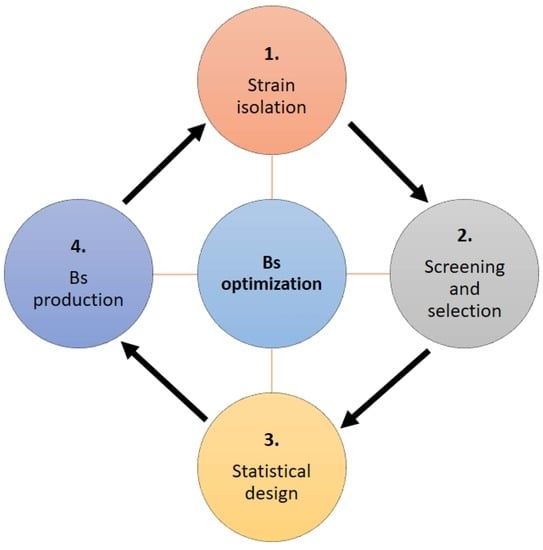
Statistical Design, a Powerful Tool for Optimizing Biosurfactant Production: A Review
Abstract
:1. Introduction
2. Factors Affecting Bs Production
2.1. Strains, Bs Classification and Metabolism
2.2. Effect of Carbon and Nitrogen Sources
2.3. The Effect of Trace Elements on Bs Production
2.4. Physicochemical Factors Affecting Bs Production: The Effect of pH, Temperature, and Shaking
3. Statistical Design, an Efficient Tool for Bs Production Optimization
4. Two-Level Factorial Designs
4.1. Plackett–Burman
4.2. Other Two-Level Factorial Designs
5. Response Surface Methodology (RSM)
5.1. Central Composite Design
5.2. Box–Behnken
6. Modified Gompertz Equation
7. Mixed Strategies
7.1. Analytical Hierarchy Process (AHP)
7.2. Artificial Neural Network
8. Improving Downstream Processes
9. Concluding Remarks
Funding
Conflicts of Interest
References
- Abalos, A.; Maximo, F.; Manresa, M.A.; Bastida, J. Utilization of response surface methodology to optimize the culture media for the production of rhamnolipids by Pseudomonas aeruginosa AT10. J. Chem. Technol. Biotechnol. 2002, 77, 777–784. [Google Scholar] [CrossRef]
- Banat, I.M.; Franzetti, A.; Gandolfi, I.; Bestetti, G.; Martinotti, M.G.; Fracchia, L.; Smyth, T.J.; Marchant, R. Microbial biosurfactants production, applications and future potential. Appl. Microbiol. Biotechnol. 2010, 87, 427–444. [Google Scholar] [CrossRef] [PubMed]
- Abbasi, H.; Sharafi, H.; Alidost, L.; Bodagh, A.; Zahiri, H.S.; Noghabi, K.A. Response surface optimization of biosurfactant produced by Pseudomonas aeruginosa ma01 isolated from spoiled apples. Prep. Biochem. Biotechnol. 2013, 43, 398–414. [Google Scholar] [CrossRef] [PubMed]
- Reis, R.S.; Pacheco, G.J.; Pereira, A.G.; Freire, D.M.G. Biosurfactants: Production and Applications. In Biodegradation-Life of Science; Chamy, R., Rosenkranz, F., Eds.; InTech: Rijeka, Croatia, 2013. [Google Scholar] [Green Version]
- Varjani, S.J.; Upasani, V.N. Critical review on biosurfactant analysis, purification and characterization using rhamnolipid as a model biosurfactant. Bioresour. Technol. 2017, 232, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Gudiña, E.J.; Teixeira, J.A.; Rodrigues, L.R. Biosurfactants produced by marine microorganisms with therapeutic applications. Mar. Drugs 2016, 14, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ismail, W.; Shammary, S.A.; El-Sayed, W.S.; Obuekwe, C.; El Nayal, A.M.; Raheem, A.S.A.; Al-Humam, A. Stimulation of rhamnolipid biosurfactants production in Pseudomonas aeruginosa AK6U by organosulfur compounds provided as sulfur sources. Biotechnol. Rep. 2015, 7, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Vecino, X.; Cruz, J.M.; Moldes, A.B.; Rodrigues, L.R. Biosurfactants in cosmetic formulations: Trends and challenges. Crit. Rev. Biotechnol. 2017, 37, 911–923. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, Z.; Crowley, D.; Marina, N.; Jha, S.K. Estimation of biosurfactant yield produced by Klebsiella sp. FKOD36 bacteria using artificial neural network approach. Meas. J. Int. Meas. Confed. 2016, 81, 163–173. [Google Scholar] [CrossRef]
- Chen, J.; Huang, P.T.; Zhang, K.Y.; Ding, F.R. Isolation of biosurfactant producers, optimization and properties of biosurfactant produced by Acinetobacter sp. from petroleum-contaminated soil. J. Appl. Microbiol. 2012, 112, 660–671. [Google Scholar] [CrossRef] [PubMed]
- Pereira, J.F.B.; Gudiña, E.J.; Costa, R.; Vitorino, R.; Teixeira, J.A.; Coutinho, J.A.P.; Rodrigues, L.R. Optimization and characterization of biosurfactant production by Bacillus subtilis isolates towards microbial enhanced oil recovery applications. Fuel 2013, 111, 259–268. [Google Scholar] [CrossRef] [Green Version]
- El-Sheshtawy, H.S.; Doheim, M.M. Selection of Pseudomonas aeruginosa for biosurfactant production and studies of its antimicrobial activity. Egypt. J. Pet. 2014, 23, 1–6. [Google Scholar] [CrossRef]
- Díaz De Rienzo, M.A.; Kamalanathan, I.D.; Martin, P.J. Comparative study of the production of rhamnolipid biosurfactants by B. thailandensis E264 and P. aeruginosa ATCC 9027 using foam fractionation. Process Biochem. 2016, 51, 820–827. [Google Scholar] [CrossRef]
- Fontes, G.C.; Amaral, P.F.; Nele, M.; Coelho, M.A. Factorial design to optimize biosurfactant production by Yarrowia lipolytica. J. Biomed. Biotechnol. 2010, 2010, 821306. [Google Scholar] [CrossRef] [PubMed]
- Heryani, H.; Putra, M.D. Kinetic study and modeling of biosurfactant production using Bacillus sp. Electron. J. Biotechnol. 2017, 27, 49–54. [Google Scholar] [CrossRef]
- Hu, X.; Wang, C.; Wang, P. Optimization and characterization of biosurfactant production from marine Vibrio sp. strain 3B-2. Front. Microbiol. 2015, 6, 976. [Google Scholar] [CrossRef] [PubMed]
- Santos, D.K.F.; Rufino, R.D.; Luna, J.M.; Santos, V.A.; Sarubbo, L.A. Biosurfactants: Multifunctional biomolecules of the 21st century. Int. J. Mol. Sci. 2016, 17, 401. [Google Scholar] [CrossRef] [PubMed]
- Roldán-Carrillo, T.; Martínez-García, X.; Zapata-Peñasco, I.; Castorena-Cortés, G.; Reyes-Avila, J.; Mayol-Castillo, M.; Olguín-Lora, P. Evaluation of the effect of nutrient ratios on biosurfactant production by Serratia marcescens using a Box-Behnken design. Colloids Surf. B 2011, 86, 384–389. [Google Scholar] [CrossRef] [PubMed]
- Mutalik, S.R.; Vaidya, B.K.; Joshi, R.M.; Desai, K.M.; Nene, S.N. Use of response surface optimization for the production of biosurfactant from Rhodococcus spp. MTCC 2574. Bioresour. Technol. 2008, 99, 7875–7880. [Google Scholar] [CrossRef] [PubMed]
- Das, P.; Mukherjee, S.; Sen, R. Genetic regulations of the biosynthesis of microbial surfactants: An overview. Biotechnol. Genet. Eng. Rev. 2008, 25, 165–186. [Google Scholar] [CrossRef] [PubMed]
- George, S.; Jayachandran, K. Production and characterization of rhamnolipid biosurfactant from waste frying coconut oil using a novel Pseudomonas aeruginosa D. J. Appl. Microbiol. 2013, 114, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Makkar, R.S.; Cameotra, S.S. Effects of various nutritional supplements on biosurfactant production by a strain of Bacillus subtilis at 45 °C. J. Surfactants Deterg. 2002, 5, 11–17. [Google Scholar] [CrossRef]
- Desai, J.D.; Desai, A.J. Production of Biosurfactants. In Biosurfactants: Production: Properties: Applications; Kosaric, N., Ed.; Marcel Dekker: New York, NY, USA, 1993; pp. 65–97. [Google Scholar]
- Pal, M.P.; Vaidya, B.K.; Desai, K.M.; Joshi, R.M.; Nene, S.N.; Kulkarni, B.D. Media optimization for biosurfactant production by Rhodococcus erythropolis MTCC 2794: Artificial intelligence versus a statistical approach. J. Ind. Microbiol. Biotechnol. 2009, 36, 747–756. [Google Scholar] [CrossRef] [PubMed]
- Joshi, S.; Yadav, S.; Nerurkar, A.; Desai, A.J. Statistical optimization of medium components for the production of biosurfactant by Bacillus licheniformis K51. J. Microbiol. Biotechnol. 2007, 17, 313–319. [Google Scholar] [PubMed]
- Praharyawan, S.; Susilaningsih, D.; Syamsu, K. Statistical screening of medium components by Plackett-Burman experimental design for biosurfactant production by Indonesian indigenous Bacillus sp. DSW17. Asian J. Microbiol. Biotechnol. Environ. Sci. 2013, 15, 805–813. [Google Scholar]
- Wei, Y.H.; Lai, C.C.; Chang, J.S. Using Taguchi experimental design methods to optimize trace element composition for enhanced surfactin production by Bacillus subtilis ATCC 21332. Process Biochem. 2007, 42, 40–45. [Google Scholar] [CrossRef]
- Najafi, A.R.; Rahimpour, M.R.; Jahanmiri, A.H.; Roostaazad, R.; Arabian, D.; Ghobadi, Z. Enhancing biosurfactant production from an indigenous strain of Bacillus mycoides by optimizing the growth conditions using a response surface methodology. Chem. Eng. J. 2010, 163, 188–194. [Google Scholar] [CrossRef]
- Santos, D.K.F.; Brandão, Y.B.; Rufino, R.D.; Luna, J.M.; Salgueiro, A.A.; Santos, V.A.; Sarubbo, L.A. Optimization of cultural conditions for biosurfactant production from Candida lipolytica. Biocatal. Agric. Biotechnol. 2014, 3, 48–57. [Google Scholar] [CrossRef]
- Quinn, G.P.; Keough, M. Experimental Design and Data Analysis for Biologists, 6th ed.; Cambridge University Press: Cambridge, UK, 2002. [Google Scholar]
- Myers, R.H.; Montgomery, D.C.; Anderson-Cook, C.M. Response Surface Methodology: Process and Product in Optimization Using Designed Experiments, 4th ed.; John Wiley and Sons, Inc.: New York, NY, USA, 2016. [Google Scholar]
- Tian, J.; Wang, J.; Zhou, Y.J.; Dong, H.-P.; Yu, L. Mixed experimental methodologies to optimize biosurfactant production conditions of Pseudomonas aeruginosa. In Proceedings of the 2010 First ACIS International Symposium on Cryptography, and Network Security, Data Mining and Knowledge Discovery, E-Commerce and Its Applications, and Embedded Systems, Qinhuangdao, China, 23–24 October 2010; pp. 358–361. [Google Scholar] [CrossRef]
- Hassan, M.; Essam, T.; Yassin, A.S.; Salama, A. Optimization of rhamnolipid production by biodegrading bacterial isolates using Plackett-Burman design. Int. J. Biol. Macromol. 2016, 82, 573–579. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Toledo, A.; Rodríguez-Vázquez, R. Response surface methodology (Box-Behnken) to improve a liquid media formulation to produce biosurfactant and phenanthrene removal by Pseudomonas putida. Ann. Microbiol. 2011, 61, 605–613. [Google Scholar] [CrossRef]
- Kumar, A.P.; Janardhan, A.; Radha, S.; Viswanath, B.; Narasimha, G. Statistical approach to optimize production of biosurfactant by Pseudomonas aeruginosa 2297. 3 Biotech 2015, 5, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.H.; Cheng, C.L.; Chien, C.C.; Wan, H.M. Enhanced di-rhamnolipid production with an indigenous isolate Pseudomonas aeruginosa J16. Process Biochem. 2008, 43, 769–774. [Google Scholar] [CrossRef]
- Fonseca, R.R.; Silva, A.J.; De Franca, F.P.; Cardoso, V.L.; Sérvulo, E.F.C. Optimizing carbon/nitrogen ratio for biosurfactant production by a Bacillus subtilis strain. Appl. Biochem. Biotechnol. 2007, 137–140, 471–486. [Google Scholar] [CrossRef]
- Zouari, R.; Ellouze-Chaabouni, S.; Ghribi-Aydi, D. Optimization of Bacillus subtilis SPB1 biosurfactant production under solid-state fermentation using by-products of a traditional olive mill factory. Achiev. Life Sci. 2014, 8, 162–169. [Google Scholar] [CrossRef]
- Mnif, I.; Ellouze-Chaabouni, S.; Ghribi, D. Optimization of inocula conditions for enhanced biosurfactant production by Bacillus subtilis SPB1, in submerged culture, using Box-Behnken Design. Probiot. Antimicrob. Proteins 2013, 5, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Seghal, K.G.; Anto, T.T.; Selvin, J.; Sabarathnam, B.; Lipton, A.P. Optimization and characterization of a new lipopeptide biosurfactant produced by marine Brevibacterium aureum MSA13 in solid state culture. Bioresour. Technol. 2010, 101, 2389–2396. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, L.; Teixeira, J.; Oliveira, R.; Van Der Mei, H.C. Response surface optimization of the medium components for the production of biosurfactants by probiotic bacteria. Process Biochem. 2006, 41, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Santos, A.P.P.; Silva, M.D.S.; Costa, E.V.L.; Rufin, R.D.; Santos, V.A.; Ramos, C.S.; Sarubbo, L.A.; Porto, A.L.F. Production and characterization of a biosurfactant produced by Streptomyces sp. DPUA 1559 isolated from lichens of the Amazon region. Braz. J. Med. Biol. Res. 2018, 51. [Google Scholar] [CrossRef] [PubMed]
- Korayem, A.S.; Abdelhafez, A.A.; Zaki, M.M.; Saleh, E.A. Optimization of biosurfactant production by Streptomyces isolated from Egyptian arid soil using Plackett–Burman design. Ann. Agric. Sci. 2015, 60, 209–217. [Google Scholar] [CrossRef]
- Mouafi, F.E.; Abo, E.M.M.; Moharam, M.E. Optimization of biosurfactant production by Bacillus brevis using response surface methodology. Biotechnol. Rep. 2016, 9, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Sivapathasekaran, C.; Mukherjee, S.; Ray, A.; Gupta, A.; Sen, R. Artificial neural network modeling and genetic algorithm based medium optimization for the improved production of marine biosurfactant. Bioresour. Technol. 2010, 101, 2884–2887. [Google Scholar] [CrossRef] [PubMed]
- Vecino Bello, X.; Devesa-Rey, R.; Cruz, J.M.; Moldes, A.B. Study of synergistic effects of Salinity, pH, and temperature on the surface-active properties of biosurfactants produced by Lactobacillus pentosus. J. Agric. Food. Chem. 2012, 60, 1258–1265. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimipour, G.; Sadeghi, H.; Zarinviarsagh, M. Statistical methodologies for the optimization of lipase and biosurfactant by Ochrobactrum intermedium strain MZV101 in an identical medium for detergent applications. Molecules 2017, 22, 1–15. [Google Scholar] [CrossRef]
- Vecino, X.; Barbosa-Pereira, L.; Devesa-Rey, R.; Cruz, J.M.; Moldes, A.B. Optimization of extraction conditions and fatty acid characterization of Lactobacillus pentosus cell-bound biosurfactant/bioemulsifier. J. Sci. Food Agric. 2014, 95, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Tjørve, K.M.C.; Tjørve, E. The use of Gompertz models in growth analyses, and new Gompertz-model approach: An addition to the Unified-Richards family. PLoS ONE 2017, 12, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Deepika, K.V.; Kalam, S.; Sridhar, R.P.; Podile, A.R.; Bramhachari, P.V. Optimization of rhamnolipid biosurfactant production by mangrove sediment bacterium Pseudomonas aeruginosa KVD-HR42 using response surface methodology. Biocatal. Agric. Biotechnol. 2016, 5, 38–47. [Google Scholar] [CrossRef]
- Rosas-Galván, N.S.; Martínez-Morales, F.; Marquina-Bahena, S.; Tinoco-Valencia, R.; Serrano-Carreón, L.; Bertrand, B.; León-Rodríguez, R.; Guzmán-Aparicio, J.; Alvaréz-Berber, L.; Trejo-Hernández, M.R. Improved production, purification and characterization of biosurfactants produced by Serratia marcescens SM3 and its isogenic SMRG-5 strain. Biotechnol. Appl. Biochem. 2018. [Google Scholar] [CrossRef] [PubMed]
- Najmi, Z.; Ebrahimipour, G.; Franzetti, A.; Banat, I.M. In situ downstream strategies for cost-effective bio/surfactant recovery. Biotechnol. Appl. Biochem. 2018. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, B.; Mayolo-Deloisa, K.; González-González, M.; Tinoco-Valencia, R.; Serrano-Carreón, L.; Martínez-Morales, F.; Trejo-Hernández, M.R.; Rito-Palomares, M. Pleurotus ostreatus laccase recovery from residual compost using aqueous two-phase systems. J. Chem. Technol. Biotechnol. 2016, 91, 2235–2242. [Google Scholar] [CrossRef]
| Microorganism | Statistical Design | Factor Evaluated | Results | Bs Nature | References |
|---|---|---|---|---|---|
| Glycolipids | |||||
| Pseudomonas aeruginosa | RSM, 24 factorial | Waste free fatty acid waste from soybean oil, NaNO3, PO42−, and FeSO2, 7H2O | Maximum yield of 18.7 g dm−1 | Mixture of rhamnolipids | [1] |
| P. aeruginosa | Single parameter, 24 BBD, CCD | Soybean oil, NaNO3, yeast extract, pH, temperature, and shaking | 47% increase from 8.6 gL−1 to 12.6 gL−1 | Possible rhamnolipid | [3] |
| Klebseilla sp. FKOD36 | Taguchi, ANN | Starch, NaNO3, temperature, petrol, pH, incubation period | Yield of 0.038 gL−1, EI 24 of 31.67% and ST of 21.6 mNm−1 | Glycolipid and/or phospholipid | [9] |
| Vibrio sp. | RSM, CCD, AHP | Glucose, sucrose, lactose, maltose, xylose, beef extract, peptone, yeast extraction, soybean meal, corn meal, (NH4)2SO4, NH4NO3, NH4Cl, NaNO3, urea | Optimization ST to 41 mNm−1 | Glycoprotien fraction detected | [16] |
| Rhodococcus erythropolis | ANN and RSM | Sucrose, yeast extract, meat peptone, and toluene | 3.5-fold | glycolipid containing trehalose | [24] |
| P. aeruginosa | PBD, SAD and CCRD | Crude oil, NaCl, (NH2)2CO3, MgSO4·7H2O, NaH2PO4, K2HPO4·3H2O, EDTA, KH2PO4, (NH4)2SO4, C3H8O3 | ST reduction of 54–27.08 mNm−1 | Glycolipid, rhamnolipid | [32] |
| Psuedomonas sp. | PBD | Carbon source, nitrogen source, C/N ratio, iron concentration, magnesium concentration, phenol toxicity, pH, temperature, shaking, sampling time | 2-5 fold increase | Glycolipid, rhamnolipid | [33] |
| Pseudomonas putida | BBD | Glucose, ammonium chloride, yeast extract | 50 mgL−1 | Glycolipid, rhamnolipid | [34] |
| P. aeruginosa | PBD and BBD | Sawdust, glycerol, groundnut husk, groundnut oil, pH, inoculum size | Reduction in surface tension from 68.72–39.11 mNm−1 | Glycolipid, rhamnolipid | [35] |
| P. aeruginosa | PBD, SAD and BBD | Glycerol, methanol, ethanol, mannitol, glucose, sucrose, starch, soybean oil, sunflower oil, NH4Cl, NH4NO3, (NH4)2SO4, urea, NaNO3 | 3089 mgL−1 | Glycolipid, rhamnolipid | [36] |
| Lipopeptides | |||||
| Bacillus sp. | Modified Gompertz Model | Glucose, ammonium sulphate | Accurate production prediction | Cyclic lipopeptide, Surfactin | [15] |
| Bacillus licheniformis | PBD and BBD | Glucose, CaCl2, H3PO4, H3BO3, CuSO4, ZnSO4, CoCl2, Na-EDTA, NaNO3, MgSO4·7H2O, KCl, MnSO4, and Na2MoO4 | 10-fold increase in Bs production and Critical micelle dilution | Cyclic lipopeptide, Surfactin | [25] |
| Bacillus sp. | PBD | Frying oil waste, sucrose FeSO4·7H2O, NaNO3, KH2PO4, K2HPO4, MgSO4·7H2O, ZnSO4·7H2O, MnSO4·4H2O, NH4NO3, and CaCl2·2H2O | 124% increase in production | Cyclic lipopeptide, Surfactin | [26] |
| Bacillus subtilis | Taguchi | K+, Mg2+, Ca2+, Fe2+, Mn2+, Na+ | 3.34 gL−1 | Cyclic lipopeptide, Surfactin | [27] |
| Bacillus mycoides | CCD | Temperature, pH, salinity, and glucose | ST reduction from 61 to 34 mNm−1 | lipopeptide | [28] |
| B. subtillis | 23 factorial design | Sucrose, NaNO3, (NH4)2SO4, (NH4)2NO3, urea, residual brewery yeast | ST reduced to 29.3 mNm−1 | Cyclic lipopeptide, Surfactin | [37] |
| B. subtilis | 23 factorial design CCD | Olive leaf residue flour, olive cake flour, inoculum size, and moisture content | 30.67 mgg−1 | Cyclic lipopeptide, Surfactin | [38] |
| B. subtilis | BBD | Primary inoculum age, secondary seed culture age, and size | 3.4 gL−1 | Cyclic lipopeptide, Surfactin | [39] |
| Brevibacterium areum MSA13 | RSM | Olive oil, ferric chloride, inoculum size, acrylamide | 3-fold | Lipopeptide, Brevifactin | [40] |
| Paritially identified | |||||
| Serratia marcescens | BBD 33 factorial | C/N, C/Fe, and C/Mg ratio | ST reduction from 66–31 mNm−1 and a yield of 4.1 gL−1 | Unidentified, Posible lipopeptide | [18] |
| Lactococcus lactis and Strepococcus thermophilus | Fractional Factorial Design, SAD and CCD | Peptone, meat extract, yeast extract, lactose, ammonium citrate and KH2PO4, Lactose, soya peptone, and sodium gylcerophosphate | 1.8 and 2.1-fold increase for Lactococcus lactis and Strepococcus thermophilus, repectively | Unidentified, possible sophorolipids | [41] |
| Rhodococcus spp. MTCC 2574 | The one-at-a- time approach and CCRD | Mannitol, yeast extract, and meat peptone | 3.2 to 10.9 gL−1 ST tension of 72 to 30 mNm−1. | unidentified, protein and carbohydrate fraction | [19] |
| Streptomyces sp. | CCRD | pH and temperature | Production yield of 1.74 gL−1 and a ST of 25.34 mNm−1 | Unidentified glycoproteic fraction | [42] |
| Unidentified | |||||
| Acinetobacter sp. YC-X 2 | One-factor RSM, CCD | Beef extract, peptone, NaCl and n-hexadecane | 57.5% increase | Unidentified | [10] |
| Yarrowia lipolytica | 24 factorial, RSM | Urea, ammonium sulfate, yeast extract, peptone, glycerol, hexadecane, olive oil, and glucose | 110.7% increase in EI24 and 108.1% decrease in ST | Unidentified | [14] |
| Candida lipolytica | 23 factorial design (RSM/CC) | Agitation, aeration, and time of the process | 0.59–7.27gL−1 yield | Unidentified | [29] |
| Streptomyces | PBD | Starch nitrate medium, molasses, peptone, Tween 80, incuabtion period, inoculum size | 13.5% increase in EI24, from 31.74–42.68% | Unidentified | [43] |
| Bacillus brevis | RSM CCD | Temperature, pH, incubation period, and glucose concentration | Emulsion of 28.8–73% | Unidentified | [44] |
| Bacillus circulans MTCC 8281 | ANN | Glucose, Urea, SrCl2, and MgSO4 | 70% increase to 4.38 gL−1 | Unidentified | [45] |
| Lactobacillus pentosus | BBD | pH, temperature, and salinity | ST reduced from 69.3–53.8 nM/m. Emulsion volume of 45.93% and emulsion stability of 100%. | Possible glycolipopeptide | [46] |
| Ochrobactrum intermedium | PBD and BBD | pH, temperature, molasses, MgSO4, Waste engine oil, Waste cooking oil, K2HPO4, Olive oil, CaCl2,Whey, Yeast extract | 1.89-fold increase in EI24 | Unidentified | [47] |
| Lactobacillus pentosus | BBD | Operation time, temperature, and salt concentration | Bs yield improved from 9.49–13.76 mgL−1 | Possible glycolipopeptide | [48] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bertrand, B.; Martínez-Morales, F.; Rosas-Galván, N.S.; Morales-Guzmán, D.; Trejo-Hernández, M.R. Statistical Design, a Powerful Tool for Optimizing Biosurfactant Production: A Review. Colloids Interfaces 2018, 2, 36. https://doi.org/10.3390/colloids2030036
Bertrand B, Martínez-Morales F, Rosas-Galván NS, Morales-Guzmán D, Trejo-Hernández MR. Statistical Design, a Powerful Tool for Optimizing Biosurfactant Production: A Review. Colloids and Interfaces. 2018; 2(3):36. https://doi.org/10.3390/colloids2030036
Chicago/Turabian StyleBertrand, Brandt, Fernando Martínez-Morales, Nashbly Sarela Rosas-Galván, Daniel Morales-Guzmán, and María R. Trejo-Hernández. 2018. "Statistical Design, a Powerful Tool for Optimizing Biosurfactant Production: A Review" Colloids and Interfaces 2, no. 3: 36. https://doi.org/10.3390/colloids2030036
APA StyleBertrand, B., Martínez-Morales, F., Rosas-Galván, N. S., Morales-Guzmán, D., & Trejo-Hernández, M. R. (2018). Statistical Design, a Powerful Tool for Optimizing Biosurfactant Production: A Review. Colloids and Interfaces, 2(3), 36. https://doi.org/10.3390/colloids2030036