sequence of the psychrophilic bacterium Tenacibaculum ovolyticum strain da5A-8 isolated from deep seawater. Genome Announc. 2016, 4, 8–10. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Pascual, D.; Lunazzi, A.; Magdelenat, G.; Rouy, Z.; Roulet, A.; Lopez-Roques, C.; Larocque, R.; Barbeyron, T.; Gobet, A.; Michel, G. The complete genome sequence of the fish pathogen Tenacibaculum maritimum provides insights into virulence mechanisms. Front. Microbiol. 2017, 8, 1542. [Google Scholar] [CrossRef] [Green Version]
- Grothusen, H.; Castillo, A.; Henríquez, P.; Navas, E.; Bohle, H.; Araya, C.; Bustamante, F.; Bustos, P.; Mancilla, M. First complete genome sequence of Tenacibaculum dicentrarchi, an emerging bacterial pathogen of salmonids. Genome Announc. 2016, 4, e01756-15. [Google Scholar] [CrossRef] [Green Version]
- Bridel, S.; Olsen, A.B.; Nilsen, H.; Bernardet, J.F.; Achaz, G.; Avendaño-Herrera, R.; Duchaud, E. Comparative genomics of Tenacibaculum dicentrarchi and “Tenacibaculum finnmarkense” highlights intricate evolution of fish-pathogenic species. Genome Biol. Evol. 2018, 10, 452–457. [Google Scholar] [CrossRef] [Green Version]
- Nowlan, J.P.; Sies, A.N.; Britney, S.R.; Cameron, A.D.S.; Siah, A.; Russell, S. Genomic Investigation into Tenacibaculum species in British Columbia, Canada. Pathogens, 2022; submitted. [Google Scholar]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef] [Green Version]
- Olsen, A.B.; Spilsberg, B.; Nilsen, H.K.; Lagesen, K.; Gulla, S.; Avendaño-Herrera, R.; Irgang, R.; Duchaud, E.; Colquhoun, D.J. Tenacibaculum piscium sp. nov., isolated from skin ulcers of sea-farmed fish, and description of Tenacibaculum finnmarkense sp. nov. with subdivision into genomovars finnmarkense and ulcerans. Int. J. Syst. Evol. Microbiol. 2020, 70, 6079–6090. [Google Scholar] [CrossRef]
- Saldarriaga-Córdoba, M.; Irgang, R.; Avendaño-Herrera, R. Comparison between genome sequences of Chilean Tenacibaculum dicentrarchi isolated from red conger eel (Genypterus chilensis) and Atlantic salmon (Salmo salar) focusing on bacterial virulence determinants. J. Fish Dis. 2021, 44, 1843–1860. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Fringuelli, E.; Savage, P.D.; Gordon, A.; Baxter, E.J.; Rodger, H.D.; Graham, D.A. Development of a quantitative real-time PCR for the detection of Tenacibaculum maritimum and its application to field samples. J. Fish Dis. 2012, 35, 579–590. [Google Scholar] [CrossRef] [PubMed]
- RStudio Team. RStudio: Integrated Development for R. Version 3.6.1 (2019-07-05) “Action of the Toes”; RStudio, PBC: Boston, MA, USA. Available online: http://www.rstudio.com/ (accessed on 10 September 2021).
- Zhang, Q.; Wang, J.; Deng, F.; Yan, Z.; Xia, Y.; Wang, Z.; Ye, J.; Deng, Y.; Zhang, Z.; Qiao, M. TqPCR: A touchdown qPCR assay with significantly improved detection sensitivity and amplification efficiency of SYBR green qPCR. PLoS ONE 2015, 10, e0132666. [Google Scholar] [CrossRef]
- Rayl, J.M.; Wellehan, J.F., Jr.; Bunick, D.; Allender, M.C. Development of reverse-transcriptase quantitative PCR assays for detection of the cytokines IL-1β, TNF-α, and IL-10 in chelonians. Cytokine 2019, 119, 16–23. [Google Scholar] [CrossRef]
- Nowlan, J.P.; Lumsden, J.S.; Russell, S. Quantitative PCR for Tenacibaculum dicentrarchi and T. finnmarkense. J. Fish Dis. 2021, 44, 655–659. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bustin, S.A.; Wittwer, C.T. MIQE: A step toward more robust and reproducible quantitative PCR. Clin. Chem. 2017, 63, 1537–1538. [Google Scholar] [CrossRef] [Green Version]
- Taylor, S.C.; Nadeau, K.; Abbasi, M.; Lachance, C.; Nguyen, M.; Fenrich, J. The ultimate qPCR experiment: Producing publication quality, reproducible data the first time. Trends Biotechnol. 2019, 37, 761–774. [Google Scholar] [CrossRef] [Green Version]
- Oxford Nanopore Technologies (ONT). Nanopore Sequencing: The Advantages of Long Reads for Genome Assembly; pp. 1–16. 2017. Available online: https://www.google.com/url?sa=t&rct=j&q=&esrc=s&source=web&cd=&cad=rja&uact=8&ved=2ahUKEwjPkviI-a_5AhXDI30KHV6WA34QFnoECAoQAw&url=https%3A%2F%2Fnanoporetech.com%2Fsites%2Fdefault%2Ffiles%2Fs3%2Fwhite-papers%2FWGS_Assembly_white_paper.pdf%3FsubmissionGuid%3D40a7546b-9e51-42e7-bde9-b5ddef3c3512&usg=AOvVaw1S_sBMns-u-WlA4qyAZoRS (accessed on 24 September 2022).
- Wynne, J.W.; Thakur, K.K.; Slinger, J.; Samsing, F.; Milligan, B.; Powell, J.F.F.; McKinnon, A.; Nekouei, O.; New, D.; Richmond, Z. Microbiome profiling reveals a microbial dysbiosis during a natural outbreak of tenacibaculosis (yellow mouth) in Atlantic salmon. Front. Microbiol. 2020, 11, 586387. [Google Scholar] [CrossRef]
- Suzuki, M.; Nakagawa, Y.; Shigeaki, H.; Yamamoto, S. Phylogenetic analysis and taxonomic study of marine Cytophaga-like bacteria: Proposal for Tenacibaculum gen. nov. with Tenacibaculum maritimum comb. nov. and Tenacibaculum ovolyticum comb. nov., and description of Tenacibaculum mesophilum sp. nov. and Tenacibaculum amylolyticum sp. nov. Int. J. Syst. Evol. Microbiol. 2001, 51, 1639–1652. [Google Scholar] [CrossRef]
- Karlsen, C.; Ottem, K.F.; Brevik, Ø.J.; Davey, M.; Sørum, H.; Winther Larsen, H.C. The environmental and host-associated bacterial microbiota of arctic seawater-farmed Atlantic salmon with ulcerative disorders. J. Fish Dis. 2017, 40, 1645–1663. [Google Scholar] [CrossRef] [PubMed]
- Llewellyn, M.S.; Leadbeater, S.; Garcia, C.; Sylvain, F.E.; Custodio, M.; Ang, K.P.; Powell, F.; Carvalho, G.R.; Creer, S.; Elliot, J. Parasitism perturbs the mucosal microbiome of Atlantic Salmon. Sci. Rep. 2017, 7, 43465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reid, K.M.; Patel, S.; Robinson, A.J.; Bu, L.; Jarungsriapisit, J.; Moore, L.J.; Salinas, I. Salmonid alphavirus infection causes skin dysbiosis in Atlantic salmon (Salmo salar L.) post-smolts. PLoS ONE 2017, 12, e0172856. [Google Scholar] [CrossRef] [Green Version]
- Wilson, T.K.; Douglas, M.; Dunn, V. First identification in Tasmania of fish pathogens Tenacibaculum dicentrarchi and T. soleae and multiplex PCR for these organisms and T. maritimum. Dis. Aquat. Org. 2019, 136, 219–226. [Google Scholar] [CrossRef] [PubMed]

| In-House Sample Name | Bacterial Designation or Most Similar NCBI BLAST Comparison | NCBI BLAST Comparison | Obtained from | Grown on | Grown at °C | |||
|---|---|---|---|---|---|---|---|---|
| 16S rDNA Amplicon Length (bp) * | Query Cover, Similarity, E-Value | |||||||
| 20-4135-2 | Tenacibaculum ovolyticum da5A-8 | 1368 | 100 | 99.9 | 0 | BC Atlantic salmon | FMM+K | 12 |
| T.mar 2.1C | Tenacibaculum maritimum NLF-15 | 1366 | 99 | 100 | 0 | BC Atlantic salmon | FMM+K | 12 |
| 20-4116-9 | Tenacibaculum dicentrarchi TdChD04 | 1409 | 99 | 98.8 | 0 | BC Atlantic salmon | FMM+K | 12 |
| 20-4106-2 | Tenacibaculum finnmarkense Tsp.2 | 1426 | 100 | 99 | 0 | BC Atlantic salmon | FMM+K | 12 |
| DSM 17995 | Tenacibaculum maritimum R-2T | NA | NA | DSMZ | MA | 30 | ||
| DSM 18103 | Tenacibaculum ovolyticum EKD 002 T | NA | NA | DSMZ | MA | 15 | ||
| DSM 18841 | Tenacibaculum gallaicum A37.1T | NA | NA | DSMZ | MA | 28 | ||
| ATCC BAA-459™ | Tenacibaculum skagerrakense D30T | NA | NA | ATCC | MA | 12 | ||
| DSM 18842 | Tenacibaculum discolor LL04 11.1.1T | NA | NA | DSMZ | MA | 28 | ||
| FP | Flavobacterium sp. | 1317 | 99 | >95 | 0 | Environmental sample | CA | 12 |
| ATCC 43844™ | Polaribacter glomeratus UQM 3055T | NA | NA | ATCC | MA | 30 | ||
| ATCC 23079™ | Flexibacter flexilis CR-63T | NA | NA | ATCC | CA | 21 | ||
| F.flex Contam | Dermacoccus sp. | 589 | >91 | >88 | <1 × 10−78 | Culture contaminate | CA | 21 |
| Pcocus | Paracoccus sp. | 1184 | 100 | >96 | 0 | Culture contaminate | FMM+K | 12 |
| Beluga HI TSA 1 | Pseudomonas sp. CC11J | 1434 | 99 | 99.8 | 0 | BC White Sturgeon | TSA/CA | 12 |
| Beluga HI TSA 2 | Flavobacterium sp. T69L.09.B.RBT.MI.W. Kidney | 1386 | 99 | 99.9 | 0 | BC White Sturgeon | TSA/CA | 12 |
| LI C4 P1 | Vibrio splendidus BST398 | 1375 | 100 | 99.7 | 0 | Environmental sample | FMM+K | 12 |
| LI C3 PCB | Pseudoalteromonas sp. NBRC 107703 | 1357 | 100 | 99.7 | 0 | Environmental sample | FMM+K | 12 |
| MS7 F1 | Celluphaga sp. W5B | 1366 | 100 | 98.7 | 0 | BC Atlantic salmon | FMM+K | 12 |
| MS5 M2 | Dokdonia sp. 6a | 1358 | 100 | 99.1 | 0 | BC Atlantic salmon | FMM+K | 12 |
| MS5 F3 | Cellulophaga baltica NN015840 | 1349 | 100 | 99.8 | 0 | BC Atlantic salmon | FMM+K | 12 |
| Aero sp. kida | Aermonas sp. | NA | NA | BC Atlantic salmon | FMM+K | 12 | ||
| V.anguill | Vibrio anguillarum 155 5RH | NA | NA | BC Atlantic salmon | FMM+K | 12 | ||
| V.aest | Vibrio aestuarianus | NA | NA | Dr. Tim Green | NA | NA | ||
| Shewn.sp | Shewnanella sp. | NA | NA | Dr. Tim Green | NA | NA | ||
| P.unid | Pseudoalternomonas udina | NA | NA | Dr. Tim Green | NA | NA | ||
| E.coliTop10 | Similar to Escherichia coli DH10B™ | NA | NA | Invitrogen Topo TA Cloning Kit | LB | 37 | ||
| Primer or Probe Name | Sequence | Tm (°C) | Length (bp) | Amplicon Length (bp) |
|---|---|---|---|---|
| Tenaci-G Fw | TRC CTT STA CAK RRG GAT ARC C | 49.7 | 22 | ~155 |
| Tenaci-G Rv | CTA TCG THG CCA TGG TAA GCC G | 65.9 | 22 | |
| OVO Probe (FAM Fluorophore) | TGT TAA TTA GAG GCA TCT | 49.2 | 18 | NA |
| Sample Type: | C1—Pre-Fish Entry | C2—One Week Post-Fish Entry | C4—During Treatment | C6—After Treatment |
|---|---|---|---|---|
| Water | 0 m (2) | 0 m (3) | 0 m (3) | 0 m (3) |
| 5 m (2) | 5 m (3) | 5 m (3) | 5 m (3) | |
| 10 m (2) | 10 m (3) | 10 m (3) | 10 m (3) | |
| Fish Tissues (Euth.) | NA | Skin (3) | Skin (3) | Skin (3) |
| Gill (3) | Gill (3) | Gill (3) | ||
| Upper Jaw (3) | Upper Jaw (3) | Upper Jaw (3) | ||
| Kidney (3) | Kidney (3) | Kidney (3) | ||
| Fish Tissues (Dead) | NA | Skin (3) | Skin (3) | Skin (3) |
| Kidney (3) | Kidney (3) | Kidney (3) | ||
| Invertebrate | Mytilus sp. (1) | Mytilus sp. (1) | Mytilus sp. (2) | Mytilus sp. (1) |
| Assay | Tenaci-G Fw Primer (µM) | Tenaci-G Rv Primer (µM) | OVO Probe (µM) | Cq Mean | Cq SD |
|---|---|---|---|---|---|
| OVO | 0.25 | 0.25 | 0.125 | 16.99 a | 0.07 |
| OVO | 0.5 | 0.5 | 0.125 | 14.84 b | 0.30 |
| OVO | 0.75 | 0.75 | 0.125 | 13.97 c | 0.28 |
| OVO | 1 | 1 | 0.125 | 13.58 d | 0.06 |
| OVO | 0.5 | 0.5 | 0.025 | NA A | NA |
| OVO | 0.5 | 0.5 | 0.05 | 19.9 B | 0.11 |
| OVO | 0.5 | 0.5 | 0.125 | 15.22 C | 0.11 |
| OVO | 0.5 | 0.5 | 0.25 | 13.61 D | 0.12 |
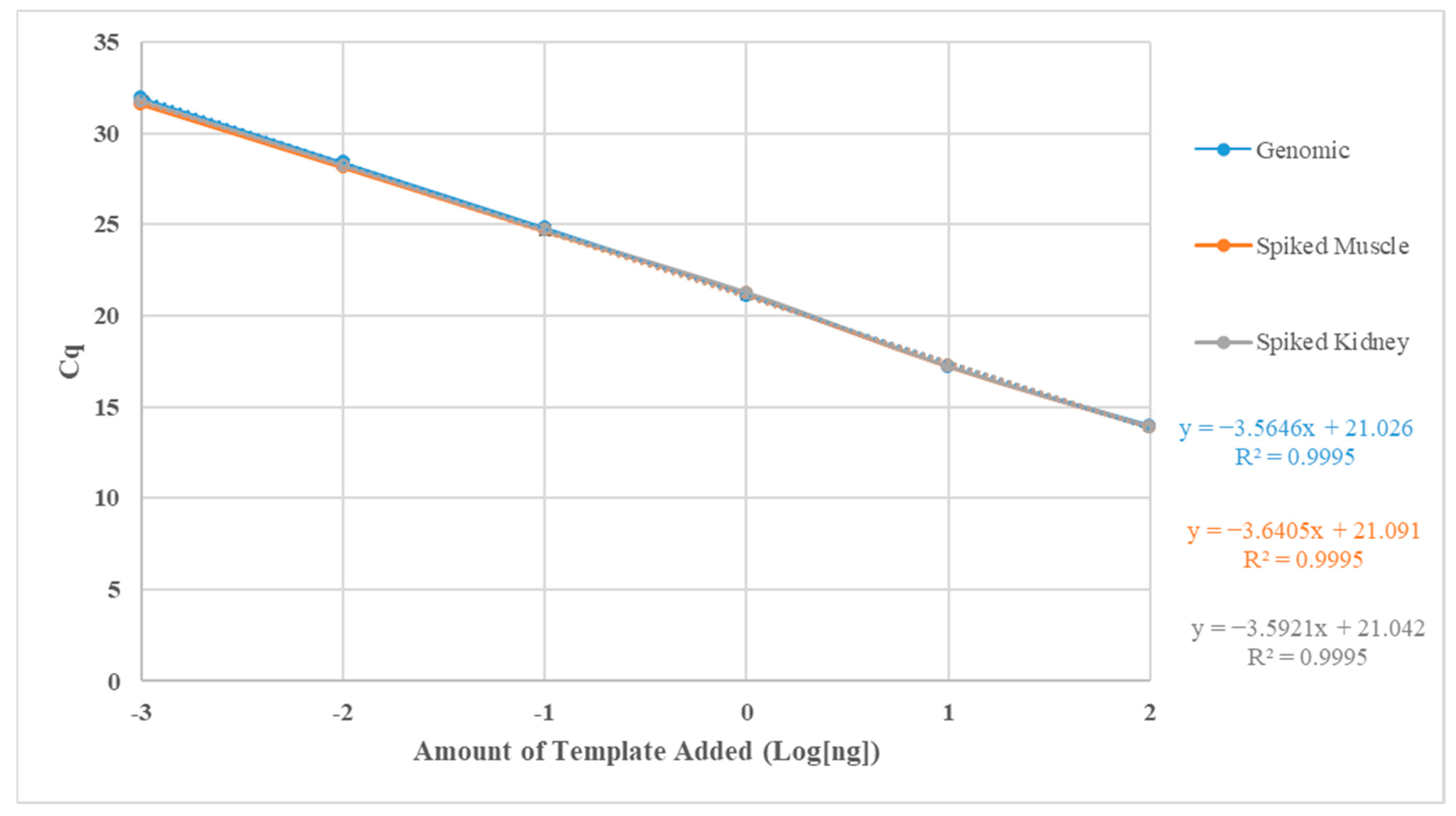
| Standard Curve | LOD | LOQD | R2 | Slope | Amplification Efficiency (%) |
|---|---|---|---|---|---|
| Genomic | 2.23 × 101–2.23 × 108 | 2.23× 102–2.23 × 107 | 0.9995 | −3.56 | 90.78 |
| Spiked (S. salar muscle DNA) | 2.23 × 101–2.23 × 108 | 2.23 × 102–2.23 × 107 | 0.9995 | −3.64 | 88.23 |
| Spiked (S. salar head kidney DNA) | 2.23 × 101–2.23 × 108 | 2.23 × 102–2.23 × 107 | 0.9995 | −3.59 | 89.84 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nowlan, J.P.; Heese, B.M.; Wilson, M.J.; Britney, S.R.; Lumsden, J.S.; Russell, S. Tenacibaculum ovolyticum 16S rDNA Quantitative-PCR Assay Development and Field Testing. Fishes 2022, 7, 303. https://doi.org/10.3390/fishes7060303
Nowlan JP, Heese BM, Wilson MJ, Britney SR, Lumsden JS, Russell S. Tenacibaculum ovolyticum 16S rDNA Quantitative-PCR Assay Development and Field Testing. Fishes. 2022; 7(6):303. https://doi.org/10.3390/fishes7060303
Chicago/Turabian StyleNowlan, Joseph P., Brianna M. Heese, Matthew J. Wilson, Scott R. Britney, John S. Lumsden, and Spencer Russell. 2022. "Tenacibaculum ovolyticum 16S rDNA Quantitative-PCR Assay Development and Field Testing" Fishes 7, no. 6: 303. https://doi.org/10.3390/fishes7060303





