Nitrogen Starvation-Responsive MicroRNAs Are Affected by Transgenerational Stress in Durum Wheat Seedlings
Abstract
1. Introduction
2. Results
2.1. Seedling Performance of Two Durum Wheat Genotypes under the Effects of Parental Water-Deficit and Heat Stress and Progeny N Starvation Stress
2.2. Durum Wheat MiRNA Expression Profile across Different Treatment Groups
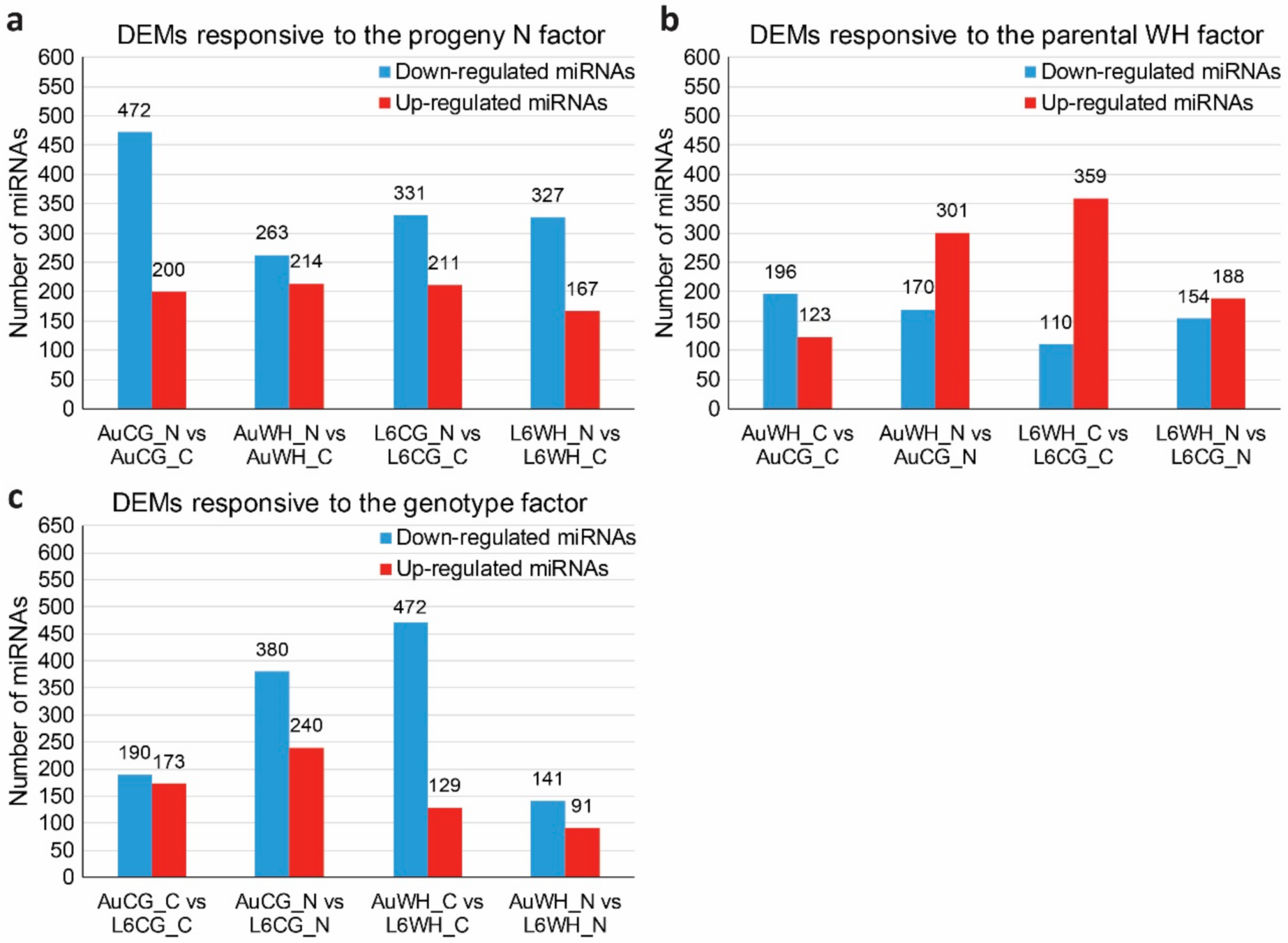
2.3. Differentially Expressed MiRNAs (DEMs) Subject to Different Factors
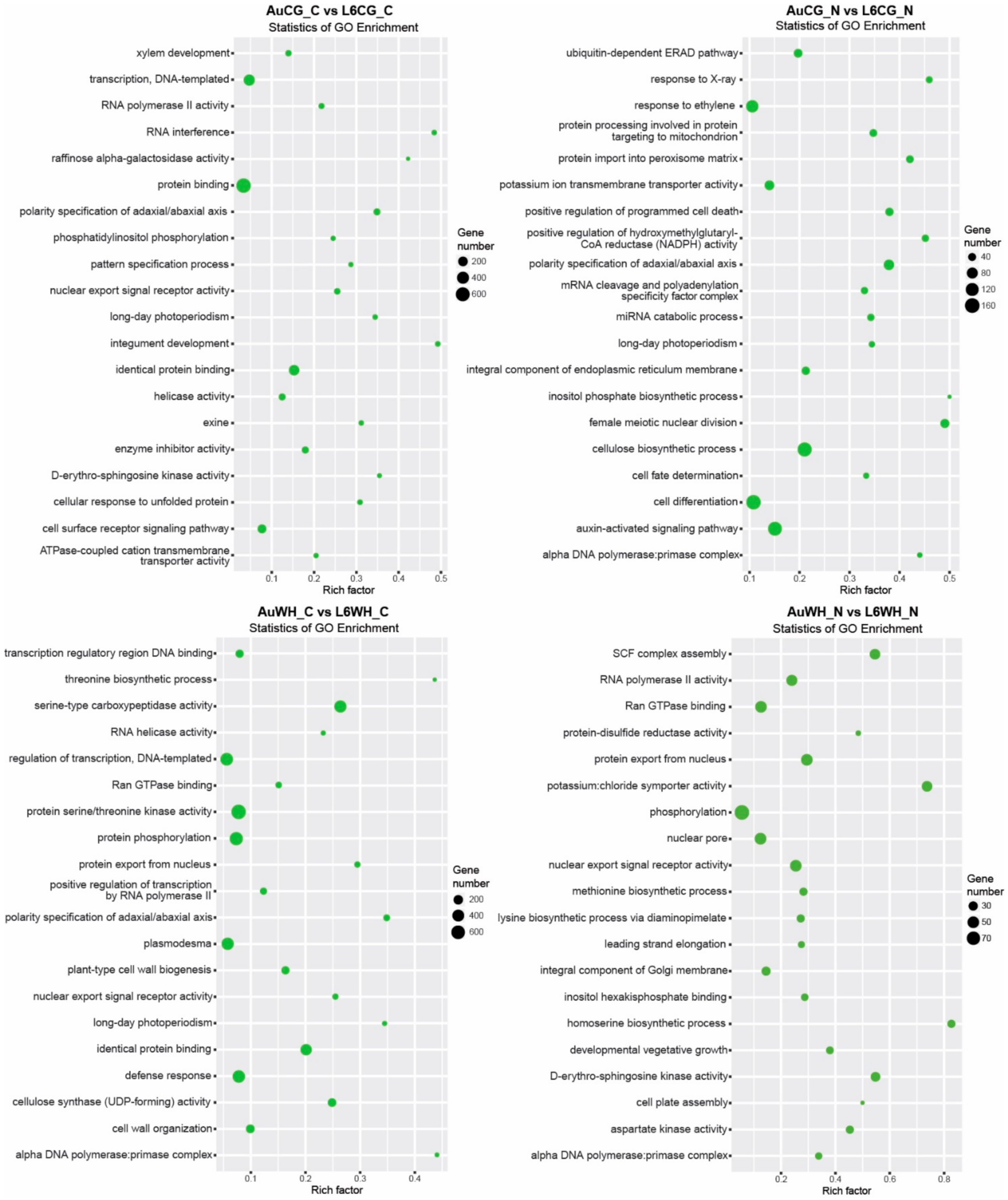
2.4. Analyses of the Functional Target Genes of DEMs
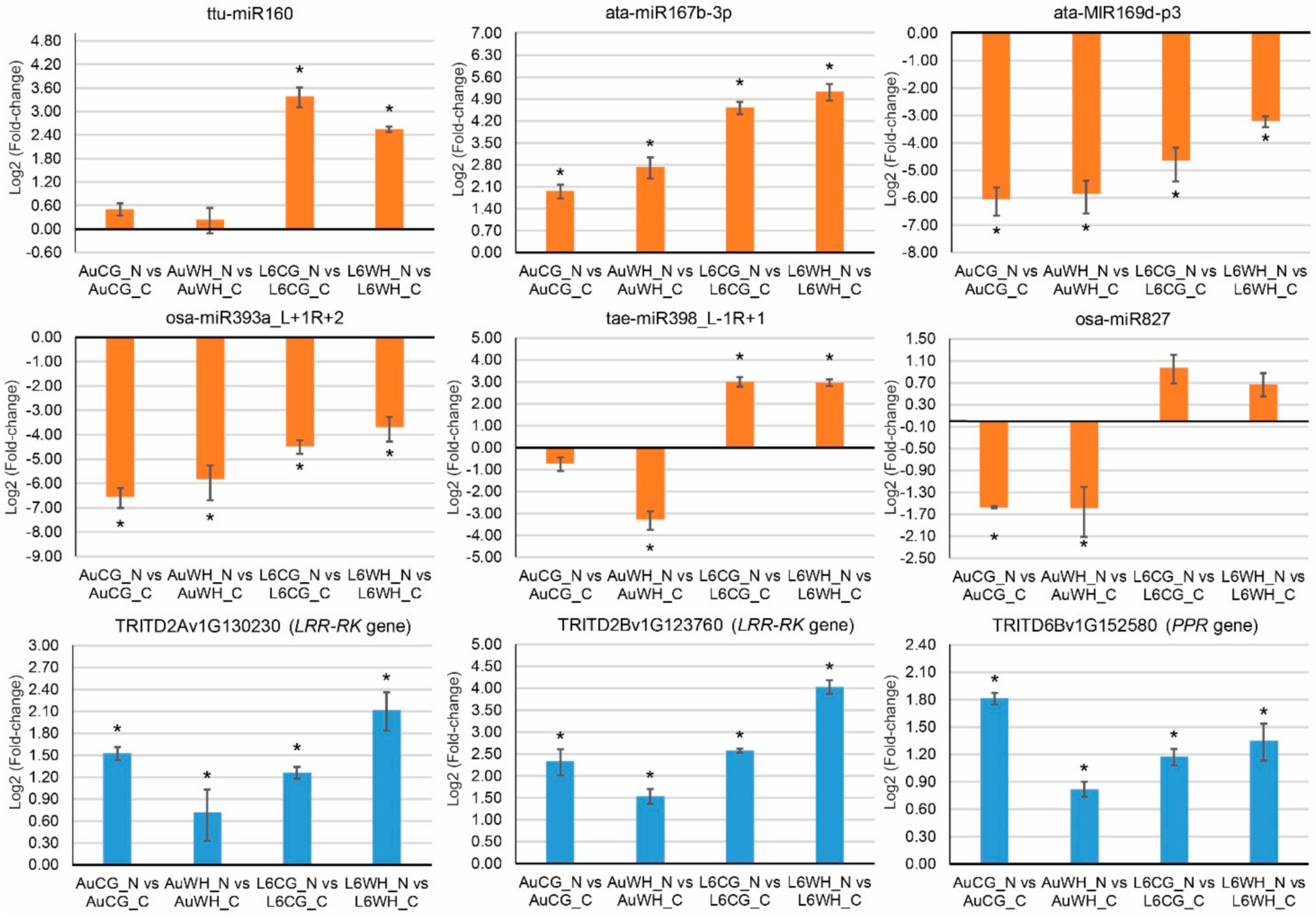
2.5. QPCR Analysis of DEMs and Their Target Genes
3. Discussion
4. Materials and Methods
4.1. Plant Growing Conditions
4.2. Seedling Measurement and Statistical Analysis
4.3. Small RNA Sequencing Analysis of Conserved and Novel Durum MiRNAs
4.4. Identification of MiRNA Target Genes and Functional Annotation
4.5. QPCR Analysis of Selected MiRNAs and Target Genes
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, H.; Searle, I.R.; Mather, D.E.; Able, A.J.; Able, J.A. Morphological, physiological and yield responses of durum wheat to pre-anthesis water-deficit stress are genotype-dependent. Crop Pasture Sci. 2015, 66, 1024–1038. [Google Scholar] [CrossRef]
- Tidiane Sall, A.; Chiari, T.; Legesse, W.; Seid-Ahmed, K.; Ortiz, R.; Van Ginkel, M.; Bassi, F.M. Durum wheat (Triticum durum Desf.): Origin, cultivation and potential expansion in Sub-Saharan Africa. Agronomy 2019, 9, 263. [Google Scholar] [CrossRef]
- Sissons, M. Role of durum wheat composition on the quality of pasta and bread. Food 2008, 2, 75–90. [Google Scholar]
- Liu, H.; Able, A.J.; Able, J.A. Genotypic performance of Australian durum under single and combined water-deficit and heat stress during reproduction. Sci. Rep. 2019, 9, 1–17. [Google Scholar] [CrossRef]
- Li, Y.-F.; Wu, Y.; Hernandez-Espinosa, N.; Peña, R.J. Heat and drought stress on durum wheat: Responses of genotypes, yield, and quality parameters. J. Cereal Sci. 2013, 57, 398–404. [Google Scholar] [CrossRef]
- Rampino, P.; Mita, G.; Fasano, P.; Borrelli, G.M.; Aprile, A.; Dalessandro, G.; De Bellis, L.; Perrotta, C. Novel durum wheat genes up-regulated in response to a combination of heat and drought stress. Plant Physiol. Biochem. 2012, 56, 72–78. [Google Scholar] [CrossRef]
- Guzmán, C.; Autrique, J.E.; Mondal, S.; Singh, R.P.; Govindan, V.; Morales-Dorantes, A.; Posadas-Romano, G.; Crossa, J.; Ammar, K.; Peña, R.J. Response to drought and heat stress on wheat quality, with special emphasis on bread-making quality, in durum wheat. Field Crops Res. 2016, 186, 157–165. [Google Scholar] [CrossRef]
- Aprile, A.; Havlickova, L.; Panna, R.; Mare, C.; Borrelli, G.M.; Marone, D.; Perrotta, C.; Rampino, P.; De Bellis, L.; Curn, V.; et al. Different stress responsive strategies to drought and heat in two durum wheat cultivars with contrasting water use efficiency. BMC Genom. 2013, 14, 1–18. [Google Scholar] [CrossRef]
- Giusti, L.; Mica, E.; Bertolini, E.; De Leonardis, A.M.; Faccioli, P.; Cattivelli, L.; Crosatti, C. microRNAs differentially modulated in response to heat and drought stress in durum wheat cultivars with contrasting water use efficiency. Funct. Integr. Genom. 2017, 17, 293–309. [Google Scholar] [CrossRef]
- Liu, H.; Bruce, D.R.; Sissons, M.; Able, A.J.; Able, J.A. Genotype-dependent changes in the phenolic content of durum under water-deficit stress. Cereal Chem. 2018, 95, 59–78. [Google Scholar] [CrossRef]
- Liu, H.; Able, A.J.; Able, J.A. Small RNAs and their targets are associated with the transgenerational effects of water-deficit stress in durum wheat. Sci. Rep. 2021, 11, 1–17. [Google Scholar]
- Liu, H.; Able, A.J.; Able, J.A. Transgenerational effects of water-deficit and heat stress on germination and seedling vigour—New insights from durum wheat microRNAs. Plants 2020, 9, 189. [Google Scholar] [CrossRef]
- Racette, K.; Zurweller, B.; Tillman, B.; Rowland, D. Transgenerational stress memory of water deficit in peanut production. Field Crops Res. 2020, 248, 107712. [Google Scholar] [CrossRef]
- Hatzig, S.V.; Nuppenau, J.-N.; Snowdon, R.J.; Schießl, S.V. Drought stress has transgenerational effects on seeds and seedlings in winter oilseed rape (Brassica napus L.). BMC Plant Biol. 2018, 18, 297. [Google Scholar] [CrossRef]
- Zheng, X.; Chen, L.; Xia, H.; Wei, H.; Lou, Q.; Li, M.; Li, T.; Luo, L. Transgenerational epimutations induced by multi-generation drought imposition mediate rice plant’s adaptation to drought condition. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef]
- Nosalewicz, A.; Siecińska, J.; Śmiech, M.; Nosalewicz, M.; Wiącek, D.; Pecio, A.; Wach, D. Transgenerational effects of temporal drought stress on spring barley morphology and functioning. Environ. Exp. Bot. 2016, 131, 120–127. [Google Scholar] [CrossRef]
- Migicovsky, Z.; Yao, Y.; Kovalchuk, I. Transgenerational phenotypic and epigenetic changes in response to heat stress in Arabidopsis thaliana. Plant Signal. Behav. 2014, 9, e27971. [Google Scholar] [CrossRef]
- Yamamoto, C.J.T.; Leite, R.G.F.; Minamiguchi, J.Y.; Braga, I.; Neto, N.B.M.; Custódio, C.C. Water-deficit tolerance induction during germination of Jalo Precoce bean (Phaseolus vulgaris L.) cultivar. Acta Physiol. Plant. 2014, 36, 2897–2904. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, Y.; Huang, B. Lipidomic reprogramming associated with drought stress priming-enhanced heat tolerance in tall fescue (Festuca arundinacea). Plant Cell Environ. 2019, 42, 947–958. [Google Scholar] [CrossRef]
- Hossain, M.A.; Li, Z.-G.; Hoque, T.S.; Burritt, D.J.; Fujita, M.; Munné-Bosch, S. Heat or cold priming-induced cross-tolerance to abiotic stresses in plants: Key regulators and possible mechanisms. Protoplasma 2018, 255, 399–412. [Google Scholar] [CrossRef]
- Tabassum, T.; Farooq, M.; Ahmad, R.; Zohaib, A.; Wahid, A. Seed priming and transgenerational drought memory improves tolerance against salt stress in bread wheat. Plant Physiol. Biochem. 2017, 118, 362–369. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Wu, X.; Li, C.; Li, M.; Xiong, T.; Tang, Y. Dry matter and nitrogen accumulation, partitioning, and translocation in synthetic-derived wheat cultivars under nitrogen deficiency at the post-jointing stage. Field Crops Res. 2020, 248, 107720. [Google Scholar] [CrossRef]
- Zuluaga, D.L.; De Paola, D.; Janni, M.; Curci, P.L.; Sonnante, G. Durum wheat miRNAs in response to nitrogen starvation at the grain filling stage. PLoS ONE 2017, 12, e0183253. [Google Scholar]
- Modhej, A.; Naderi, A.; Emam, Y.; Aynehband, A.; Normohamadi, G. Effects of post-anthesis heat stress and nitrogen levels on grain yield in wheat (T. durum and T. aestivum) genotypes. Int. J. Plant Prod. 2012, 2, 257–268. [Google Scholar]
- Zuluaga, D.L.; Liuzzi, V.; Curci, P.L.; Sonnante, G. MicroRNAs in durum wheat seedlings under chronic and short-term nitrogen stress. Funct. Integr. Genom. 2018, 18, 645–657. [Google Scholar] [CrossRef] [PubMed]
- Ercoli, L.; Lulli, L.; Mariotti, M.; Masoni, A.; Arduini, I. Post-anthesis dry matter and nitrogen dynamics in durum wheat as affected by nitrogen supply and soil water availability. Eur. J. Agron. 2008, 28, 138–147. [Google Scholar] [CrossRef]
- Živčák, M.; Olšovská, K.; Slamka, P.; Galambošová, J.; Rataj, V.; Shao, H.; Brestič, M. Application of chlorophyll fluorescence performance indices to assess the wheat photosynthetic functions influenced by nitrogen deficiency. Plant Soil Environ. 2015, 60, 210–215. [Google Scholar] [CrossRef]
- Wang, J.; Song, K.; Sun, L.; Qin, Q.; Sun, Y.; Pan, J.; Xue, Y. Morphological and transcriptome analysis of wheat seedlings response to low nitrogen stress. Plants 2019, 8, 98. [Google Scholar] [CrossRef]
- Guo, T.; Xuan, H.; Yang, Y.; Wang, L.; Wei, L.; Wang, Y.; Kang, G. Transcription analysis of genes encoding the wheat root transporter NRT1 and NRT2 families during nitrogen starvation. J. Plant Growth Regul. 2014, 33, 837–848. [Google Scholar] [CrossRef]
- Sinha, S.K.; Rani, M.; Bansal, N.; Venkatesh, K.; Mandal, P. Nitrate starvation induced changes in root system architecture, carbon: Nitrogen metabolism, and miRNA expression in nitrogen-responsive wheat genotypes. Appl. Biochem. Biotechnol. 2015, 177, 1299–1312. [Google Scholar] [CrossRef]
- Curci, P.L.; Cigliano, R.A.; Zuluaga, D.L.; Janni, M.; Sanseverino, W.; Sonnante, G. Transcriptomic response of durum wheat to nitrogen starvation. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef]
- Ruuska, S.A.; Lewis, D.C.; Kennedy, G.; Furbank, R.T.; Jenkins, C.L.; Tabe, L.M. Large scale transcriptome analysis of the effects of nitrogen nutrition on accumulation of stem carbohydrate reserves in reproductive stage wheat. Plant Mol. Biol. Report. 2008, 66, 15–32. [Google Scholar] [CrossRef]
- Gao, S.; Guo, C.; Zhang, Y.; Zhang, F.; Du, X.; Gu, J.; Xiao, K. Wheat microRNA member TaMIR444a is nitrogen deprivation-responsive and involves plant adaptation to the nitrogen-starvation stress. Plant Mol. Biol. Rep. 2016, 34, 931–946. [Google Scholar] [CrossRef]
- Tiwari, J.K.; Buckseth, T.; Zinta, R.; Saraswati, A.; Singh, R.K.; Rawat, S.; Chakrabarti, S.K. Genome-wide identification and characterization of microRNAs by small RNA sequencing for low nitrogen stress in potato. PLoS ONE 2020, 15, e0233076. [Google Scholar] [CrossRef]
- Zhao, Y.; Xu, K.; Liu, G.; Li, S.; Zhao, S.; Liu, X.; Yang, X.; Xiao, K. Global identification and characterization of miRNA family members responsive to potassium deprivation in wheat (Triticum aestivum L.). Sci. Rep. 2020, 10, 1–13. [Google Scholar]
- Liu, H.; Able, A.J.; Able, J.A. SMARTER de-stressed cereal breeding. Trends Plant Sci. 2016, 21, 909–925. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Allen, E.; Xie, Z.X.; Gustafson, A.M.; Carrington, J.C. MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 2005, 121, 207–221. [Google Scholar] [CrossRef]
- Mallory, A.C.; Bartel, D.P.; Bartel, B. MicroRNA-directed regulation of Arabidopsis Auxin Response Factor17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 2005, 17, 1360–1375. [Google Scholar] [CrossRef]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef]
- Budak, H.; Zhang, B. MicroRNAs in model and complex organisms. Funct. Integr. Genom. 2017, 17, 121–124. [Google Scholar] [CrossRef] [PubMed]
- Budak, H.; Akpinar, B.A. Plant miRNAs: Biogenesis, organization and origins. Funct. Integr. Genom. 2015, 15, 523–531. [Google Scholar] [CrossRef] [PubMed]
- Alptekin, B.; Langridge, P.; Budak, H. Abiotic stress miRNomes in the Triticeae. Funct. Integr. Genom. 2016, 17, 145–170. [Google Scholar] [CrossRef] [PubMed]
- Budak, H.; Kantar, M.; Bulut, R.; Akpinar, B.A. Stress responsive miRNAs and isomiRs in cereals. Plant Sci. 2015, 235, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhang, B. MicroRNAs in control of plant development. J. Cell. Physiol. 2016, 231, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B. MicroRNA: A new target for improving plant tolerance to abiotic stress. J. Exp. Bot. 2015, 66, 1749–1761. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wang, Q. MicroRNA-based biotechnology for plant improvement. J. Cell. Physiol. 2015, 230, 1–15. [Google Scholar] [CrossRef]
- Liu, H.; Able, A.J.; Able, J.A. Water-deficit stress responsive microRNAs and their targets in four durum wheat genotypes. Funct. Integr. Genom. 2017, 17, 237–251. [Google Scholar] [CrossRef]
- Liu, H.; Able, A.J.; Able, J.A. Genotypic water-deficit stress responses in durum wheat: Association between physiological traits, microRNA regulatory modules and yield components. Funct. Plant Biol. 2017, 44, 538–551. [Google Scholar] [CrossRef]
- De Paola, D.; Zuluaga, D.L.; Sonnante, G. The miRNAome of durum wheat: Isolation and characterisation of conserved and novel microRNAs and their target genes. BMC Genom. 2016, 17, 505. [Google Scholar] [CrossRef]
- Liu, H.; Searle, I.R.; Watson-Haigh, N.S.; Baumann, U.; Mather, D.E.; Able, A.J.; Able, J.A. Genome-wide identification of microRNAs in leaves and the developing head of four durum genotypes during water deficit stress. PLoS ONE 2015, 10, e0142799. [Google Scholar] [CrossRef]
- Akpinar, B.A.; Kantar, M.; Budak, H. Root precursors of microRNAs in wild emmer and modern wheats show major differences in response to drought stress. Funct. Integr. Genom. 2015, 15, 587–598. [Google Scholar] [CrossRef]
- Liu, H.; Able, A.J.; Able, J.A. Multi-omics analysis of small RNA, transcriptome, and degradome in T. turgidum—Regulatory networks of grain development and abiotic stress response. Int. J. Mol. 2020, 21, 7772. [Google Scholar] [CrossRef]
- Liu, H.; Able, A.J.; Able, J.A. Integrated analysis of small RNA, transcriptome, and degradome sequencing reveals the water-deficit and heat stress response network in durum wheat. Int. J. Mol. 2020, 21, 6017. [Google Scholar] [CrossRef]
- Barnabás, B.; Jäger, K.; Fehér, A. The effect of drought and heat stress on reproductive processes in cereals. Plant Cell Environ. 2008, 31, 11–38. [Google Scholar] [CrossRef]
- Akpinar, B.A.; Lucas, S.J.; Budak, H. Genomics approaches for crop improvement against abiotic stress. Sci. World J. 2013. [Google Scholar] [CrossRef]
- Peleg, Z.; Blumwald, E. Hormone balance and abiotic stress tolerance in crop plants. Curr. Opin. Plant Biol. 2011, 14, 290–295. [Google Scholar] [CrossRef]
- Crisp, P.A.; Ganguly, D.; Eichten, S.R.; Borevitz, J.O.; Pogson, B.J. Reconsidering plant memory: Intersections between stress recovery, RNA turnover, and epigenetics. Sci. Adv. 2016, 2, e1501340. [Google Scholar] [CrossRef]
- Wijewardana, C.; Reddy, K.R.; Krutz, L.J.; Gao, W.; Bellaloui, N. Drought stress has transgenerational effects on soybean seed germination and seedling vigor. PLoS ONE 2019, 14, e0214977. [Google Scholar] [CrossRef]
- Zheng, X.; Chen, L.; Li, M.; Lou, Q.; Xia, H.; Wang, P.; Li, T.; Liu, H.; Luo, L. Transgenerational variations in DNA methylation induced by drought stress in two rice varieties with distinguished difference to drought resistance. PLoS ONE 2013, 8, e80253. [Google Scholar] [CrossRef]
- Zuluaga, D.L.; Sonnante, G. The use of nitrogen and its regulation in cereals: Structural genes, transcription factors, and the role of miRNAs. Plants 2019, 8, 294. [Google Scholar] [CrossRef]
- Feng, Q.; Yang, C.; Lin, X.; Wang, J.; Ou, X.; Zhang, C.; Chen, Y.; Liu, B. Salt and alkaline stress induced transgenerational alteration in DNA methylation of rice (‘Oryza sativa’). Aust. J. Crop Sci. 2012, 6, 877–883. [Google Scholar]
- Perez, I.B.; Brown, P.J. The role of ROS signaling in cross-tolerance: From model to crop. Front. Plant Sci. 2014, 5, 754. [Google Scholar] [CrossRef]
- Liu, Q.; Zhang, Y.-C.; Wang, C.-Y.; Luo, Y.-C.; Huang, Q.-J.; Chen, S.-Y.; Zhou, H.; Qu, L.-H.; Chen, Y.-Q. Expression analysis of phytohormone-regulated microRNAs in rice, implying their regulation roles in plant hormone signaling. FEBS Lett. 2009, 583, 723–728. [Google Scholar] [CrossRef]
- Yamamuro, C.; Zhu, J.-K.; Yang, Z. Epigenetic modifications and plant hormone action. Mol. Plant 2016, 9, 57–70. [Google Scholar] [CrossRef]
- Wang, Y.; Li, K.; Chen, L.; Zou, Y.; Liu, H.; Tian, Y.; Li, D.; Wang, R.; Zhao, F.; Ferguson, B.J.; et al. microRNA167-directed regulation of the auxin response factors, GmARF8a and GmARF8b, is required for soybean nodulation and lateral root development. Plant Physiol. 2015, 168, 984–999. [Google Scholar] [CrossRef]
- Schommer, C.; Palatnik, J.F.; Aggarwal, P.; Chételat, A.; Cubas, P.; Farmer, E.E.; Nath, U.; Weigel, D. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 2008, 6, e230. [Google Scholar] [CrossRef]
- Hernández, Y.; Sanan-Mishra, N. miRNA mediated regulation of NAC transcription factors in plant development and environment stress response. Plant Gene 2017, 11, 190–198. [Google Scholar] [CrossRef]
- Devi, S.R.; Madhav, M.; Kumar, G.R.; Goel, A.; Umakanth, B.; Jahnavi, B.; Viraktamath, B. Identification of abiotic stress miRNA transcription factor binding motifs (TFBMs) in rice. Gene 2013, 531, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Kapoor, A.; Zhu, J.-K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 2006, 18, 2051–2065. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Ding, Y.; Liu, H. MiR398 and plant stress responses. Physiol. Plant 2011, 143, 1–9. [Google Scholar] [CrossRef]
- Zhu, H.; Chen, C.; Zeng, J.; Yun, Z.; Liu, Y.; Qu, H.; Jiang, Y.; Duan, X.; Xia, R. MicroRNA528, a hub regulator modulating ROS homeostasis via targeting of a diverse set of genes encoding copper-containing proteins in monocots. New Phytol. 2020, 225, 385–399. [Google Scholar] [CrossRef]
- Fileccia, V.; Bertolini, E.; Ruisi, P.; Giambalvo, D.; Frenda, A.S.; Cannarozzi, G.; Tadele, Z.; Crosatti, C.; Martinelli, F. Identification and characterization of durum wheat microRNAs in leaf and root tissues. Funct. Integr. Genom. 2017, 17, 583–598. [Google Scholar] [CrossRef]
- Danquah, A.; de Zelicourt, A.; Colcombet, J.; Hirt, H. The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol. Adv. 2014, 32, 40–52. [Google Scholar] [CrossRef]
- Pitzschke, A.; Schikora, A.; Hirt, H. MAPK cascade signalling networks in plant defence. Curr. Opin. Plant Biol. 2009, 12, 421–426. [Google Scholar] [CrossRef]
- Tabata, R.; Sumida, K.; Yoshii, T.; Ohyama, K.; Shinohara, H.; Matsubayashi, Y. Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science 2014, 346, 343–346. [Google Scholar] [CrossRef]
- Xu, G.; Fan, X.; Miller, A.J. Plant nitrogen assimilation and use efficiency. Annu. Rev. Plant Biol. 2012, 63, 153–182. [Google Scholar] [CrossRef]
- White, P.J. Long-distance transport in the xylem and phloem. In Marschner’s Mineral Nutrition of Higher Plants; Elsevier: Armsterdam, The Netherlands, 2012; pp. 49–70. [Google Scholar]
- Lacombe, B.; Achard, P. Long-distance transport of phytohormones through the plant vascular system. Curr. Opin. Plant Biol. 2016, 34, 1–8. [Google Scholar] [CrossRef]
- Liu, F.; Andersen, M.N.; Jensen, C.R. Loss of pod set caused by drought stress is associated with water status and ABA content of reproductive structures in soybean. Funct. Plant Biol. 2003, 30, 271–280. [Google Scholar] [CrossRef]
- Liu, F.; Jensen, C.R.; Andersen, M.N. A review of drought adaptation in crop plants: Changes in vegetative and reproductive physiology induced by ABA-based chemical signals. Aust. J. Agric. Res. 2005, 56, 1245–1252. [Google Scholar] [CrossRef]
- Zhang, C.-X.; Feng, B.-H.; Chen, T.-T.; Fu, W.-M.; Li, H.-B.; Li, G.-Y.; Jin, Q.-Y.; Tao, L.-X.; Fu, G.-F. Heat stress-reduced kernel weight in rice at anthesis is associated with impaired source-sink relationship and sugars allocation. Environ. Exp. Bot. 2018, 155, 718–733. [Google Scholar] [CrossRef]
- Li, J.-Y.; Fu, Y.-L.; Pike, S.M.; Bao, J.; Tian, W.; Zhang, Y.; Chen, C.-Z.; Zhang, Y.; Li, H.-M.; Huang, J. The Arabidopsis nitrate transporter NRT1. 8 functions in nitrate removal from the xylem sap and mediates cadmium tolerance. Plant Cell 2010, 22, 1633–1646. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ouyang, J.; Wang, Y.-Y.; Hu, R.; Xia, K.; Duan, J.; Wang, Y.; Tsay, Y.-F.; Zhang, M. Disruption of the rice nitrate transporter OsNPF2. 2 hinders root-to-shoot nitrate transport and vascular development. Sci. Rep. 2015, 5, 1–10. [Google Scholar]
- Hsu, P.-K.; Tsay, Y.-F. Two phloem nitrate transporters, NRT1. 11 and NRT1. 12, are important for redistributing xylem-borne nitrate to enhance plant growth. Plant Physiol. 2013, 163, 844–856. [Google Scholar] [CrossRef]
- Boursiac, Y.; Léran, S.; Corratgé-Faillie, C.; Gojon, A.; Krouk, G.; Lacombe, B. ABA transport and transporters. Trends Plant Sci. 2013, 18, 325–333. [Google Scholar] [CrossRef]
- Glass, A.D.; Britto, D.T.; Kaiser, B.N.; Kinghorn, J.R.; Kronzucker, H.J.; Kumar, A.; Okamoto, M.; Rawat, S.; Siddiqi, M.; Unkles, S.E. The regulation of nitrate and ammonium transport systems in plants. J. Exp. Bot. 2002, 53, 855–864. [Google Scholar] [CrossRef]
- Miller, A.J.; Fan, X.; Orsel, M.; Smith, S.J.; Wells, D.M. Nitrate transport and signalling. J. Exp. Bot. 2007, 58, 2297–2306. [Google Scholar] [CrossRef]
- Zheng, D.; Han, X.; An, Y.; Guo, H.; Xia, X.; Yin, W. The nitrate transporter NRT2. 1 functions in the ethylene response to nitrate deficiency in Arabidopsis. Plant Cell Environ. 2013, 36, 1328–1337. [Google Scholar] [CrossRef]
- Remans, T.; Nacry, P.; Pervent, M.; Girin, T.; Tillard, P.; Lepetit, M.; Gojon, A. A central role for the nitrate transporter NRT2. 1 in the integrated morphological and physiological responses of the root system to nitrogen limitation in Arabidopsis. Plant Physiol. 2006, 140, 909–921. [Google Scholar] [CrossRef]
- Barkan, A.; Small, I. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 2014, 65, 415–442. [Google Scholar] [CrossRef]
- Huang, J.; Lu, G.; Liu, L.; Raihan, M.S.; Xu, J.; Jian, L.; Zhao, L.; Tran, T.M.; Zhang, Q.; Liu, J. The kernel size-related quantitative trait locus qkw9 encodes a pentatricopeptide repeat protein that aaffects photosynthesis and grain filling. Plant Physiol. 2020, 183, 1696–1709. [Google Scholar] [CrossRef]
- Wu, M.; Ren, Y.; Cai, M.; Wang, Y.; Zhu, S.; Zhu, J.; Hao, Y.; Teng, X.; Zhu, X.; Jing, R. Rice FLOURY ENDOSPERM 10 encodes a pentatricopeptide repeat protein that is essential for the trans-splicing of mitochondrial nad1 intron 1 and endosperm development. New Phytol. 2019, 223, 736–750. [Google Scholar] [CrossRef]
- Liu, Y.-J.; Xiu, Z.-H.; Meeley, R.; Tan, B.-C. Empty pericarp5 encodes a pentatricopeptide repeat protein that is required for mitochondrial RNA editing and seed development in maize. Plant Cell 2013, 25, 868–883. [Google Scholar] [CrossRef]
- Wu, L.; Wu, J.; Liu, Y.; Gong, X.; Xu, J.; Lin, D.; Dong, Y. The rice pentatricopeptide repeat gene TCD10 is needed for chloroplast development under cold stress. Rice 2016, 9, 1–13. [Google Scholar] [CrossRef]



| Treatment Group | Seedling Height (cm) | Leaf Number | Shoot Fresh Weight (g) | Shoot Dry Weight (g) | Root Fresh Weight (g) | Root Dry Weight (g) | Primary Root Length (cm) | Chlorophyll Content (SPAD Units) |
|---|---|---|---|---|---|---|---|---|
| AuCG_C | 36.92 ± 0.77 | 5.42 ± 0.15 | 1.658 ± 0.058 | 0.211 ± 0.009 | 1.327 ± 0.026 | 0.155 ± 0.004 | 27.62 ± 0.46 | 48.33 ± 0.70 |
| AuCG_N | 24.17 ± 0.42 | 3.17 ± 0.25 | 0.605 ± 0.022 | 0.087 ± 0.003 | 0.779 ± 0.018 | 0.094 ± 0.001 | 23.80 ± 0.34 | 38.38 ± 0.57 |
| % Reduction | 34.5% | 41.5% | 63.5% | 60.7% | 41.3% | 39.3% | 13.8% | 20.6% |
| AuWH_C | 35.93 ± 0.53 | 5.33 ± 0.17 | 1.630 ± 0.060 | 0.214 ± 0.006 | 1.328 ± 0.027 | 0.154 ± 0.003 | 28.77 ± 0.34 | 49.18 ± 0.55 |
| AuWH_N | 24.13 ± 0.50 | 3.25 ± 0.17 | 0.640 ± 0.019 | 0.089 ± 0.001 | 0.804 ± 0.023 | 0.097 ± 0.002 | 25.52 ± 0.41 | 40.28 ± 0.55 |
| % Reduction | 32.8% | 39.1% | 60.7% | 58.4% | 39.4% | 37.1% | 11.3% | 18.1% |
| F pr. Parent treatment | 0.381 | 1.000 | 0.940 | 0.667 | 0.584 | 0.876 | 0.001 | 0.032 |
| F pr. Progeny treatment | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| F pr. Parent × Progeny treatment | 0.413 | 0.663 | 0.481 | 0.412 | 0.613 | 0.447 | 0.476 | 0.388 |
| l.s.d Parent treatment | n.a 1 | n.a | n.a | n.a | n.a | n.a | 0.813 | 1.242 |
| l.s.d Progeny treatment | 1.184 | 0.393 | 0.092 | 0.011 | 0.049 | 0.005 | 0.813 | 1.242 |
| l.s.d Parent × Progeny treatment | n.a | n.a | n.a | n.a | n.a | n.a | n.a | n.a |
| Treatment Group | Seedling Height (cm) | Leaf Number | Shoot Fresh Weight (g) | Shoot Dry Weight (g) | Root Fresh Weight (g) | Root Dry Weight (g) | Primary Root Length (cm) | Chlorophyll Content (SPAD Units) |
|---|---|---|---|---|---|---|---|---|
| L6CG_C | 35.47 ± 0.45 | 5.00 ± 0.22 | 1.556 ± 0.040 | 0.205 ± 0.005 | 1.313 ± 0.024 | 0.152 ± 0.002 | 27.13 ± 0.35 | 47.33 ± 0.73 |
| L6CG_N | 22.73 ± 0.57 | 2.83 ± 0.17 | 0.562 ± 0.010 | 0.076 ± 0.002 | 0.755 ± 0.019 | 0.090 ± 0.002 | 23.28 ± 0.37 | 35.97 ± 0.60 |
| % Reduction | 35.9% | 43.3% | 63.9% | 63.1% | 42.5% | 40.9% | 14.2% | 24.0% |
| L6WH_C | 34.42 ± 0.57 | 5.00 ± 0.18 | 1.502 ± 0.046 | 0.199 ± 0.006 | 1.303 ± 0.021 | 0.152 ± 0.003 | 27.83 ± 0.31 | 46.28 ± 0.56 |
| L6WH_N | 22.60 ± 0.63 | 2.75 ± 0.11 | 0.517 ± 0.011 | 0.072 ± 0.001 | 0.735 ± 0.022 | 0.088 ± 0.002 | 24.00 ± 0.31 | 34.90 ± 0.62 |
| % Reduction | 34.3% | 45.0% | 65.6% | 63.9% | 43.6% | 41.7% | 13.8% | 24.6% |
| F pr. Parent treatment | 0.301 | 0.815 | 0.133 | 0.211 | 0.507 | 0.704 | 0.047 | 0.108 |
| F pr. Progeny treatment | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| F pr. Parent × Progeny treatment | 0.421 | 0.815 | 0.893 | 0.759 | 0.826 | 0.799 | 0.980 | 0.990 |
| F pr. Parent treatment | n.a 1 | n.a | n.a | n.a | n.a | n.a | 0.697 | n.a |
| F pr. Progeny treatment | 1.164 | 0.367 | 0.066 | 0.009 | 0.045 | 0.005 | 0.697 | 1.313 |
| F pr. Parent × Progeny treatment | n.a | n.a | n.a | n.a | n.a | n.a | n.a | n.a |
| Pathway ID | KEGG Pathway Description | S Gene Number | TS Gene Number | B Gene Number | TB Gene Number |
|---|---|---|---|---|---|
| AuCG_N vs. AuCG_C | |||||
| ko04010 | MAPK signaling pathway | 286 | 1783 | 2171 | 50,492 |
| ko04620 | Toll-like receptor signaling pathway | 167 | 1783 | 1298 | 50,492 |
| ko04624 | Toll and Imd signaling pathway | 126 | 1783 | 633 | 50,492 |
| ko05145 | Toxoplasmosis | 118 | 1783 | 1046 | 50,492 |
| ko04621 | NOD-like receptor signaling pathway | 104 | 1783 | 1111 | 50,492 |
| AuWH_N vs. AuWH_C | |||||
| ko04010 | MAPK signaling pathway | 101 | 1556 | 2171 | 50,492 |
| ko04070 | Phosphatidylinositol signaling system | 91 | 1556 | 1224 | 50,492 |
| ko04141 | Protein processing in endoplasmic reticulum | 89 | 1556 | 2406 | 50,492 |
| ko00562 | Inositol phosphate metabolism | 84 | 1556 | 989 | 50,492 |
| ko00230 | Purine metabolism | 84 | 1556 | 1483 | 50,492 |
| L6CG_N vs. L6CG_C | |||||
| ko04141 | Protein processing in endoplasmic reticulum | 170 | 1451 | 2406 | 50,492 |
| ko04070 | Phosphatidylinositol signaling system | 116 | 1451 | 1224 | 50,492 |
| ko00562 | Inositol phosphate metabolism | 85 | 1451 | 989 | 50,492 |
| ko00250 | Alanine, aspartate and glutamate metabolism | 73 | 1451 | 542 | 50,492 |
| ko04144 | Endocytosis | 72 | 1451 | 1720 | 50,492 |
| L6WH_N vs. L6WH_C | |||||
| ko04010 | MAPK signaling pathway | 326 | 1905 | 2171 | 50,492 |
| ko04620 | Toll-like receptor signaling pathway | 186 | 1905 | 1298 | 50,492 |
| ko05145 | Toxoplasmosis | 126 | 1905 | 1046 | 50,492 |
| ko04141 | Protein processing in endoplasmic reticulum | 125 | 1905 | 2406 | 50,492 |
| ko04624 | Toll and Imd signaling pathway | 122 | 1905 | 633 | 50,492 |
| Pathway ID | KEGG Pathway Description | S Gene Number | TS Gene Number | B Gene Number | TB Gene Number |
|---|---|---|---|---|---|
| AuWH_C vs. AuCG_C | |||||
| ko04141 | Protein processing in endoplasmic reticulum | 87 | 789 | 2406 | 50,492 |
| ko04010 | MAPK signaling pathway | 77 | 789 | 2171 | 50,492 |
| ko03015 | mRNA surveillance pathway | 70 | 789 | 1365 | 50,492 |
| ko03040 | Spliceosome | 68 | 789 | 1911 | 50,492 |
| ko04144 | Endocytosis | 60 | 789 | 1720 | 50,492 |
| AuWH_N vs. AuCG_N | |||||
| ko04010 | MAPK signaling pathway | 80 | 1031 | 2171 | 50,492 |
| ko04070 | Phosphatidylinositol signaling system | 64 | 1031 | 1224 | 50,492 |
| ko00562 | Inositol phosphate metabolism | 56 | 1031 | 989 | 50,492 |
| ko04146 | Peroxisome | 53 | 1031 | 1272 | 50,492 |
| ko03013 | RNA transport | 53 | 1031 | 1670 | 50,492 |
| L6WH_C vs. L6CG_C | |||||
| ko04010 | MAPK signaling pathway | 258 | 1429 | 2171 | 50,492 |
| ko04620 | Toll-like receptor signaling pathway | 141 | 1429 | 1298 | 50,492 |
| ko03040 | Spliceosome | 109 | 1429 | 1911 | 50,492 |
| ko04624 | Toll and Imd signaling pathway | 101 | 1429 | 633 | 50,492 |
| ko04621 | NOD-like receptor signaling pathway | 100 | 1429 | 1111 | 50,492 |
| L6WH_N vs. L6CG_N | |||||
| ko03018 | RNA degradation | 85 | 1187 | 1274 | 50,492 |
| ko04070 | Phosphatidylinositol signaling system | 82 | 1187 | 1224 | 50,492 |
| ko04141 | Protein processing in endoplasmic reticulum | 73 | 1187 | 2406 | 50,492 |
| ko03040 | Spliceosome | 65 | 1187 | 1911 | 50,492 |
| ko03008 | Ribosome biogenesis in eukaryotes | 52 | 1187 | 871 | 50,492 |
| Pathway ID | KEGG Pathway Description | S Gene Number | TS Gene Number | B Gene Number | TB Gene Number |
|---|---|---|---|---|---|
| AuCG_C vs. L6CG_C | |||||
| ko04010 | MAPK signaling pathway | 91 | 922 | 2171 | 50,492 |
| ko00230 | Purine metabolism | 78 | 922 | 1483 | 50,492 |
| ko04141 | Protein processing in endoplasmic reticulum | 76 | 922 | 2406 | 50,492 |
| ko03015 | mRNA surveillance pathway | 75 | 922 | 1365 | 50,492 |
| ko00240 | Pyrimidine metabolism | 68 | 922 | 1168 | 50,492 |
| AuCG_N vs. L6CG_N | |||||
| ko04141 | Protein processing in endoplasmic reticulum | 196 | 1969 | 2406 | 50,492 |
| ko04010 | MAPK signaling pathway | 123 | 1969 | 2171 | 50,492 |
| ko03018 | RNA degradation | 94 | 1969 | 1274 | 50,492 |
| ko00230 | Purine metabolism | 84 | 1969 | 1483 | 50,492 |
| ko03040 | Spliceosome | 84 | 1969 | 1911 | 50,492 |
| AuWH_C vs. L6WH_C | |||||
| ko04010 | MAPK signaling pathway | 284 | 1457 | 2171 | 50,492 |
| ko04620 | Toll-like receptor signaling pathway | 145 | 1457 | 1298 | 50,492 |
| ko04624 | Toll and Imd signaling pathway | 108 | 1457 | 633 | 50,492 |
| ko04621 | NOD-like receptor signaling pathway | 108 | 1457 | 1111 | 50,492 |
| ko03040 | Spliceosome | 101 | 1457 | 1911 | 50,492 |
| AuWH_N vs. L6WH_N | |||||
| ko00240 | Pyrimidine metabolism | 66 | 693 | 1168 | 50,492 |
| ko00230 | Purine metabolism | 66 | 693 | 1483 | 50,492 |
| ko03020 | RNA polymerase | 46 | 693 | 469 | 50,492 |
| ko00270 | Cysteine and methionine metabolism | 46 | 693 | 1355 | 50,492 |
| ko02010 | ABC transporters | 42 | 693 | 1530 | 50,492 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Able, A.J.; Able, J.A. Nitrogen Starvation-Responsive MicroRNAs Are Affected by Transgenerational Stress in Durum Wheat Seedlings. Plants 2021, 10, 826. https://doi.org/10.3390/plants10050826
Liu H, Able AJ, Able JA. Nitrogen Starvation-Responsive MicroRNAs Are Affected by Transgenerational Stress in Durum Wheat Seedlings. Plants. 2021; 10(5):826. https://doi.org/10.3390/plants10050826
Chicago/Turabian StyleLiu, Haipei, Amanda J. Able, and Jason A. Able. 2021. "Nitrogen Starvation-Responsive MicroRNAs Are Affected by Transgenerational Stress in Durum Wheat Seedlings" Plants 10, no. 5: 826. https://doi.org/10.3390/plants10050826
APA StyleLiu, H., Able, A. J., & Able, J. A. (2021). Nitrogen Starvation-Responsive MicroRNAs Are Affected by Transgenerational Stress in Durum Wheat Seedlings. Plants, 10(5), 826. https://doi.org/10.3390/plants10050826








