An Improved K-Means Algorithm Based on Evidence Distance
Abstract
:1. Introduction
2. Related Theories
2.1. Traditional K-Means Algorithm
| Algorithm 1 The traditional k-means algorithm. |
| Input: data set, k value Output: divided into k clusters
|
2.2. D-S Evidence Theory
3. Algorithm Design
3.1. Algorithm Idea Description
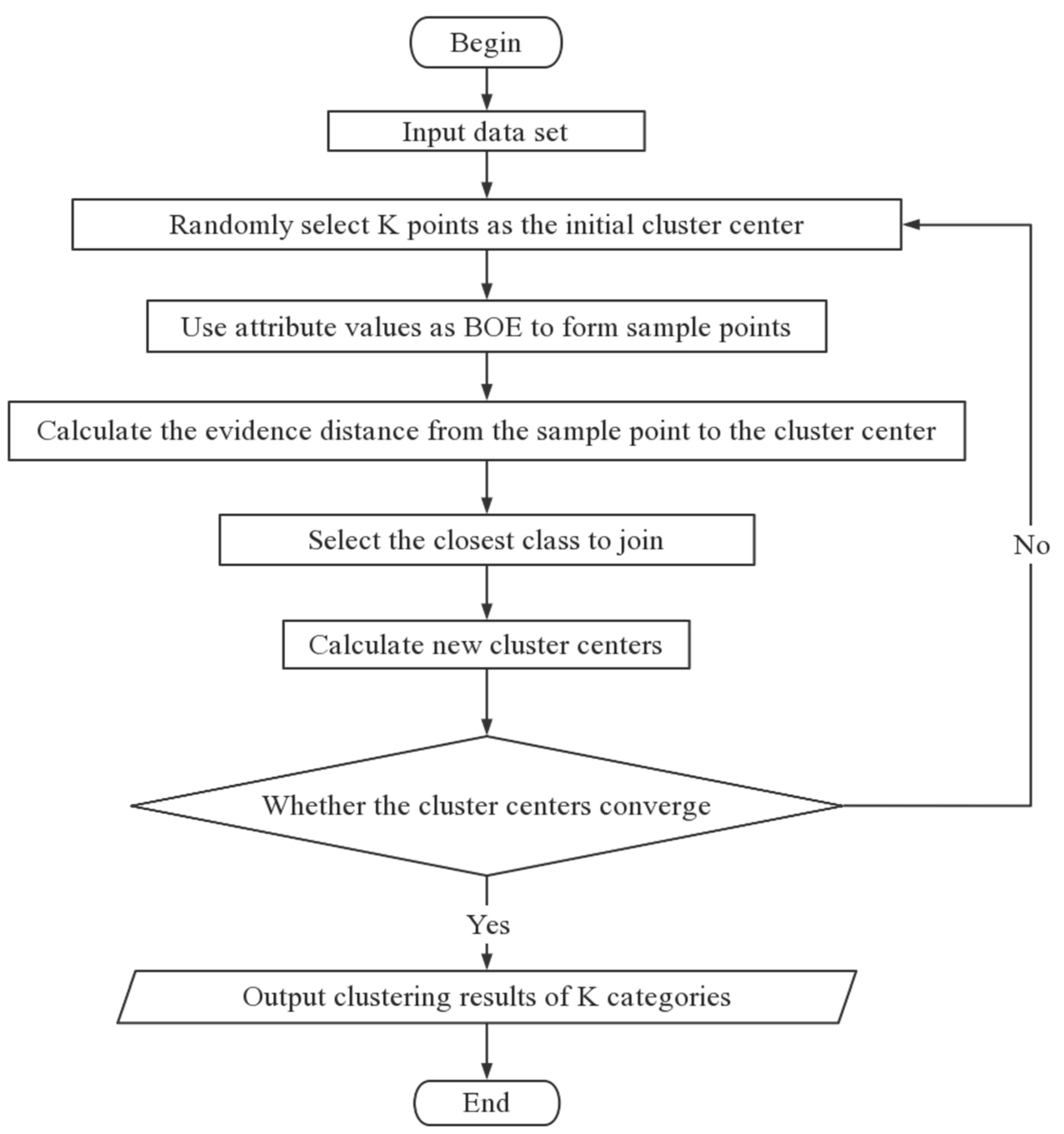
3.2. Algorithm Flow
| Algorithm 2 the k-means algorithm based on evidence distance. |
| Input: data set, k value Output: clustering results
|
4. Experiment
4.1. Experiment Preparation
4.1.1. Experimental Data Set
4.1.2. Experimental Evaluation Indicators
- (1)
- Adjusted Rand index
- (2)
- Silhouette Coefficient
- (3)
- Number of iterations
4.2. Experimental Procedure
- (1)
- Import the iris data set and enter the cluster category k value.
- (2)
- The traditional k-means method and the improved k-means method are used for clustering, respectively.
- (3)
- Perform clustering 10 times, find the average value, and output the ARI, contour coefficient, and number of iterations as the final result.
- (4)
- Compare the experimental results of the improved algorithm and the traditional algorithm.
- (5)
- Use wine, breast cancer, and other data sets for verification.
5. Results and Analysis
5.1. Iris Data Set Test Results
5.2. Validation Results of Other Data Sets
5.3. Algorithm Comparison
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Barua, H.B.; Mondal, K.C. A comprehensive survey on cloud data mining (CDM) frameworks and algorithms. ACM Comput. Surv. 2019, 52, 1–62. [Google Scholar] [CrossRef] [Green Version]
- Atluri, G.; Karpatne, A.; Kumar, V. Spatio-temporal data mining: A survey of problems and methods. ACM Comput. Surv. 2018, 51, 1–41. [Google Scholar] [CrossRef]
- Fei, X.; Tian, G.; Lima, S. Research on data mining algorithm based on neural network and particle swarm optimization. J. Intell. Fuzzy Syst. 2018, 35, 2921–2926. [Google Scholar] [CrossRef]
- Manda, P. Data mining powered by the gene ontology. Wiley Interdisciplinary Reviews. Data Min. Knowl. Discov. 2020, 10, e1359. [Google Scholar]
- Duggirala, H.J.; Tonning, J.M.; Smith, E.; Bright, R.A.; Baker, J.D.; Ball, R.; Bell, C.; Bright-Ponte, S.J.; Botsis, T.; Bouri, K.; et al. Use of data mining at the Food and Drug Administration. J. Am. Med. Inform. Assoc. 2016, 23, 428–434. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Hao, L.; Fan, L. Optimization and improvement of data mining algorithm based on efficient incremental kernel fuzzy clustering for large data. Clust. Comput. 2019, 22, 3001–3010. [Google Scholar] [CrossRef]
- Yu, W. Challenges and reflections of big data mining Based on mobile internet customers. Agro. Food Ind. Hi Tech. 2017, 28, 3221–3224. [Google Scholar]
- Feng, Z.; Zhu, Y. A Survey on Trajectory Data Mining: Techniques and Applications. IEEE Access 2017, 4, 2056–2067. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, Q.; Hung, C.C.; Yang, F. Credibilistic clustering algorithms via alternating cluster estimation. J. Intell. Manuf. 2017, 28, 727–738. [Google Scholar] [CrossRef]
- Bulut, H.; Onan, A.; Korukoğlu, S. An improved ant-based algorithm based on heaps merging and fuzzy c-means for clustering cancer gene expression data. Sādhanā 2020, 45, 1–17. [Google Scholar] [CrossRef]
- Mbyamm Kiki, M.J.; Zhang, J.; Kouassi, B.A. MapReduce FCM clustering set algorithm. Clust. Comput. 2021, 24, 489–500. [Google Scholar] [CrossRef]
- Cao, L.; Liu, Y.; Wang, D.; Wang, T.; Fu, C. A Novel Density Peak Fuzzy Clustering Algorithm for Moving Vehicles Using Traffic Ra-dar. Electronics 2019, 9, 46. [Google Scholar] [CrossRef] [Green Version]
- Gao, W. Improved Ant Colony Clustering Algorithm and Its Performance Study. Comput. Intell. Neurosci. 2016, 2016, 4835932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi, D.; Xian, F. Kernel-based fuzzy c-means clustering algorithm based on genetic algorithm. Neurocomputing 2016, 188, 233–238. [Google Scholar]
- Kuo, R.J.; Mei, C.H.; Zulvia, F.E.; Tsai, C.Y. An application of a metaheuristic algorithm-based clustering ensemble method to APP customer segmentation. Neurocomputing 2016, 205, 116–129. [Google Scholar] [CrossRef]
- Zhan, Q.; Jiang, Y.; Xia, K.; Xue, J.; Hu, W.; Lin, H.; Liu, Y. Epileptic EEG Detection Using a Multi-View Fuzzy Clustering Algorithm with Multi-Medoid. IEEE Access 2019, 7, 152990–152997. [Google Scholar] [CrossRef]
- Ismkhan, H. I-k-means-plus: An iterative clustering algorithm based on an enhanced version of the k-means. Pattern Recognition: J. Pattern. Recognit. Soc. 2018, 79, 402–413. [Google Scholar] [CrossRef]
- Sinaga, K.P.; Hussain, I.; Yang, M.S. Entropy K-Means Clustering with Feature Reduction Under Unknown Number of Clusters. IEEE Access 2021, 9, 67736–67751. [Google Scholar] [CrossRef]
- Wang, X.; Bai, Y. The global Minmax k-means algorithm. Springerplus 2016, 5, 1665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aggarwal, S.; Singh, P. Cuckoo, Bat and Krill Herd based k-means++ clustering algorithms. Clust. Comput. 2018, 22, 14169–14180. [Google Scholar] [CrossRef]
- Yin, C.; Zhang, S. Parallel implementing improved k-means applied for image retrieval and anomaly detection. Multimed. Tools. Appl. 2017, 76, 16911–16927. [Google Scholar] [CrossRef]
- Yu, S.; Chu, S.; Wang, C.; Chan, Y.; Chang, T. Two improved k-means algorithms. Appl. Soft Comput. 2018, 68, 747–755. [Google Scholar] [CrossRef]
- Prasada, M.; Tripathia, S.; Dahalb, K. Unsupervised feature selection and cluster center initialization based arbitrary shaped clusters for intrusion detection. Comput. Secur. 2020, 99, 102062. [Google Scholar] [CrossRef]
- Tang, Z.K.; Zhu, Z.Y.; Yang, Y.; Caihong, L.; Lian, L. D-K-means algorithm based on distance and density. Appl. Res. Comp. 2020, 37, 1719–1723. [Google Scholar]
- Zilong, W.; Jin, L.; Yafei, S. Improved K-means algorithm based on distance and weight. Comp. Eng. Appl. 2020, 56, 87–94. [Google Scholar]
- Wang, Y.; Luo, X.; Zhang, J.; Zhao, Z.; Zhang, J. An Improved Algorithm of K-means Based on Evolutionary Computation. Intell. Autom. Soft Comput. 2020, 26, 961–971. [Google Scholar] [CrossRef]
- Zhao, W.L.; Deng, C.H.; Ngo, C.W. k-means: A revisit. Neurocomputing 2018, 291, 195–206. [Google Scholar] [CrossRef]
- Qi, J.; Yu, Y.; Wang, L.; Liu, J.; Wang, Y. An effective and efficient hierarchical K-means clustering algorithm. Int. J. Distrib. Sens. Netw. 2017, 13, 1550147717728627. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Qi, X.; Chen, L.; Chen, F.; Cheng, G. Quantum-inspired ant lion optimized hybrid k-means for cluster analysis and intrusion detection. Knowl. Based. Syst. 2020, 203, 106167. [Google Scholar] [CrossRef]
- Zhang, G.; Zhang, C.; Zhang, H. Improved K-means algorithm based on density canopy. Knowl. Based. Syst. 2018, 145, 289–297. [Google Scholar] [CrossRef]
- Fred, A.L.; Jain, A.K. Data clustering using evidence accumulation. In Proceedings of the 2002 International Conference on Pattern Recognition, Quebec City, QC, Canada, 11–15 August 2002. [Google Scholar]
- Li, F.; Qian, Y.; Wang, J.; Liang, J. Multigranulation information fusion: A Dempster-Shafer evidence theory-based clustering ensemble method. Inf. Sci. 2017, 378, 389–409. [Google Scholar] [CrossRef]
- Yu, H.; Chen, L.; Yao, J. A three-way density peak clustering method based on evidence theory. Knowl.-Based Syst. 2020, 211, 106532. [Google Scholar] [CrossRef]
- Fränti, P.; Sieranoja, S. K-means properties on six clustering benchmark datasets. Appl. Intell. 2018, 48, 4743–4759. [Google Scholar] [CrossRef]
- Giannella, C.R. Instability results for Euclidean distance, nearest neighbor search on high dimensional Gaussian data. Inf. Process. Lett. 2021, 169, 106115. [Google Scholar] [CrossRef]
- Drusvyatskiy, D.; Lee, H.L.; Ottaviani, G.; Thomas, R.R. The Euclidean distance degree of orthogonally invariant matrix varieties. Isr. J. Math. 2017, 221, 291–316. [Google Scholar] [CrossRef]
- Morin, L.; Gilormini, P.; Derrien, K. Generalized Euclidean distances for elasticity tensors. J. Elast. 2020, 138, 221–232. [Google Scholar] [CrossRef]
- Subba Rao, T. Classification, Parameter Estimation and State Estimation-an Engineering Approach Using MATLAB; John Wiley & Sons, Ltd.: West Sussex, UK, 2011; Volume 32, p. 194. [Google Scholar]
- Dempster, A.P. Upper and Lower Probabilities Induced by a Multivalued Mapping. In Classic Works Dempster–Shafer Theory Belief Functions; Springer: Berlin/Heidelberg, Germany, 1966; Volume 38, pp. 57–72. [Google Scholar]
- Shafer, G. A Mathematical Theory of Evidence; Princeton University Press: Princeton, NJ, USA, 1976; Volume 42. [Google Scholar]
- Tang, Y.; Wu, D.; Liu, Z. A new approach for generation of generalized basic probability assignment in the evidence theory. Pattern Anal. Appl. 2021, 24, 1007–1023. [Google Scholar] [CrossRef]
- Gong, Y.; Su, X.; Qian, H.; Yang, N. Research on fault diagnosis methods for the reactor coolant system of nuclear power plant based on D-S evidence theory. Ann. Nucl. Energy 2018, 112, 395–399. [Google Scholar] [CrossRef]
- Deng, X.; Liu, Q.; Deng, Y.; Mahadevan, S. An improved method to construct basic probability assignment based on the confusion matrix for classification problem. Inf. Sci. 2016, 340, 250–261. [Google Scholar] [CrossRef]
- Yuan, K.; Deng, Y. Conflict evidence management in fault diagnosis. Int. J. Mach. Learn. Cybern. 2019, 10, 121–130. [Google Scholar] [CrossRef]
- Li, M.; Hu, Y.; Zhang, Q.; Deng, Y. A novel distance function of D numbers and its application in product engineering. Eng. Appl. Artif. Intell. 2016, 47, 61–67. [Google Scholar] [CrossRef]
- Mo, H.; Lu, X.; Deng, Y. A generalized evidence distance. J. Syst. Eng. Electron. 2016, 27, 470–476. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Xiao, F.; Deng, X.; Fei, L.; Deng, Y. Weighted evidence combination based on distance of evidence and entropy function. Int. J. Distrib. Sens. Netw. 2016, 12, 3218784. [Google Scholar] [CrossRef] [Green Version]
- Qiaoling, W.; Fei, Q.; Youhao, J. Improved K-means algorithm based on aggregation distance parameter. Int. J. Comput. Appl. 2019, 39, 2586–2590. [Google Scholar]
- Khan, M.H.; Farid, M.S.; Grzegorzek, M. Spatiotemporal features of human motion for gait recognition. Signal Image Video Process. 2018, 13, 369–377. [Google Scholar] [CrossRef]











| Data Set | Number of Samples | Feature Number | Number of Categories |
|---|---|---|---|
| Iris | 150 | 4 | 3 |
| Wine | 178 | 13 | 3 |
| Breast_cancer | 699 | 10 | 2 |
| Digits | 1797 | 5 | 9 |
| Pima | 768 | 8 | 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, A.; Hua, Z.; Shi, Y.; Tang, Y.; Miao, L. An Improved K-Means Algorithm Based on Evidence Distance. Entropy 2021, 23, 1550. https://doi.org/10.3390/e23111550
Zhu A, Hua Z, Shi Y, Tang Y, Miao L. An Improved K-Means Algorithm Based on Evidence Distance. Entropy. 2021; 23(11):1550. https://doi.org/10.3390/e23111550
Chicago/Turabian StyleZhu, Ailin, Zexi Hua, Yu Shi, Yongchuan Tang, and Lingwei Miao. 2021. "An Improved K-Means Algorithm Based on Evidence Distance" Entropy 23, no. 11: 1550. https://doi.org/10.3390/e23111550
APA StyleZhu, A., Hua, Z., Shi, Y., Tang, Y., & Miao, L. (2021). An Improved K-Means Algorithm Based on Evidence Distance. Entropy, 23(11), 1550. https://doi.org/10.3390/e23111550






