Cellular Competency during Development Alters Evolutionary Dynamics in an Artificial Embryogeny Model
Abstract
1. Introduction
2. Methods
2.1. Creating Populations for Evolution
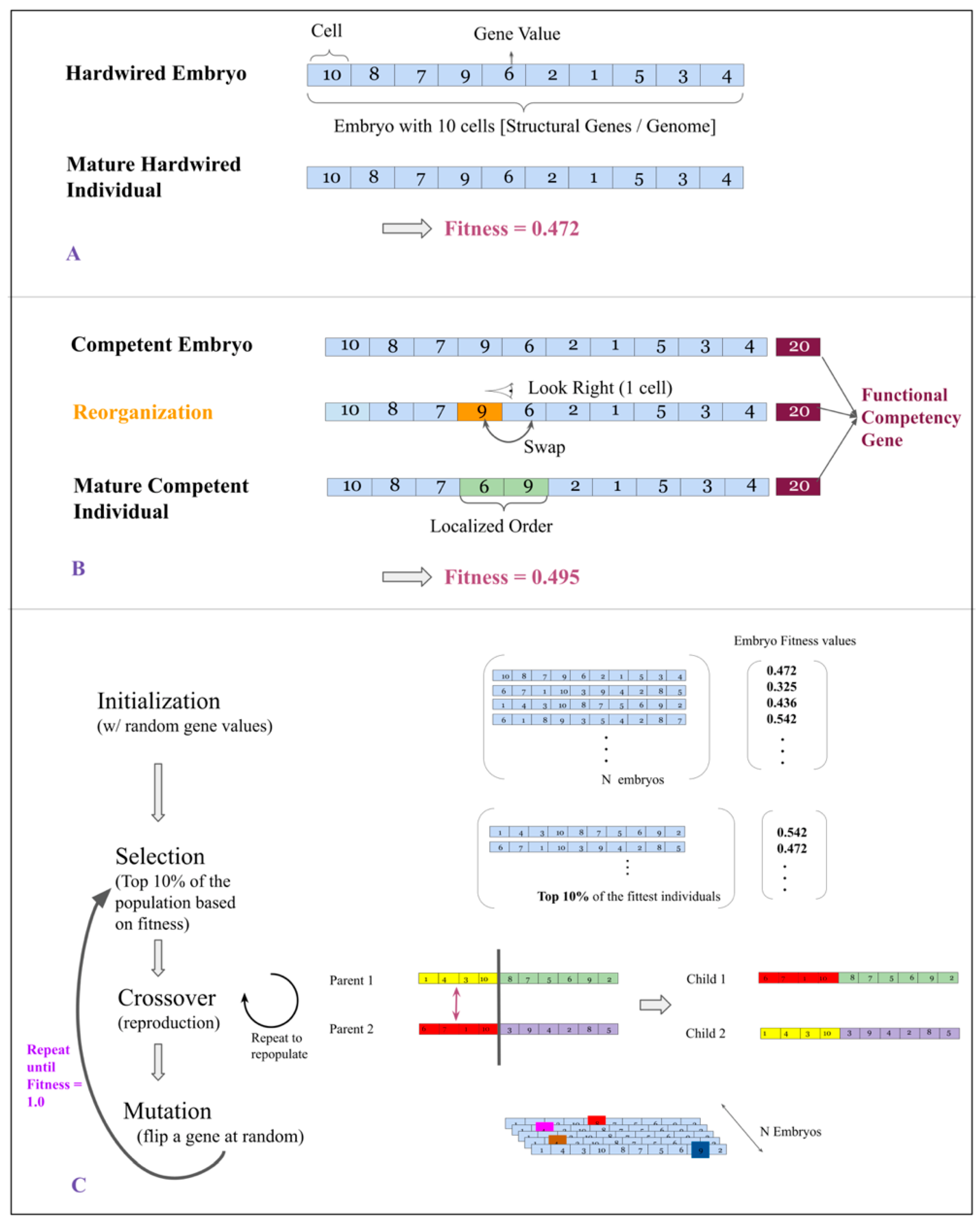
2.2. Hardwired and Competent Embryos
2.3. Developmental Cycle
2.4. Fitness of Embryos and Individuals
2.5. Competency Level
2.6. Genetic Algorithm
- Selection: The fittest 10% of individuals in a population are selected to move on to the next generation. Selection in a population is based on its individuals’ phenotypic fitness.
- Cross-Over: In order to repopulate a population back to its original strength, we carry out a process of reproduction called cross-over. It occurs as follows: Two individuals are involved, each of these are split at a random location along their length. One half of Individual 1 is swapped with the same half of Individual 2 to give rise to two children. Figure 1 contains an illustration of this process.
- Mutation: The repopulated population is subjected to random point mutations. We set the probability of an individual receiving a point mutation to be 0.6.
3. Results
3.1. A Minimal System for Investigating Effects of Cellular Competency on Evolution
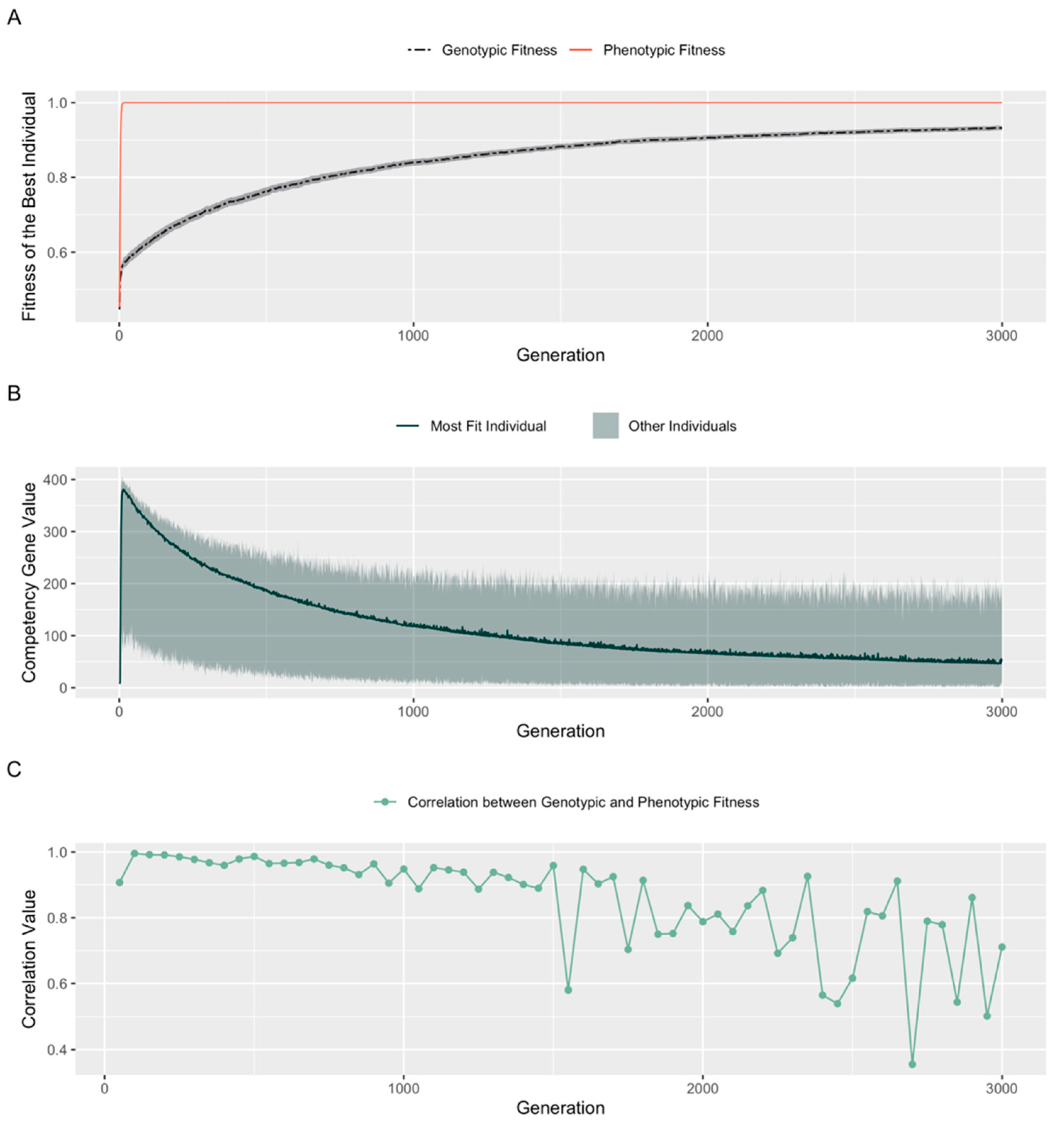
3.2. Developmental Competency Accelerates Evolutionary Search
3.3. Competent Individuals Take over Mixed Populations
3.4. Evolution Results in a High, Constant Level of Competency
3.5. A Fitness Penalty for Competency Leads to Continued Improvement of an Embryo’s Structural Genes
4. Discussion
4.1. Genotypic vs. Phenotypic Fitness: Cellular Competencies and Learning
4.2. Limitations of the Study
4.3. The Role of Cellular Competency in Evolution
4.4. The Paradox of Robustness: Why the Animal with the Worst Genome Has the Best Anatomy
4.5. Genes Can Specify Direct Features, or Problem-Solving Behaviors
4.6. Where the Hard Work Is Done: An Intelligence Ratchet
4.7. The Costs of Competency
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huizinga, J.; Stanley, K.O.; Clune, J. The Emergence of Canalization and Evolvability in an Open-Ended, Interactive Evolutionary System. Artif. Life 2018, 24, 157–181. [Google Scholar] [CrossRef] [PubMed]
- Cheney, N.; Bongard, J.C.; Lipson, H. Evolving Soft Robots in Tight Spaces. In Proceedings of the Gecco’15: Proceedings of the 2015 Genetic and Evolutionary Computation Conference, Madrid, Spain, 11–15 July 2015; pp. 935–942. [Google Scholar]
- Auerbach, J.E.; Bongard, J.C. Evolving Complete Robots with CPPN-NEAT: The Utility of Recurrent Connections. In Proceedings of the Gecco-2011: Proceedings of the 13th Annual Genetic and Evolutionary Computation Conference, Dublin, Ireland, 12–16 July 2011; pp. 1475–1482. [Google Scholar]
- Clune, J.; Beckmann, B.E.; Ofria, C.; Pennock, R.T. Evolving Coordinated Quadruped Gaits with the HyperNEAT Generative Encoding. In Proceedings of the 2009 IEEE Congress on Evolutionary Computation, Trondheim, Norway, 18–21 May 2009; Volumes 1–5, pp. 2764–2771. [Google Scholar] [CrossRef]
- Lai, G.; Leymarie, F.F.; Latham, W.; Arita, T.; Suzuki, R. Virtual Creature Morphology—A Review. Comput. Graph. Forum 2021, 40, 659–681. [Google Scholar] [CrossRef]
- Miller, J.F. Evolving a Self-Repairing, Self-Regulating, French Flag Organism; Springer: Berlin/Heidelberg, Germany, 2004; pp. 129–139. [Google Scholar]
- Hampton, A.N.; Adami, C. Evolution of robust developmental neural networks. Artif. Life IX 2004, 438–443. [Google Scholar]
- Stanley, K.O.; Miikkulainen, R. A taxonomy for artificial embryogeny. Artif. Life 2003, 9, 93–130. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.F. Evolving developmental programs for adaptation, morphogenesis, and self-repair. Lect. Notes Artif. Intell. 2003, 2801, 256–265. [Google Scholar]
- Astor, J.C.; Adami, C. A developmental model for the evolution of artificial neural networks. Artif. Life 2000, 6, 189–218. [Google Scholar] [CrossRef] [PubMed]
- Gruau, F. Genetic Synthesis of Modular Neural Networks. In Proceedings of the 5th International Conference on Genetic Algorithms, ICGA-93, Champaign, IL, USA, 1 June 1993; pp. 318–325. [Google Scholar]
- Gruau, F. Cellular Encoding of Genetic Neural Networks; Laboratoire de l’Informatique du Parallilisme: Lyon, France, 1992. [Google Scholar]
- Kitano, H. Designing neural networks using genetic algorithms with a graph generation system. Complex Syst. 1990, 4, 461–476. [Google Scholar]
- Newman, S.A. Inherency of Form and Function in Animal Development and Evolution. Front. Physiol. 2019, 10, 702. [Google Scholar] [CrossRef]
- Newman, S.A. Inherency and homomorphy in the evolution of development. Curr. Opin. Genet. Dev. 2019, 57, 1–8. [Google Scholar] [CrossRef]
- Newman, S.A. Inherency. In Evolutionary Developmental Biology: A Reference Guide; Nuno de la Rosa, L., Müller, G., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 1–12. [Google Scholar]
- Beloussov, L.V. Mechano-geometric generative rules of morphogenesis. Biol. Bull. 2012, 39, 119–126. [Google Scholar] [CrossRef]
- Beloussov, L.V. Mechanically based generative laws of morphogenesis. Phys. Biol. 2008, 5, 015009. [Google Scholar] [CrossRef]
- Levin, M.; Pietak, A.M.; Bischof, J. Planarian regeneration as a model of anatomical homeostasis: Recent progress in biophysical and computational approaches. Semin. Cell Dev. Biol. 2019, 87, 125–144. [Google Scholar] [CrossRef]
- Oviedo, N.J.; Beane, W.S. Regeneration: The origin of cancer or a possible cure? Semin. Cell Dev. Biol. 2009, 20, 557–564. [Google Scholar] [CrossRef]
- Lobo, D.; Solano, M.; Bubenik, G.A.; Levin, M. A linear-encoding model explains the variability of the target morphology in regeneration. J. R. Soc. Interface 2014, 11, 20130918. [Google Scholar] [CrossRef]
- Mathews, J.; Levin, M. The body electric 2.0: Recent advances in developmental bioelectricity for regenerative and synthetic bioengineering. Curr. Opin. Biotechnol. 2018, 52, 134–144. [Google Scholar] [CrossRef]
- Levin, M. The wisdom of the body: Future techniques and approaches to morphogenetic fields in regenerative medicine, developmental biology and cancer. Regen. Med. 2011, 6, 667–673. [Google Scholar] [CrossRef]
- Levin, M. Collective Intelligence of Morphogenesis as a Teleonomic Process. PsyArxiv 2022. [Google Scholar] [CrossRef]
- Harris, A.K. The need for a concept of shape homeostasis. Biosystems 2018, 173, 65–72. [Google Scholar] [CrossRef]
- Davies, J.; Levin, M. Synthetic morphology via active and agential matter. Nat. Bioeng. 2022. preprint. [Google Scholar] [CrossRef]
- Fields, C.; Levin, M. Competency in Navigating Arbitrary Spaces as an Invariant for Analyzing Cognition in Diverse Embodiments. Entropy 2022, 24, 819. [Google Scholar] [CrossRef]
- Clawson, W.P.; Levin, M. Endless Forms Most Beautiful: Teleonomy and the bioengineering of chimeric and synthetic organisms. Biol. J. Linn. Soc. 2022, in press. [CrossRef]
- Birnbaum, K.D.; Sanchez Alvarado, A. Slicing across kingdoms: Regeneration in plants and animals. Cell 2008, 132, 697–710. [Google Scholar] [CrossRef] [PubMed]
- Vandenberg, L.N.; Adams, D.S.; Levin, M. Normalized shape and location of perturbed craniofacial structures in the Xenopus tadpole reveal an innate ability to achieve correct morphology. Dev. Dyn. 2012, 241, 863–878. [Google Scholar] [CrossRef] [PubMed]
- Pinet, K.; McLaughlin, K.A. Mechanisms of physiological tissue remodeling in animals: Manipulating tissue, organ, and organism morphology. Dev. Biol. 2019, 451, 134–145. [Google Scholar] [CrossRef] [PubMed]
- Pinet, K.; Deolankar, M.; Leung, B.; McLaughlin, K.A. Adaptive correction of craniofacial defects in pre-metamorphic Xenopus laevis tadpoles involves thyroid hormone-independent tissue remodeling. Development 2019, 146, dev175893. [Google Scholar] [CrossRef]
- Blackiston, D.J.; Levin, M. Ectopic eyes outside the head in Xenopus tadpoles provide sensory data for light-mediated learning. J. Exp. Biol. 2013, 216, 1031–1040. [Google Scholar] [CrossRef]
- Pezzulo, G.; Levin, M. Top-down models in biology: Explanation and control of complex living systems above the molecular level. J. R. Soc. Interface 2016, 13, 20160555. [Google Scholar] [CrossRef]
- Pezzulo, G.; Levin, M. Re-membering the body: Applications of computational neuroscience to the top-down control of regeneration of limbs and other complex organs. Integr. Biol. 2015, 7, 1487–1517. [Google Scholar] [CrossRef]
- Kirschner, M.; Gerhart, J. Evolvability. Proc. Natl. Acad. Sci. USA 1998, 95, 8420–8427. [Google Scholar] [CrossRef]
- Alberch, P. From genes to phenotype: Dynamical systems and evolvability. Genetica 1991, 84, 5–11. [Google Scholar] [CrossRef]
- Draghi, J.A.; Parsons, T.L.; Wagner, G.P.; Plotkin, J.B. Mutational robustness can facilitate adaptation. Nature 2010, 463, 353–355. [Google Scholar] [CrossRef]
- Wagner, G.P.; Pavlicev, M.; Cheverud, J.M. The road to modularity. Nat. Rev. Genet. 2007, 8, 921–931. [Google Scholar] [CrossRef]
- Schlosser, G.; Wagner, G.P. Modularity in Development and Evolution; University of Chicago Press: Chicago, IL, USA, 2004. [Google Scholar]
- Calabretta, R.; Ferdinando, A.D.; Wagner, G.P.; Parisi, D. What does it take to evolve behaviorally complex organisms? Biosystems 2003, 69, 245–262. [Google Scholar] [CrossRef]
- Muller, V.C.; Hoffmann, M. What Is Morphological Computation? On How the Body Contributes to Cognition and Control. Artif. Life 2017, 23, 1–24. [Google Scholar] [CrossRef]
- Pfeifer, R.; Iida, F.; Lungarella, M. Cognition from the bottom up: On biological inspiration, body morphology, and soft materials. Trends Cogn. Sci. 2014, 18, 404–413. [Google Scholar] [CrossRef]
- Zahedi, K.; Ay, N. Quantifying Morphological Computation. Entropy 2013, 15, 1887–1995. [Google Scholar]
- Corucci, F.; Cheney, N.; Lipson, H.; Laschi, C.; Bongard, J.C. Material properties affect evolution’s ability to exploit morphological computation in growing soft-bodied creatures. In Proceedings of the The Fifteenth International Conference on the Synthesis and Simulation of Living Systems (ALIFE XV), Cancún, Mexico, 4–8 July 2016. [Google Scholar]
- Kriegman, S.; Cheney, N.; Bongard, J. How morphological development can guide evolution. arXiv 2017, arXiv:1711.07387. [Google Scholar] [CrossRef]
- Niehrs, C. On growth and form: A Cartesian coordinate system of Wnt and BMP signaling specifies bilaterian body axes. Development 2010, 137, 845–857. [Google Scholar] [CrossRef]
- Frankel, J. Positional information in cells and organisms. Trends Cell Biol. 1992, 2, 256–260. [Google Scholar] [CrossRef]
- Astrachan, O. Bubble sort: An archaeological algorithmic analysis. ACM SIGCSE Bull. 2003, 35, 1–5. [Google Scholar] [CrossRef]
- Jablonka, E.; Lamb, M.J.; Avital, E. ‘Lamarckian’ mechanisms in darwinian evolution. Trends Ecol. Evol. 1998, 13, 206–210. [Google Scholar] [CrossRef] [PubMed]
- Jablonka, E.; Lamb, M.J.; Zeligowski, A. Evolution in Four Dimensions: Genetic, Epigenetic, Behavioral, and Symbolic Variation in the History of Life; Revised Edition; A Bradford Book; The MIT Press: Cambridge, MA, USA, 2014; p. xii. 563p. [Google Scholar]
- Lyon, P.; Keijzer, F.; Arendt, D.; Levin, M. Reframing cognition: Getting down to biological basics. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2021, 376, 20190750. [Google Scholar] [CrossRef] [PubMed]
- Levin, M.; Keijzer, F.; Lyon, P.; Arendt, D. Uncovering cognitive similarities and differences, conservation and innovation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2021, 376, 20200458. [Google Scholar] [CrossRef] [PubMed]
- Lyon, P. Of what is “minimal cognition” the half-baked version? Adapt. Behav. 2020, 28, 407–424. [Google Scholar] [CrossRef]
- Lyon, P. The cognitive cell: Bacterial behavior reconsidered. Front. Microbiol. 2015, 6, 264. [Google Scholar] [CrossRef]
- Keijzer, F.; van Duijn, M.; Lyon, P. What nervous systems do: Early evolution, input-output, and the skin brain thesis. Adapt. Behav. 2013, 21, 67–85. [Google Scholar] [CrossRef]
- Lyon, P. The biogenic approach to cognition. Cogn. Process. 2006, 7, 11–29. [Google Scholar] [CrossRef]
- Hinton, G.E.; Nowlan, J. How learning can guide evolution. Complex Syst. 1987, 1, 495–502. [Google Scholar]
- Baldwin, J.M. A new factor in evolution. Am. Nat. 1896, 30, 441–445. [Google Scholar] [CrossRef]
- Kouvaris, K.; Clune, J.; Kounios, L.; Brede, M.; Watson, R.A. How evolution learns to generalise: Using the principles of learning theory to understand the evolution of developmental organisation. PLoS Comput. Biol. 2017, 13, e1005358. [Google Scholar] [CrossRef]
- Livnat, A.; Papadimitriou, C. Evolution and Learning: Used Together, Fused Together. A Response to Watson and Szathmary. Trends Ecol. Evol. 2016, 31, 894–896. [Google Scholar] [CrossRef]
- Sznajder, B.; Sabelis, M.W.; Egas, M. How Adaptive Learning Affects Evolution: Reviewing Theory on the Baldwin Effect. Evol. Biol. 2012, 39, 301–310. [Google Scholar] [CrossRef]
- Paenke, I.; Kawecki, T.J.; Sendhoff, B. The Influence of Learning on Evolution: A Mathematical Framework. Artif. Life 2009, 15, 227–245. [Google Scholar] [CrossRef][Green Version]
- Dopazo, H.; Gordon, M.B.; Perazzo, R.; Risau-Gusman, S. A model for the interaction of learning and evolution. Bull. Math. Biol. 2001, 63, 117–134. [Google Scholar] [CrossRef]
- Ackley, D.; Litman, N. Interactions Between Learning and Evolution. In Proceedings of the Second Conference on Artificial Life, Santa Fe, NM, USA, 5–9 February 1990; pp. 487–509. [Google Scholar]
- Farmer, J.D.; Packard, N.H. Evolution, Games and Learning—Models for Adaptation in Machines and Nature—Proceedings of the 5th Annual International-Conference of the Center for Nonlinear Studies, Los-Alamos, Nm, May 20–24, 1985—Introduction. Physical D 1986, 22, R7–R12. [Google Scholar] [CrossRef]
- Thorpe, W.H. Animal Learning and Evolution. Nature 1945, 156, 46. [Google Scholar] [CrossRef]
- Rojo-Laguna, J.I.; Garcia-Cabot, S.; Salo, E. Tissue transplantation in planarians: A useful tool for molecular analysis of pattern formation. Semin. Cell Dev. Biol. 2018, 87, 116–124. [Google Scholar] [CrossRef]
- French, V. Pattern regulation and regeneration. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 1981, 295, 601–617. [Google Scholar]
- Bryant, S.V.; Iten, L.E. Intercalary and supernumerary regeneration in regenerating the mature limbs of Notophthalmus viridescens. J. Exp. Zool. 1977, 202, 1–16. [Google Scholar] [CrossRef]
- French, V.; Bryant, P.J.; Bryant, S.V. Pattern regulation in epimorphic fields. Science 1976, 193, 969–981. [Google Scholar] [CrossRef]
- Levin, M. Technological Approach to Mind Everywhere: An Experimentally-Grounded Framework for Understanding Diverse Bodies and Minds. Front. Syst. Neurosci. 2022, 16, 768201. [Google Scholar] [CrossRef] [PubMed]
- Levin, M. The Computational Boundary of a “Self”: Developmental Bioelectricity Drives Multicellularity and Scale-Free Cognition. Front. Psychol. 2019, 10, 2688. [Google Scholar] [CrossRef] [PubMed]
- Tschantz, A.; Barca, L.; Maisto, D.; Buckley, C.L.; Seth, A.K.; Pezzulo, G. Simulating homeostatic, allostatic and goal-directed forms of interoceptive control using active inference. Biol. Psychol. 2022, 169, 108266. [Google Scholar] [CrossRef] [PubMed]
- Deans, C. Biological Prescience: The Role of Anticipation in Organismal Processes. Front. Physiol. 2021, 12, 672457. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, Y.; Kaneko, K. Homeorhesis in Waddington’s landscape by epigenetic feedback regulation. Phys. Rev. Res. 2020, 2, 023083. [Google Scholar] [CrossRef]
- Colditz, I.G. A consideration of physiological regulation from the perspective of Bayesian enactivism. Physiol. Behav. 2020, 214, 112758. [Google Scholar] [CrossRef]
- Houri-Zeevi, L.; Rechavi, O. A Matter of Time: Small RNAs Regulate the Duration of Epigenetic Inheritance. Trends Genet. 2017, 33, 46–57. [Google Scholar] [CrossRef]
- Neuhof, M.; Levin, M.; Rechavi, O. Vertically- and horizontally-transmitted memories—The fading boundaries between regeneration and inheritance in planaria. Biol. Open 2016, 5, 1177–1188. [Google Scholar] [CrossRef]
- Rechavi, O.; Houri-Ze’evi, L.; Anava, S.; Goh, W.S.; Kerk, S.Y.; Hannon, G.J.; Hobert, O. Starvation-induced transgenerational inheritance of small RNAs in C. elegans. Cell 2014, 158, 277–287. [Google Scholar] [CrossRef]
- Rechavi, O. Guest list or black list: Heritable small RNAs as immunogenic memories. Trends Cell Biol. 2014, 24, 212–220. [Google Scholar] [CrossRef][Green Version]
- Anava, S.; Posner, R.; Rechavi, O. The soft genome. Worm 2014, 3, e989798. [Google Scholar] [CrossRef]
- Dongen, S.V. Fluctuating asymmetry and developmental instability in evolutionary biology: Past, present and future. J. Evol. Biol. 2006, 19, 1727–1743. [Google Scholar] [CrossRef]
- Lens, L.; Van Dongen, S.; Kark, S.; Matthysen, E. Fluctuating asymmetry as an indicator of fitness: Can we bridge the gap between studies? Biol. Rev. Camb. Philos. Soc. 2002, 77, 27–38. [Google Scholar] [CrossRef]
- Enquist, M.; Arak, A. Symmetry, beauty and evolution. Nature 1994, 372, 169–172. [Google Scholar] [CrossRef]
- Zaidel, D.W.; Aarde, S.M.; Baig, K. Appearance of symmetry, beauty, and health in human faces. Brain Cogn. 2005, 57, 261–263. [Google Scholar] [CrossRef]
- Zaidel, D.W.; Cohen, J.A. The face, beauty, and symmetry: Perceiving asymmetry in beautiful faces. Int. J. Neurosci. 2005, 115, 1165–1173. [Google Scholar] [CrossRef]
- Frank, S.A. Evolutionary design of regulatory control. I. A robust control theory analysis of tradeoffs. J. Theor. Biol. 2019, 463, 121–137. [Google Scholar] [CrossRef]
- Frank, S.A. Evolutionary design of regulatory control. II. Robust error-correcting feedback increases genetic and phenotypic variability. J. Theor. Biol. 2019, 468, 72–81. [Google Scholar] [CrossRef]
- Frank, S.A. Evolution of robustness and cellular stochasticity of gene expression. PLoS Biol. 2013, 11, e1001578. [Google Scholar] [CrossRef]
- Frank, S.A. Natural selection. III. Selection versus transmission and the levels of selection. J. Evol. Biol. 2012, 25, 227–243. [Google Scholar] [CrossRef]
- Frank, S.A. Maladaptation and the paradox of robustness in evolution. PLoS ONE 2007, 2, e1021. [Google Scholar] [CrossRef] [PubMed]
- Bailon-Zambrano, R.; Sucharov, J.; Mumme-Monheit, A.; Murry, M.; Stenzel, A.; Pulvino, A.T.; Mitchell, J.M.; Colborn, K.L.; Nichols, J.T. Variable paralog expression underlies phenotype variation. Elife 2022, 11, e79247. [Google Scholar] [CrossRef] [PubMed]
- Nichols, J.T.; Blanco-Sanchez, B.; Brooks, E.P.; Parthasarathy, R.; Dowd, J.; Subramanian, A.; Nachtrab, G.; Poss, K.D.; Schilling, T.F.; Kimmel, C.B. Ligament versus bone cell identity in the zebrafish hyoid skeleton is regulated by mef2ca. Development 2016, 143, 4430–4440. [Google Scholar] [CrossRef] [PubMed]
- Sucharov, J.; Ray, K.; Brooks, E.P.; Nichols, J.T. Selective breeding modifies mef2ca mutant incomplete penetrance by tuning the opposing Notch pathway. PLoS Genet. 2019, 15, e1008507. [Google Scholar] [CrossRef] [PubMed]
- Oviedo, N.J.; Morokuma, J.; Walentek, P.; Kema, I.P.; Gu, M.B.; Ahn, J.M.; Hwang, J.S.; Gojobori, T.; Levin, M. Long-range neural and gap junction protein-mediated cues control polarity during planarian regeneration. Dev. Biol. 2010, 339, 188–199. [Google Scholar] [CrossRef] [PubMed]
- Nogi, T.; Levin, M. Characterization of innexin gene expression and functional roles of gap-junctional communication in planarian regeneration. Dev. Biol. 2005, 287, 314–335. [Google Scholar] [CrossRef]
- Durant, F.; Morokuma, J.; Fields, C.; Williams, K.; Adams, D.S.; Levin, M. Long-Term, Stochastic Editing of Regenerative Anatomy via Targeting Endogenous Bioelectric Gradients. Biophys. J. 2017, 112, 2231–2243. [Google Scholar] [CrossRef]
- Levin, M. Bioelectric signaling: Reprogrammable circuits underlying embryogenesis, regeneration, and cancer. Cell 2021, 184, 1971–1989. [Google Scholar] [CrossRef]
- Harris, M.P. Bioelectric signaling as a unique regulator of development and regeneration. Development 2021, 148, dev180794. [Google Scholar] [CrossRef]
- Levin, M.; Pezzulo, G.; Finkelstein, J.M. Endogenous Bioelectric Signaling Networks: Exploiting Voltage Gradients for Control of Growth and Form. Annu. Rev. Biomed. Eng. 2017, 19, 353–387. [Google Scholar] [CrossRef]
- Sullivan, K.G.; Emmons-Bell, M.; Levin, M. Physiological inputs regulate species-specific anatomy during embryogenesis and regeneration. Commun. Integr. Biol. 2016, 9, e1192733. [Google Scholar] [CrossRef]
- Emmons-Bell, M.; Durant, F.; Hammelman, J.; Bessonov, N.; Volpert, V.; Morokuma, J.; Pinet, K.; Adams, D.S.; Pietak, A.; Lobo, D.; et al. Gap Junctional Blockade Stochastically Induces Different Species-Specific Head Anatomies in Genetically Wild-Type Girardia dorotocephala Flatworms. Int. J. Mol. Sci. 2015, 16, 27865–27896. [Google Scholar] [CrossRef]
- West-Eberhard, M.J. Evolution in the light of developmental and cell biology, and vice versa. Proc. Natl. Acad. Sci. USA 1998, 95, 8417–8419. [Google Scholar] [CrossRef]
- West-Eberhard, M.J. Phenotypic accommodation: Adaptive innovation due to developmental plasticity. J. Exp. Zool. B Mol. Dev. Evol. 2005, 304, 610–618. [Google Scholar] [CrossRef]
- West-Eberhard, M.J. Developmental plasticity and the origin of species differences. Proc. Natl. Acad. Sci. USA 2005, 102 (Suppl. 1), 6543–6549. [Google Scholar] [CrossRef]
- Kauffman, S.A. The Origins of Order: Self Organization and Selection in Evolution; Oxford University Press: New York, NY, USA, 1993. [Google Scholar]
- Kauffman, S.A.; Johnsen, S. Coevolution to the edge of chaos: Coupled fitness landscapes, poised states, and coevolutionary avalanches. J. Theor. Biol. 1991, 149, 467–505. [Google Scholar] [CrossRef]
- Shmulevich, I.; Kauffman, S.A. Activities and sensitivities in boolean network models. Phys. Rev. Lett. 2004, 93, 048701. [Google Scholar] [CrossRef]
- Watson, R.; Levin, M.; Buckley, C.L. Design for an individual: Connectionist approaches to the evolutionary transitions in individuality. Front. Ecol. Evol. 2022, 10, 823588. [Google Scholar] [CrossRef]
- Buckley, C.L.; Watson, R.A. Natural Induction. 2023; manuscript in preparation. [Google Scholar]
- Watson, R.A.; Mills, R.; Buckley, C.L.; Kouvaris, K.; Jackson, A.; Powers, S.T.; Cox, C.; Tudge, S.; Davies, A.; Kounios, L.; et al. Evolutionary Connectionism: Algorithmic Principles Underlying the Evolution of Biological Organisation in Evo-Devo, Evo-Eco and Evolutionary Transitions. Evol. Biol. 2016, 43, 553–581. [Google Scholar] [CrossRef]
- Karve, S.; Wagner, A. Multiple Novel Traits without Immediate Benefits Originate in Bacteria Evolving on Single Antibiotics. Mol. Biol. Evol. 2022, 39, msab341. [Google Scholar] [CrossRef]
- Calcagni, G. The geometry of learning. J. Math. Psychol. 2018, 84, 74–88. [Google Scholar] [CrossRef]
- Payne, J.L.; Moore, J.H.; Wagner, A. Robustness, evolvability, and the logic of genetic regulation. Artif. Life 2014, 20, 111–126. [Google Scholar] [CrossRef] [PubMed]
- Raman, K.; Wagner, A. The evolvability of programmable hardware. J. R. Soc. Interface 2011, 8, 269–281. [Google Scholar] [CrossRef] [PubMed]
- Calcott, B.; Levy, A.; Siegal, M.L.; Soyer, O.S.; Wagner, A. Engineering and Biology: Counsel for a Continued Relationship. Biol. Theory 2015, 10, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Wagner, A.; Rosen, W. Spaces of the possible: Universal Darwinism and the wall between technological and biological innovation. J. R. Soc. Interface 2014, 11, 20131190. [Google Scholar] [CrossRef] [PubMed]
- Wagner, A. Arrival of the Fittest: Solving Evolution’s Greatest Puzzle; Current: New York, NY, USA, 2014. [Google Scholar]
- Boyle, E.A.; Li, Y.I.; Pritchard, J.K. An Expanded View of Complex Traits: From Polygenic to Omnigenic. Cell 2017, 169, 1177–1186. [Google Scholar] [CrossRef]
- Baluska, F.; Miller, W.B.; Reber, A.S. Cellular and evolutionary perspectives on organismal cognition: From unicellular to multicellular organisms. Biol. J. Linn. Soc. 2022, blac005. [Google Scholar] [CrossRef]
- Reber, A.S.; Baluska, F. Cognition in some surprising places. Biochem. Biophys. Res. Commun. 2021, 564, 150–157. [Google Scholar] [CrossRef]
- Baluska, F.; Reber, A.S. Cellular and organismal agency—Not based on genes: A comment on Baverstock. Prog. Biophys. Mol. Biol. 2021, 167, 161–162. [Google Scholar] [CrossRef]
- Manicka, S.; Levin, M. Modeling somatic computation with non-neural bioelectric networks. Sci. Rep. 2019, 9, 18612. [Google Scholar] [CrossRef]
- Manicka, S.; Levin, M. Minimal Developmental Computation: A Causal Network Approach to Understand Morphogenetic Pattern Formation. Entropy 2022, 24, 107. [Google Scholar] [CrossRef]
- Bates, E. Ion Channels in Development and Cancer. Annu. Rev. Cell Dev. Biol. 2015, 31, 231–247. [Google Scholar] [CrossRef]
- Biswas, S.; Manicka, S.; Hoel, E.; Levin, M. Gene Regulatory Networks Exhibit Several Kinds of Memory: Quantification of Memory in Biological and Random Transcriptional Networks. iScience 2021, 24, 102131. [Google Scholar] [CrossRef]
- Watson, R.A.; Buckley, C.L.; Mills, R.; Davies, A. Associative memory in gene regulation networks. In Proceedings of the Artificial Life Conference XII, Odense, Denmark, 19–23 August 2010; pp. 194–201. [Google Scholar]
- Smith-Ferguson, J.; Beekman, M. Who needs a brain? Slime moulds, behavioural ecology and minimal cognition. Adapt. Behav. 2020, 28, 465–478. [Google Scholar] [CrossRef]
- Baluska, F.; Reber, A.S.; Miller, W.B., Jr. Cellular sentience as the primary source of biological order and evolution. Biosystems 2022, 218, 104694. [Google Scholar] [CrossRef]
- Baluška, F.; Levin, M. On Having No Head: Cognition throughout Biological Systems. Front. Psychol. 2016, 7, 902. [Google Scholar] [CrossRef]
- Levin, M. Life, death, and self: Fundamental questions of primitive cognition viewed through the lens of body plasticity and synthetic organisms. Biochem. Biophys. Res. Commun. 2020, 564, 114–133. [Google Scholar] [CrossRef]
- Goodwin, B.C. A cognitive view of biological process. J. Soc. Biol. Struct. 1978, 1, 117–125. [Google Scholar] [CrossRef]
- Goodwin, B.C. Cognitive Biology. Commun. Cogn. 1977, 10, 87–91. [Google Scholar]
- Goodwin, B.C. On some relationships between embryogenesis and cognition. Theor. Theory 1976, 10, 33–44. [Google Scholar]
- O’Donnell, S.; Bulova, S.J.; DeLeon, S.; Khodak, P.; Miller, S.; Sulger, E. Distributed cognition and social brains: Reductions in mushroom body investment accompanied the origins of sociality in wasps (Hymenoptera: Vespidae). Proc. Biol. Sci. 2015, 282, 20150791. [Google Scholar] [CrossRef] [PubMed]
- Thompson, B.; Kirby, S.; Smith, K. Culture shapes the evolution of cognition. Proc. Natl. Acad. Sci. USA 2016, 113, 4530–4535. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.M. The Importance of the Nervous System in the Evolution of Animal Flight. Evolution 1952, 6, 127–129. [Google Scholar] [CrossRef]
- Keijzer, F.A. Evolutionary convergence and biologically embodied cognition. Interface Focus 2017, 7, 20160123. [Google Scholar] [CrossRef] [PubMed]
- Keijzer, F.; Arnellos, A. The animal sensorimotor organization: A challenge for the environmental complexity thesis. Biol. Philos. 2017, 32, 421–441. [Google Scholar] [CrossRef] [PubMed]
- Keijzer, F. Moving and sensing without input and output: Early nervous systems and the origins of the animal sensorimotor organization. Biol. Philos. 2015, 30, 311–331. [Google Scholar] [CrossRef]
- Jekely, G.; Keijzer, F.; Godfrey-Smith, P. An option space for early neural evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370, 20150181. [Google Scholar] [CrossRef]
- De Wiljes, O.O.; van Elburg, R.A.; Biehl, M.; Keijzer, F.A. Modeling spontaneous activity across an excitable epithelium: Support for a coordination scenario of early neural evolution. Front. Comput. Neurosci. 2015, 9, 110. [Google Scholar] [CrossRef]
- Bongard, J.; Zykov, V.; Lipson, H. Resilient machines through continuous self-modeling. Science 2006, 314, 1118–1121. [Google Scholar] [CrossRef]
- Pfeifer, R.; Iida, F.; Bongard, J. New robotics: Design principles for intelligent systems. Artif. Life 2005, 11, 99–120. [Google Scholar] [CrossRef]
- Sole, R.; Amor, D.R.; Duran-Nebreda, S.; Conde-Pueyo, N.; Carbonell-Ballestero, M.; Montanez, R. Synthetic collective intelligence. Biosystems 2016, 148, 47–61. [Google Scholar] [CrossRef]
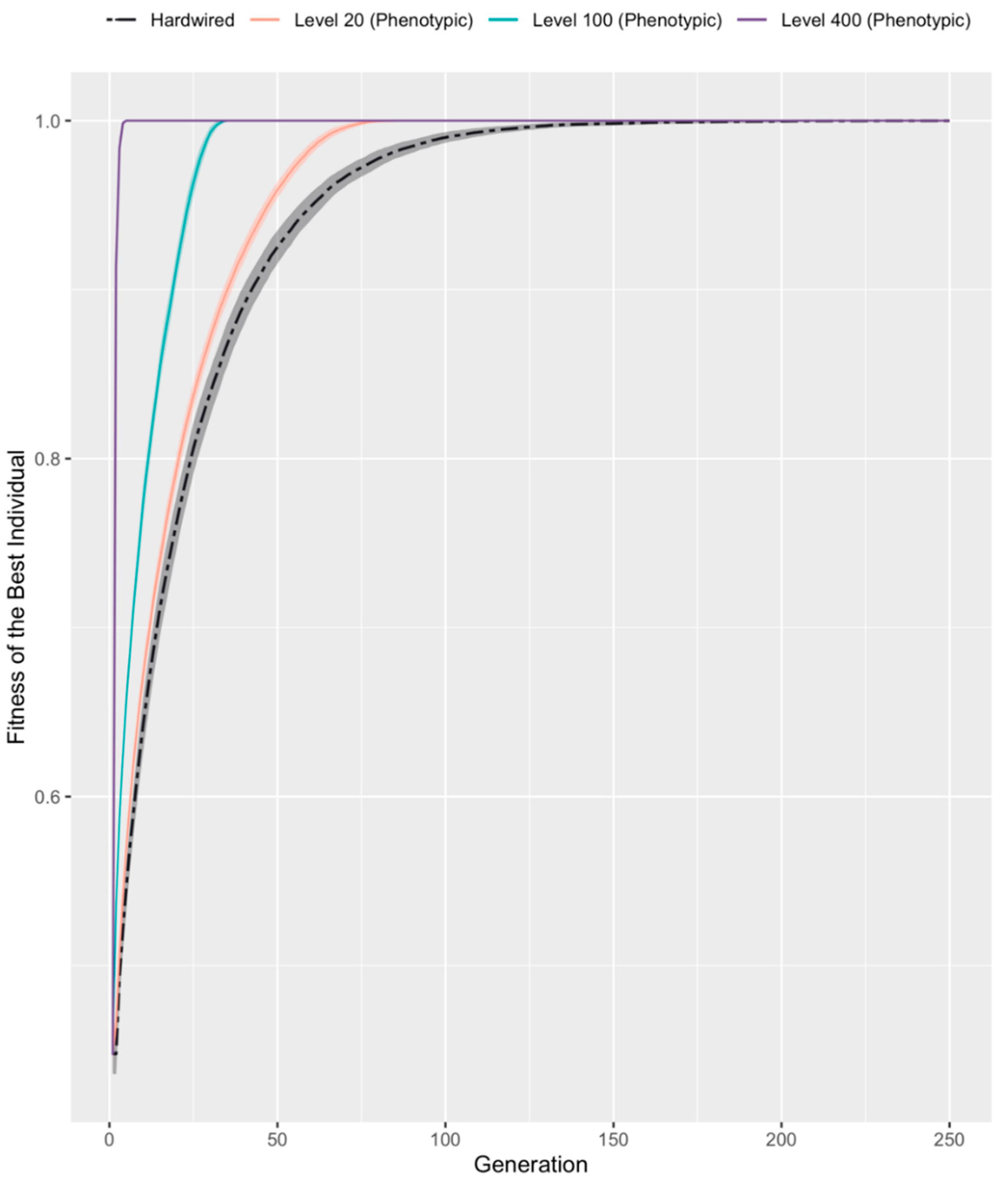

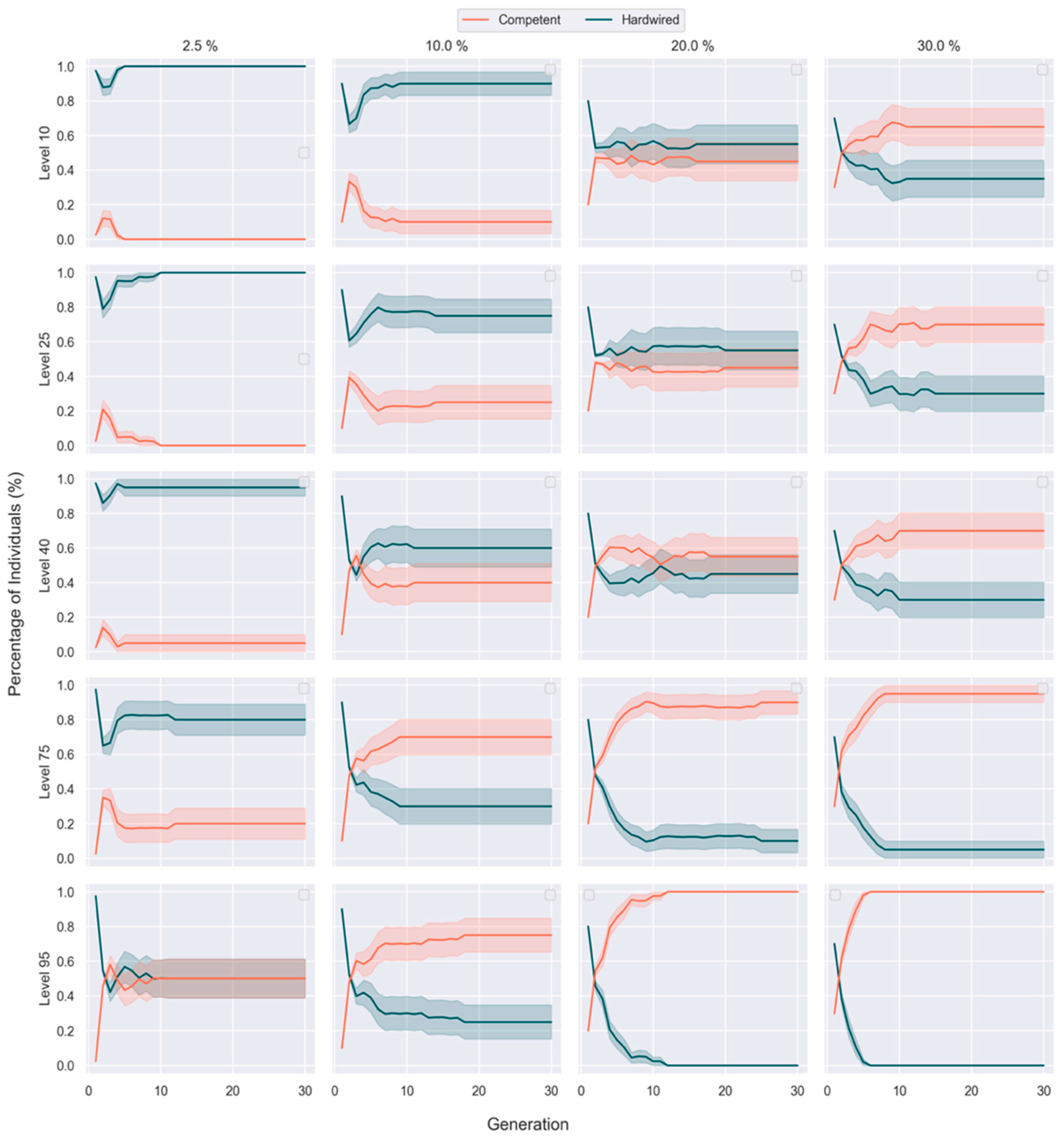

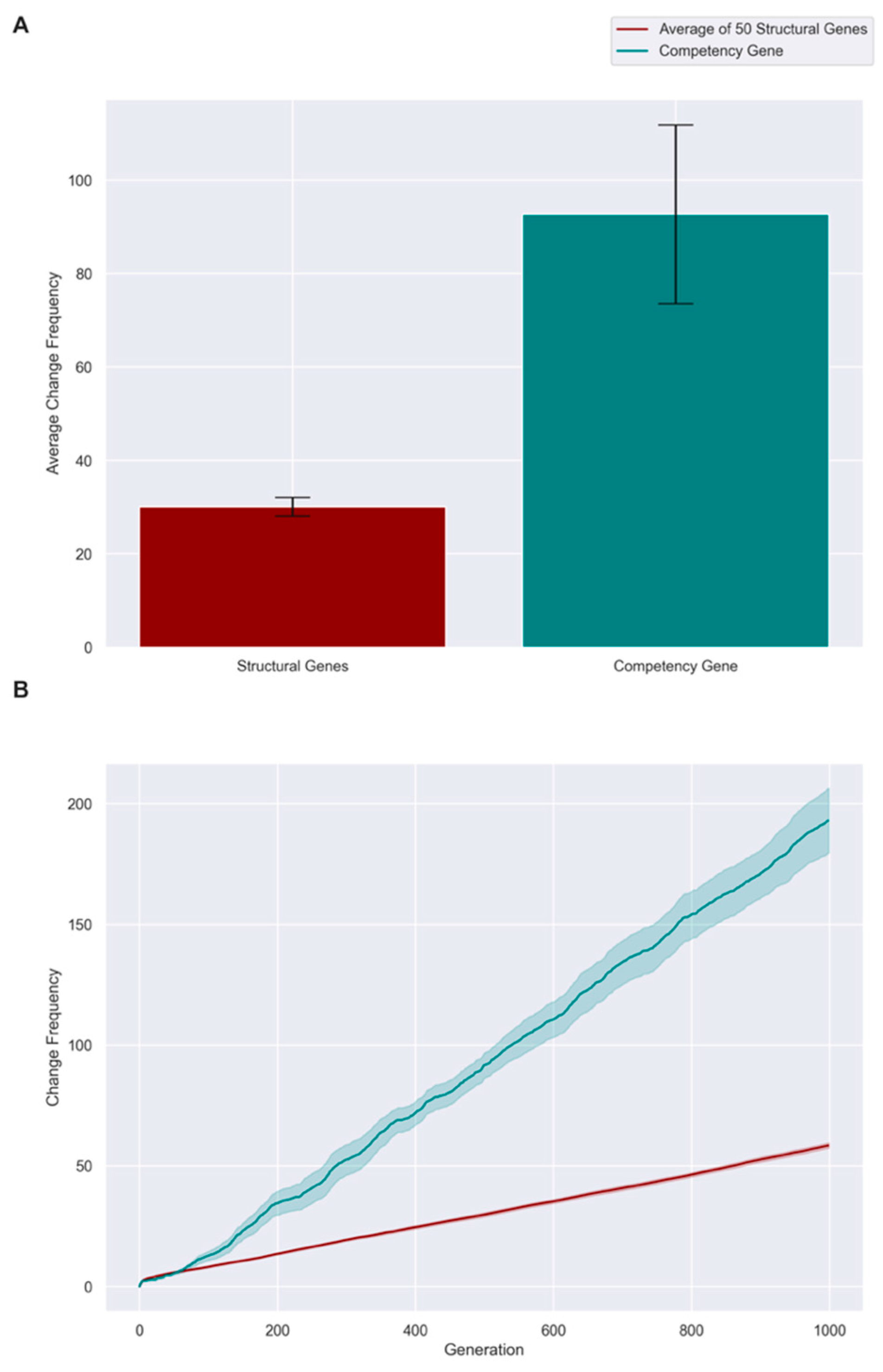
| Competency Level | Fitness Threshold | |||||
|---|---|---|---|---|---|---|
| 0.65 | 0.75 | 0.8 | 0.9 | 0.97 | 1.0 | |
| No competency (Hardwired) | 10 | 18 | 24 | 42 | 72 | 250 |
| Level 20 | 9 | 16 | 21 | 36 | 55 | 93 |
| Level 100 | 5 | 9 | 12 | 19 | 26 | 37 |
| Level 400 | 2 | 2 | 2 | 2 | 3 | 5 |
| Competency Level | Percentage of Competent Embryos | |||
|---|---|---|---|---|
| 2.5% | 10% | 20% | 30% | |
| Level 10 | x | x | x | 3 |
| Level 25 | x | x | x | 3 |
| Level 40 | x | x | 3 | 3 |
| Level 75 | x | 3 | 3 | 2 |
| Level 95 | x | 3 | 2 | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shreesha, L.; Levin, M. Cellular Competency during Development Alters Evolutionary Dynamics in an Artificial Embryogeny Model. Entropy 2023, 25, 131. https://doi.org/10.3390/e25010131
Shreesha L, Levin M. Cellular Competency during Development Alters Evolutionary Dynamics in an Artificial Embryogeny Model. Entropy. 2023; 25(1):131. https://doi.org/10.3390/e25010131
Chicago/Turabian StyleShreesha, Lakshwin, and Michael Levin. 2023. "Cellular Competency during Development Alters Evolutionary Dynamics in an Artificial Embryogeny Model" Entropy 25, no. 1: 131. https://doi.org/10.3390/e25010131
APA StyleShreesha, L., & Levin, M. (2023). Cellular Competency during Development Alters Evolutionary Dynamics in an Artificial Embryogeny Model. Entropy, 25(1), 131. https://doi.org/10.3390/e25010131







