Spatio-Temporal Fractal Dimension Analysis from Resting State EEG Signals in Parkinson’s Disease
Abstract
:1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. EEG Acquisition and Processing
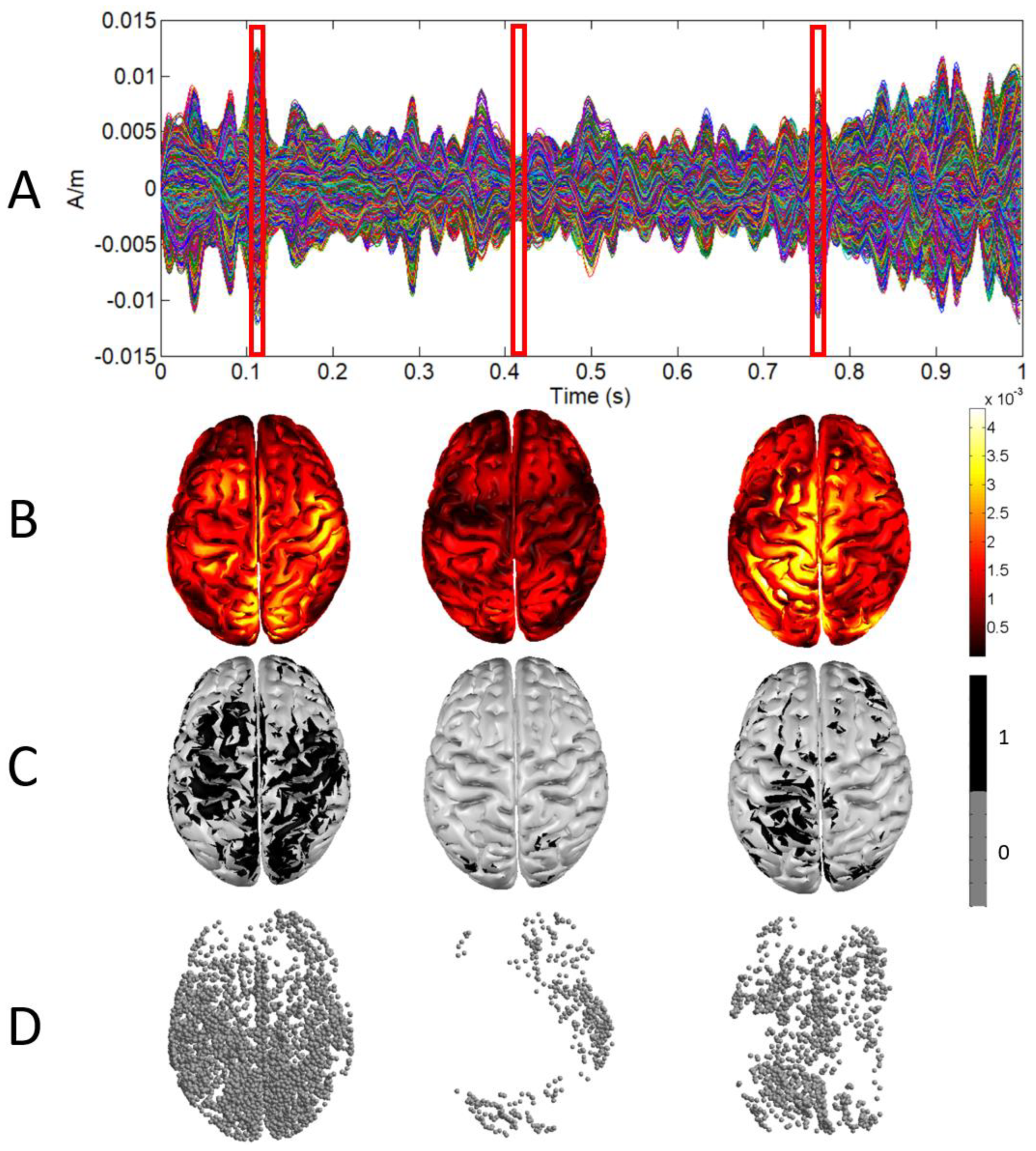
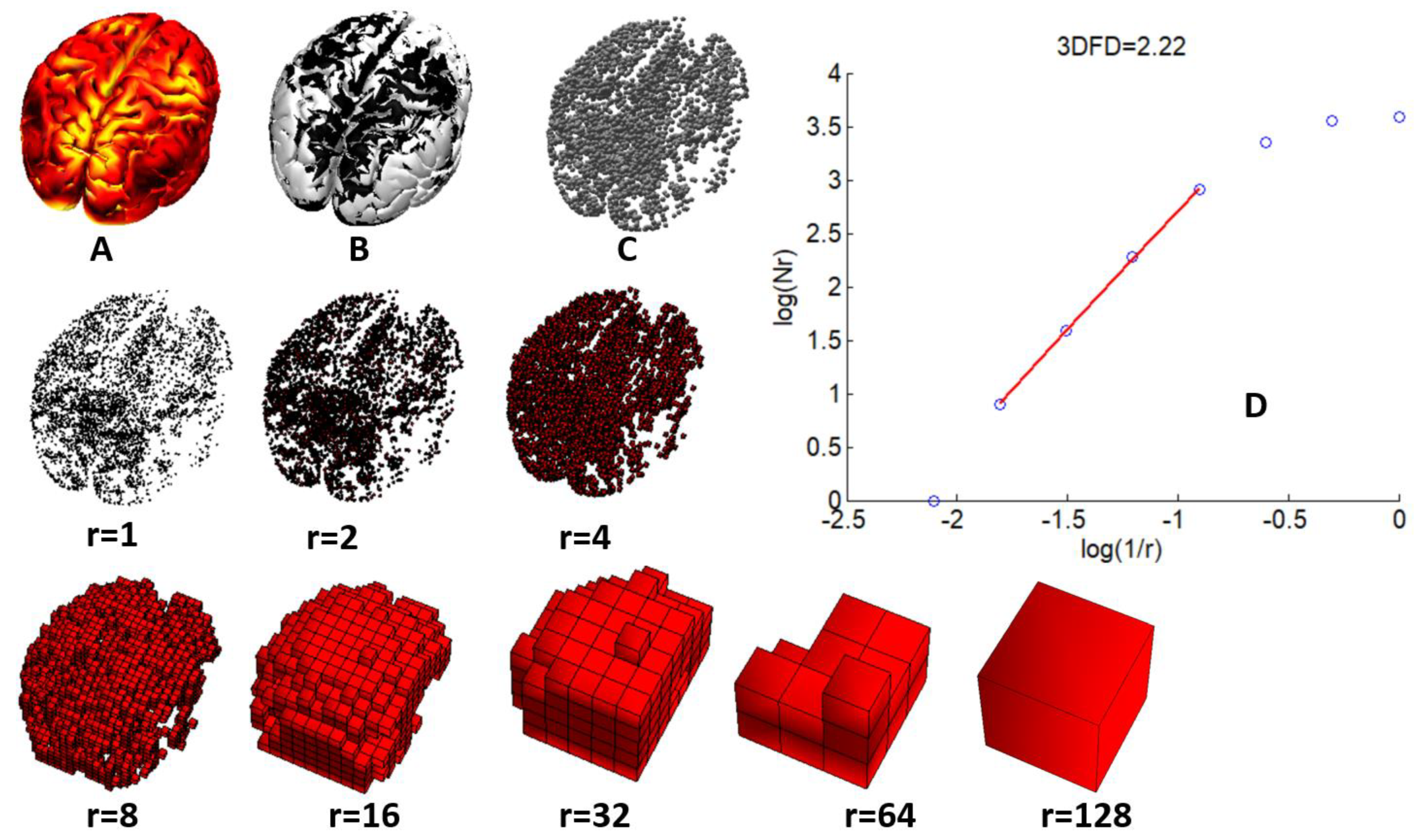
2.3. Fractal Dimension Computation
2.4. Statistical Analysis
3. Results
3.1. Demographic Results
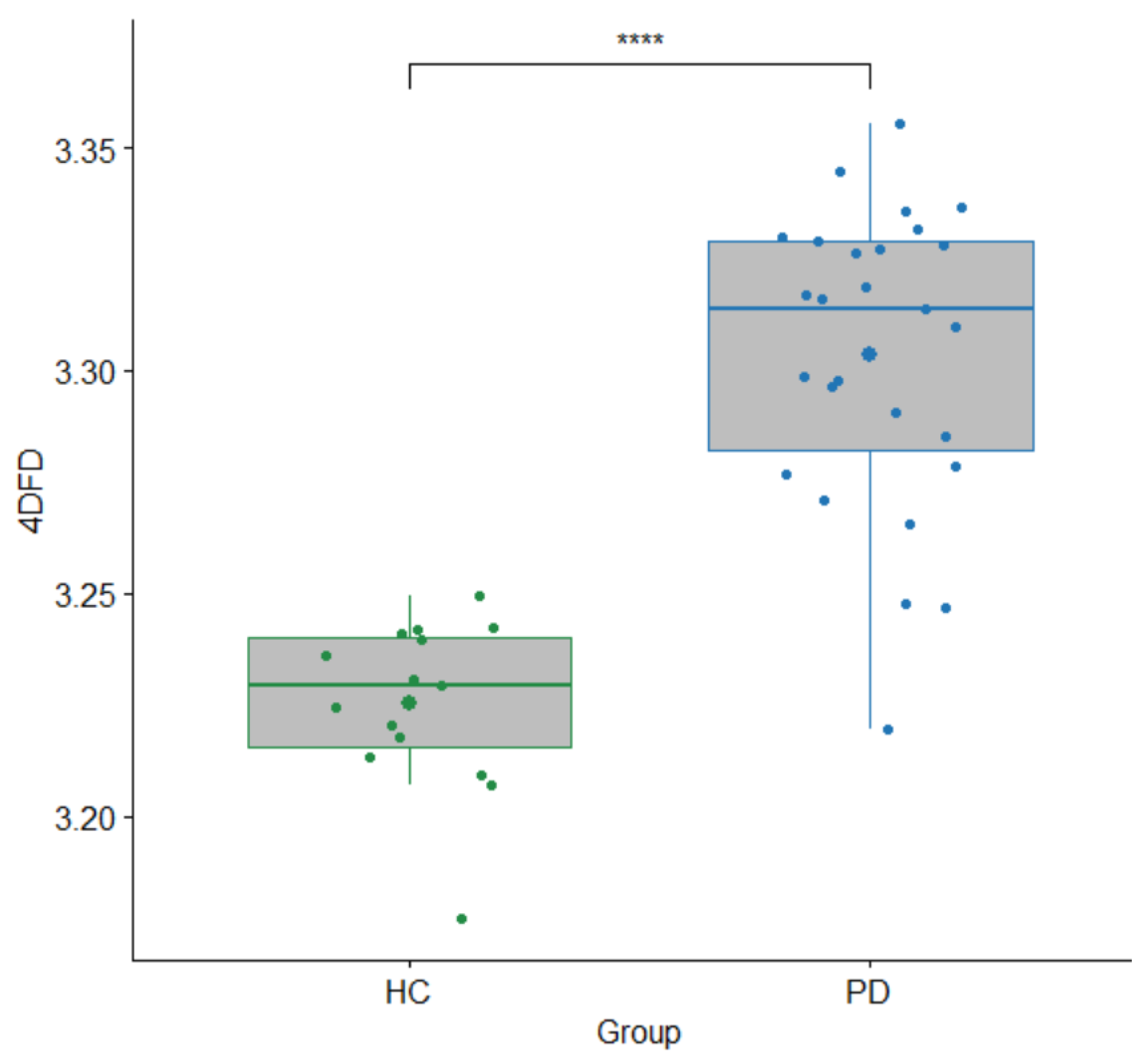
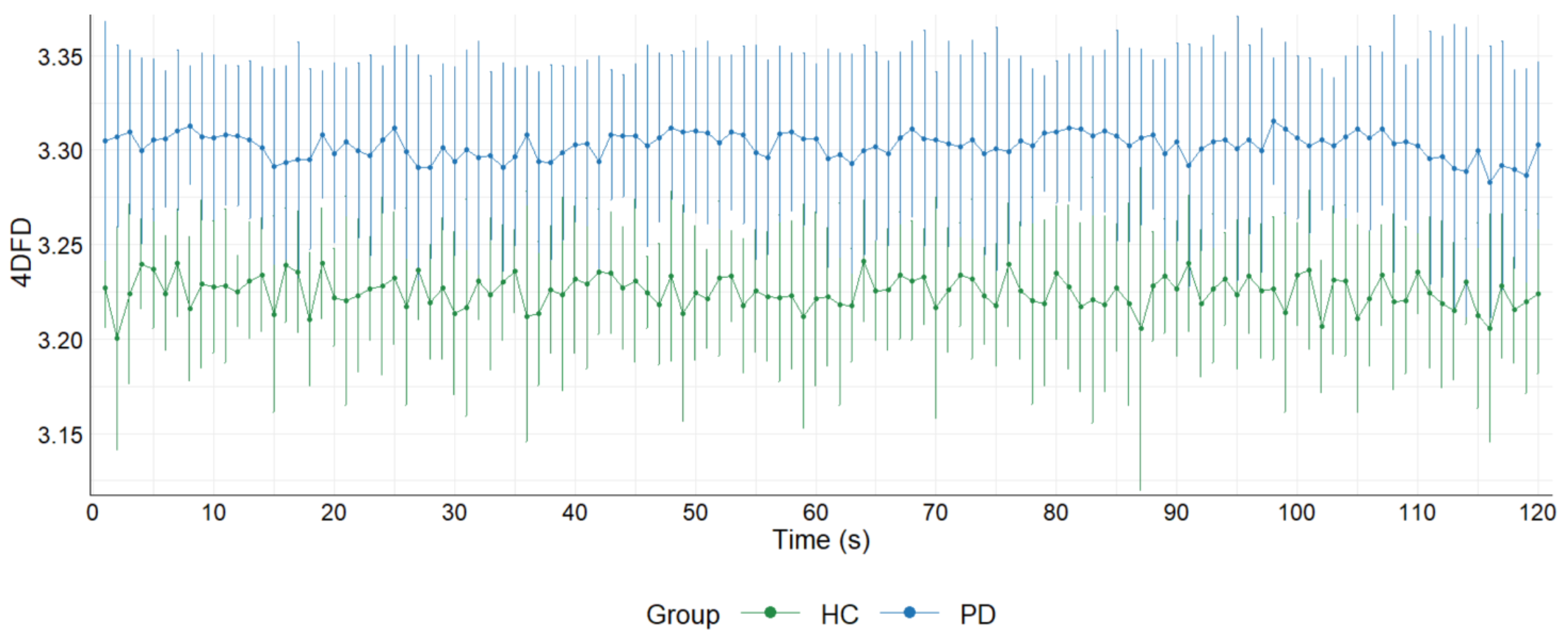
3.2. 4DFD Comparison between HC and PD
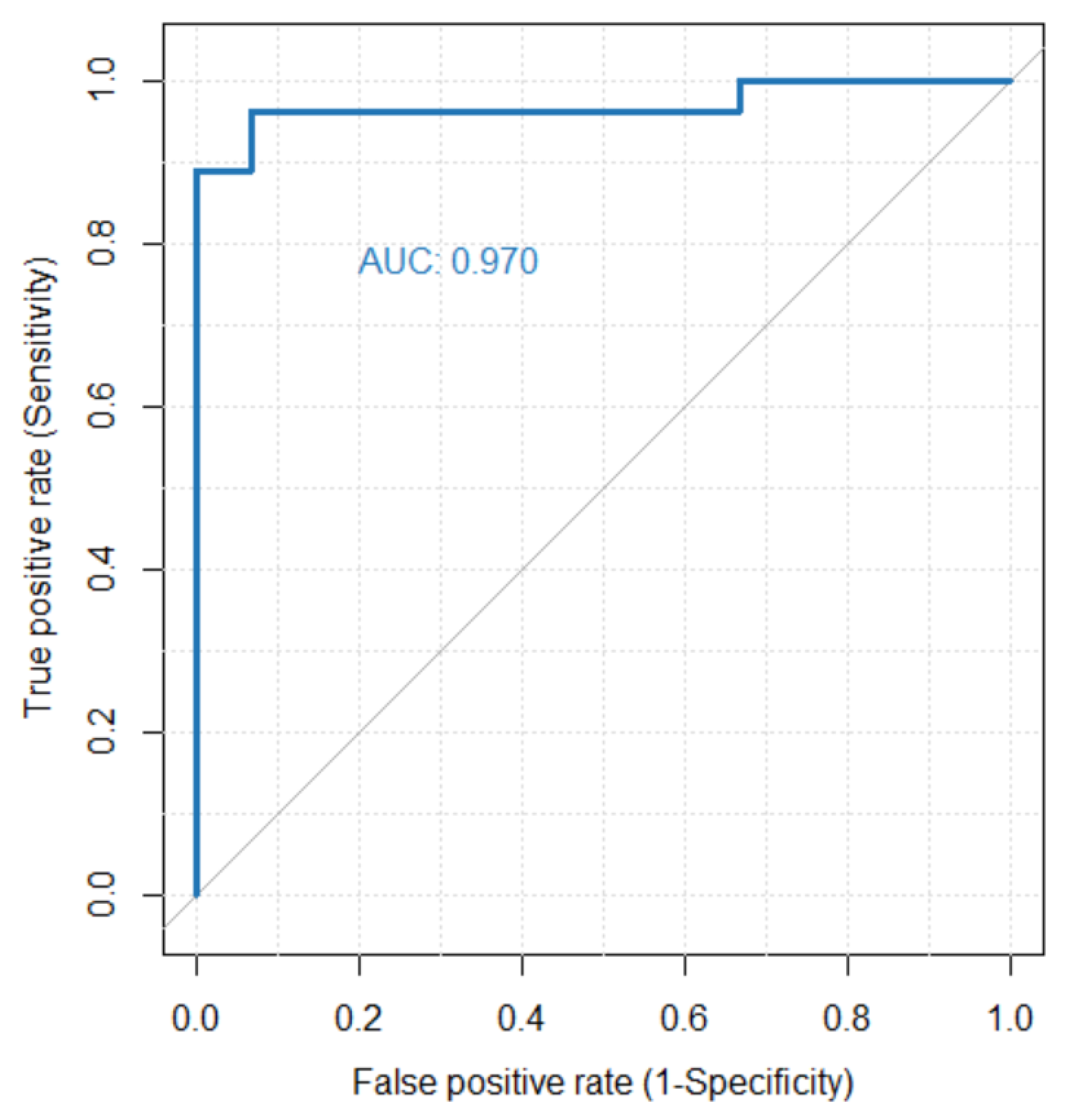
3.3. 4DFD as Classifier for PD
3.4. Correlations between 4DFD and Motor and Neuropsychological Scores
4. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Mandelbrot, B.B. The Fractal Geometry of Nature; WH Freeman Company: New York, NY, USA, 1983; Volume 51, ISBN 0716711869. [Google Scholar]
- Ziukelis, E.T.; Mak, E.; Dounavi, M.E.; Su, L.; O’Brien, J.T. Fractal dimension of the brain in neurodegenerative disease and dementia: A systematic review. Ageing Res. Rev. 2022, 79, 101651. [Google Scholar] [CrossRef]
- Lau, Z.J.; Pham, T.; Chen, S.H.A.; Makowski, D. Brain entropy, fractal dimensions and predictability: A review of complexity measures for EEG in healthy and neuropsychiatric populations. Eur. J. Neurosci. 2022, 56, 5047–5069. [Google Scholar] [CrossRef]
- Grassberger, P.; Procaccia, I. Measuring the strangeness of strange attractors. Phys. D Nonlinear Phenom. 1983, 9, 189–208. [Google Scholar] [CrossRef]
- Higuchi, T. Approach to an irregular time series on the basis of the fractal theory. Phys. D Nonlinear Phenom. 1988, 31, 277–283. [Google Scholar] [CrossRef]
- Richman, J.S.; Moorman, J.R. Physiological time-series analysis using approximate entropy and sample entropy. Am. J. Physiol. Circ. Physiol. 2000, 278, H2039–H2049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz de Miras, J.; Soler, F.; Iglesias-Parro, S.; Ibáñez-Molina, A.J.; Casali, A.G.; Laureys, S.; Massimini, M.; Esteban, F.J.; Navas, J.; Langa, J.A. Fractal dimension analysis of states of consciousness and unconsciousness using transcranial magnetic stimulation. Comput. Methods Programs Biomed. 2019, 175, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Valls-Solé, J.; Valldeoriola, F. Neurophysiological correlate of clinical signs in Parkinson’s disease. Clin. Neurophysiol. 2002, 113, 792–805. [Google Scholar] [CrossRef] [PubMed]
- Jankovic, J. Parkinson’s disease: Clinical features and diagnosis. J. Neurol. Neurosurg. Psychiatry 2008, 79, 368–376. [Google Scholar] [CrossRef] [Green Version]
- Kalia, L.V.; Lang, A.E. Parkinson’s disease. Lancet 2015, 386, 896–912. [Google Scholar] [CrossRef]
- Müller, V.; Lutzenberger, W.; Pulvermüller, F.; Mohr, B.; Birbaumer, N. Investigation of brain dynamics in Parkinson’s disease by methods derived from nonlinear dynamics. Exp. Brain Res. 2001, 136, 103–110. [Google Scholar] [CrossRef]
- Pezard, L.; Jech, R.; Růžička, E. Investigation of non-linear properties of multichannel EEG in the early stages of Parkinson’s disease. Clin. Neurophysiol. 2001, 112, 38–45. [Google Scholar] [CrossRef]
- Chung, C.C.; Kang, J.H.; Yuan, R.Y.; Wu, D.; Chen, C.C.; Chi, N.F.; Chen, P.C.; Hu, C.J. Multiscale entropy analysis of electroencephalography during sleep in patients with parkinson disease. Clin. EEG Neurosci. 2013, 44, 221–226. [Google Scholar] [CrossRef]
- Han, C.X.; Wang, J.; Yi, G.S.; Che, Y.Q. Investigation of EEG abnormalities in the early stage of Parkinson’s disease. Cogn. Neurodyn. 2013, 7, 351–359. [Google Scholar] [CrossRef] [Green Version]
- Postuma, R.B.; Berg, D.; Stern, M.; Poewe, W.; Olanow, C.W.; Oertel, W.; Obeso, J.; Marek, K.; Litvan, I.; Lang, A.E.; et al. MDS clinical diagnostic criteria for Parkinson’s disease. Mov. Disord. 2015, 30, 1591–1601. [Google Scholar] [CrossRef] [PubMed]
- Boel, J.A.; de Bie, R.M.A.; Schmand, B.A.; Dalrymple-Alford, J.C.; Marras, C.; Adler, C.H.; Goldman, J.G.; Tröster, A.I.; Burn, D.J.; Litvan, I.; et al. Level I PD-MCI Using Global Cognitive Tests and the Risk for Parkinson’s Disease Dementia. Mov. Disord. Clin. Pract. 2022, 9, 479–483. [Google Scholar] [CrossRef]
- Dubois, B.; Burn, D.; Goetz, C.; Aarsland, D.; Brown, R.G.; Broe, G.A.; Dickson, D.; Duyckaerts, C.; Cummings, J.; Gauthier, S.; et al. Diagnostic procedures for Parkinson’s disease dementia: Recommendations from the movement disorder society task force. Mov. Disord. 2007, 22, 2314–2324. [Google Scholar] [CrossRef] [PubMed]
- Goetz, C.G.; Tilley, B.C.; Shaftman, S.R.; Stebbins, G.T.; Fahn, S.; Martinez-Martin, P.; Poewe, W.; Sampaio, C.; Stern, M.B.; Dodel, R.; et al. Movement Disorder Society-sponsored revision of the Unified Parkinson’s Disease Rating Scale (MDS-UPDRS): Scale presentation and clinimetric testing results. Mov. Disord. 2008, 23, 2129–2170. [Google Scholar] [CrossRef]
- Hoehn, M.M.; Yahr, M.D. Parkinsonism. Neurology 1967, 17, 427. [Google Scholar] [CrossRef] [Green Version]
- Delorme, A.; Makeig, S. EEGLAB: An open source toolbox for analysis of single-trial EEG dynamics including independent component analysis. J. Neurosci. Methods 2004, 134, 9–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tadel, F.; Baillet, S.; Mosher, J.C.; Pantazis, D.; Leahy, R.M. Brainstorm: A user-friendly application for MEG/EEG analysis. Comput. Intell. Neurosci. 2011, 2011, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Gramfort, A.; Papadopoulo, T.; Olivi, E.; Clerc, M. OpenMEEG: Opensource software for quasistatic bioelectromagnetics. Biomed. Eng. Online 2010, 9, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fonov, V.; Evans, A.; McKinstry, R.; Almli, C.; Collins, D. Unbiased nonlinear average age-appropriate brain templates from birth to adulthood. Neuroimage 2009, 47 (Suppl. S1). [Google Scholar] [CrossRef]
- Pascual-Marqui, R.D. Standardized low-resolution brain electromagnetic tomography (sLORETA): Technical details. Methods Find. Exp. Clin. Pharmacol. 2002, 24 (Suppl. D), 5–12. [Google Scholar] [PubMed]
- Russell, D.A.; Hanson, J.D.; Ott, E. Dimension of strange attractors. Phys. Rev. Lett. 1980, 45, 1175–1178. [Google Scholar] [CrossRef]
- Ruiz de Miras, J.; Posadas, M.A.; Ibáñez-Molina, A.J.; Soriano, M.F.; Iglesias-Parro, S. Fast computation of fractal dimension for 2D, 3D and 4D data. J. Comput. Sci. 2023, 66, 101908. [Google Scholar] [CrossRef]
- Mann, H.B.; Whitney, D.R. On a Test of Whether one of Two Random Variables is Stochastically Larger than the Other. Ann. Math. Stat. 1947, 18, 50–60. [Google Scholar] [CrossRef]
- Pearson, K.X. On the criterion that a given system of deviations from the probable in the case of a correlated system of variables is such that it can be reasonably supposed to have arisen from random sampling. Lond. Edinb. Dublin Philos. Mag. J. Sci. 1900, 50, 157–175. [Google Scholar] [CrossRef] [Green Version]
- Spearman, C. The Proof and Measurement of Association between Two Things. Am. J. Psychol. 1904, 15, 72–101. [Google Scholar] [CrossRef]
- Hanley, J.A.; McNeil, B.J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 1982, 143, 29–36. [Google Scholar] [CrossRef] [Green Version]
- Yi, G.S.; Wang, J.; Deng, B.; Wei, X. Le Complexity of resting-state EEG activity in the patients with early-stage Parkinson’s disease. Cogn. Neurodyn. 2017, 11, 147–160. [Google Scholar] [CrossRef] [Green Version]
- Keller, S.M.; Gschwandtner, U.; Meyer, A.; Chaturvedi, M.; Roth, V.; Fuhr, P. Cognitive decline in Parkinson’s disease is associated with reduced complexity of EEG at baseline. Brain Commun. 2020, 2, fcaa207. [Google Scholar] [CrossRef] [PubMed]
- Pappalettera, C.; Miraglia, F.; Cotelli, M.; Rossini, P.M.; Vecchio, F. Analysis of complexity in the EEG activity of Parkinson’s disease patients by means of approximate entropy. GeroScience 2022, 44, 1599–1607. [Google Scholar] [CrossRef] [PubMed]
- Watson, G.S.; Leverenz, J.B. Profile of Cognitive Impairment in Parkinson’s Disease. Brain Pathol. 2010, 20, 640–645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leviashvili, S.; Ezra, Y.; Droby, A.; Ding, H.; Groppa, S.; Mirelman, A.; Muthuraman, M.; Maidan, I. EEG-Based Mapping of Resting-State Functional Brain Networks in Patients with Parkinson’s Disease. Biomimetics 2022, 7, 231. [Google Scholar] [CrossRef]
- Nahli, F.; Paramonov, A.; Soliman, N.F.; Aleisa, H.N.; Alkanhel, R.; Muthanna, A.; Ateya, A.A. Novel Path Counting-Based Method for Fractal Dimension Estimation of the Ultra-Dense Networks. Intell. Autom. Soft Comput. 2022, 36, 561–572. [Google Scholar] [CrossRef]
- Aljalal, M.; Aldosari, S.A.; Molinas, M.; AlSharabi, K.; Alturki, F.A. Detection of Parkinson’s disease from EEG signals using discrete wavelet transform, different entropy measures, and machine learning techniques. Sci. Rep. 2022, 12, 22547. [Google Scholar] [CrossRef]
- Zhang, R.; Jia, J.; Zhang, R. EEG analysis of Parkinson’s disease using time–frequency analysis and deep learning. Biomed. Signal Process. Control 2022, 78, 103883. [Google Scholar] [CrossRef]
- Jiang, W.; Liu, Y.; Wang, J.; Li, R.; Liu, X.; Zhang, J. Problems of the Grid Size Selection in Differential Box-Counting (DBC) Methods and an Improvement Strategy. Entropy 2022, 24, 977. [Google Scholar] [CrossRef]

| PD | HC | Test, p-Value | |
|---|---|---|---|
| N | 27 | 15 | |
| Sex (F:M) | 12:15 | 7:8 | χ2 = 0.019, p = 0.89 a |
| Age | 69.59 ± 6.74 | 67.53 ± 4.94 | U = 249.5, p = 0.22 b |
| Education | 12.74 ± 3.79 | 12.80 ± 2.95 | U = 195.0, p = 0.84 b |
| Disease duration | 10.24 ± 6.78 | ||
| Hoehn and Yahr | 2.41 ± 0.42 | ||
| UPDRS III | 39.00 ± 9.79 | ||
| MoCA | 25.01 ± 2.65 | ||
| MMP | 28.62 ± 2.45 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruiz de Miras, J.; Derchi, C.-C.; Atzori, T.; Mazza, A.; Arcuri, P.; Salvatore, A.; Navarro, J.; Saibene, F.L.; Meloni, M.; Comanducci, A. Spatio-Temporal Fractal Dimension Analysis from Resting State EEG Signals in Parkinson’s Disease. Entropy 2023, 25, 1017. https://doi.org/10.3390/e25071017
Ruiz de Miras J, Derchi C-C, Atzori T, Mazza A, Arcuri P, Salvatore A, Navarro J, Saibene FL, Meloni M, Comanducci A. Spatio-Temporal Fractal Dimension Analysis from Resting State EEG Signals in Parkinson’s Disease. Entropy. 2023; 25(7):1017. https://doi.org/10.3390/e25071017
Chicago/Turabian StyleRuiz de Miras, Juan, Chiara-Camilla Derchi, Tiziana Atzori, Alice Mazza, Pietro Arcuri, Anna Salvatore, Jorge Navarro, Francesca Lea Saibene, Mario Meloni, and Angela Comanducci. 2023. "Spatio-Temporal Fractal Dimension Analysis from Resting State EEG Signals in Parkinson’s Disease" Entropy 25, no. 7: 1017. https://doi.org/10.3390/e25071017






