Discovering Low-Dimensional Descriptions of Multineuronal Dependencies
Abstract
1. Introduction
2. Materials and Methods
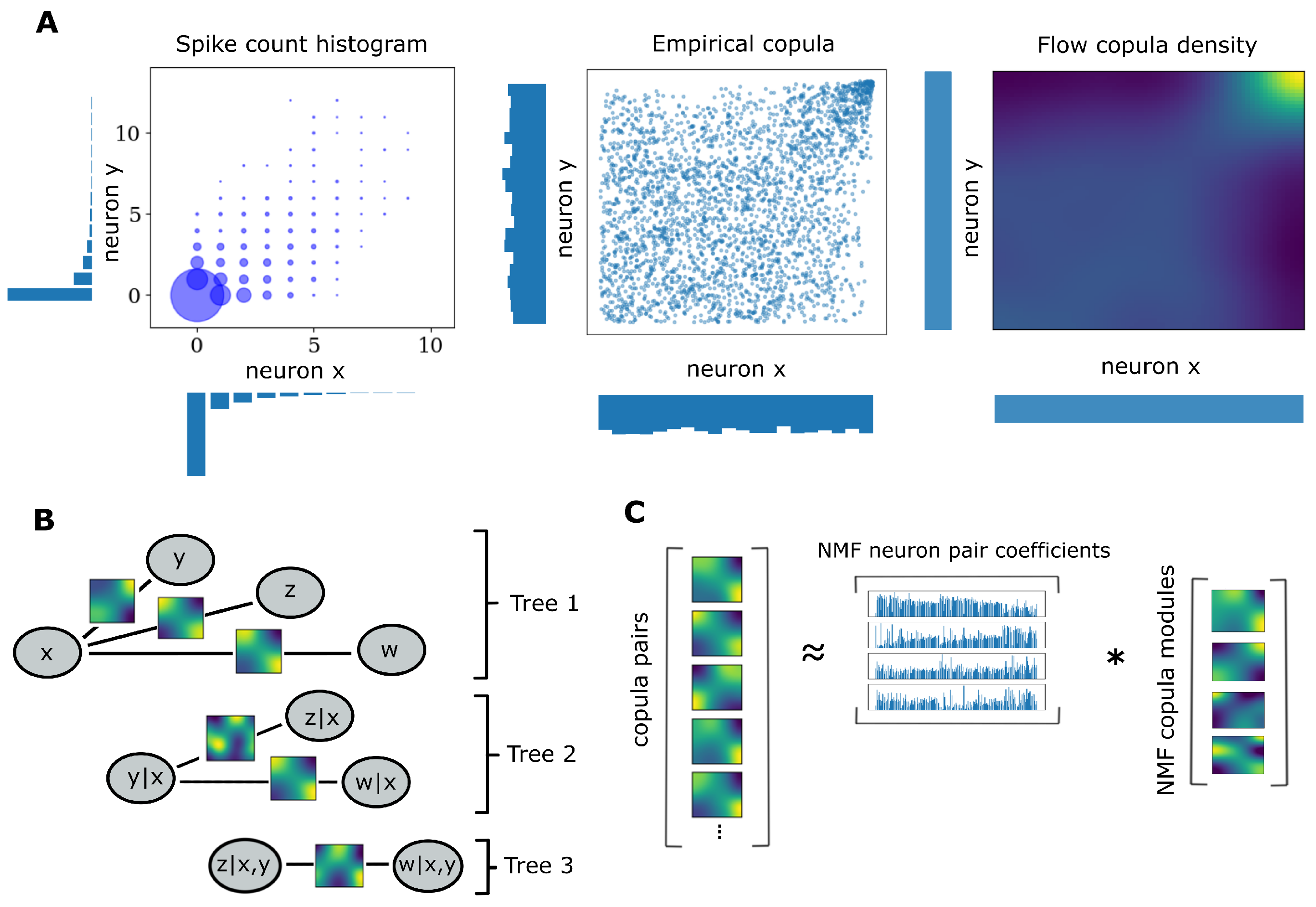
2.1. Copulas
2.2. Pair Copula Constructions
2.3. Copula Flows
2.4. Sequential Estimation and Model Selection
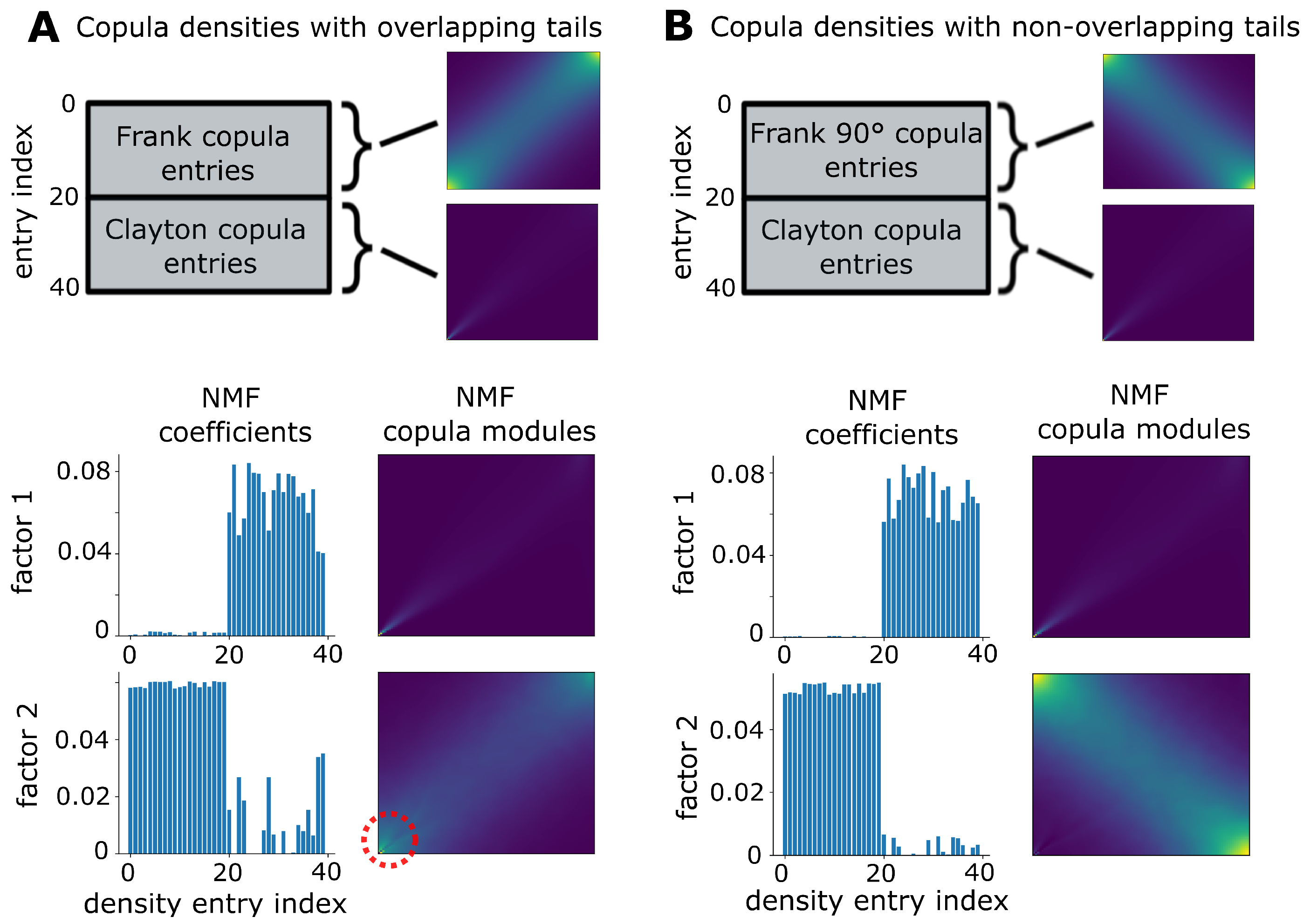
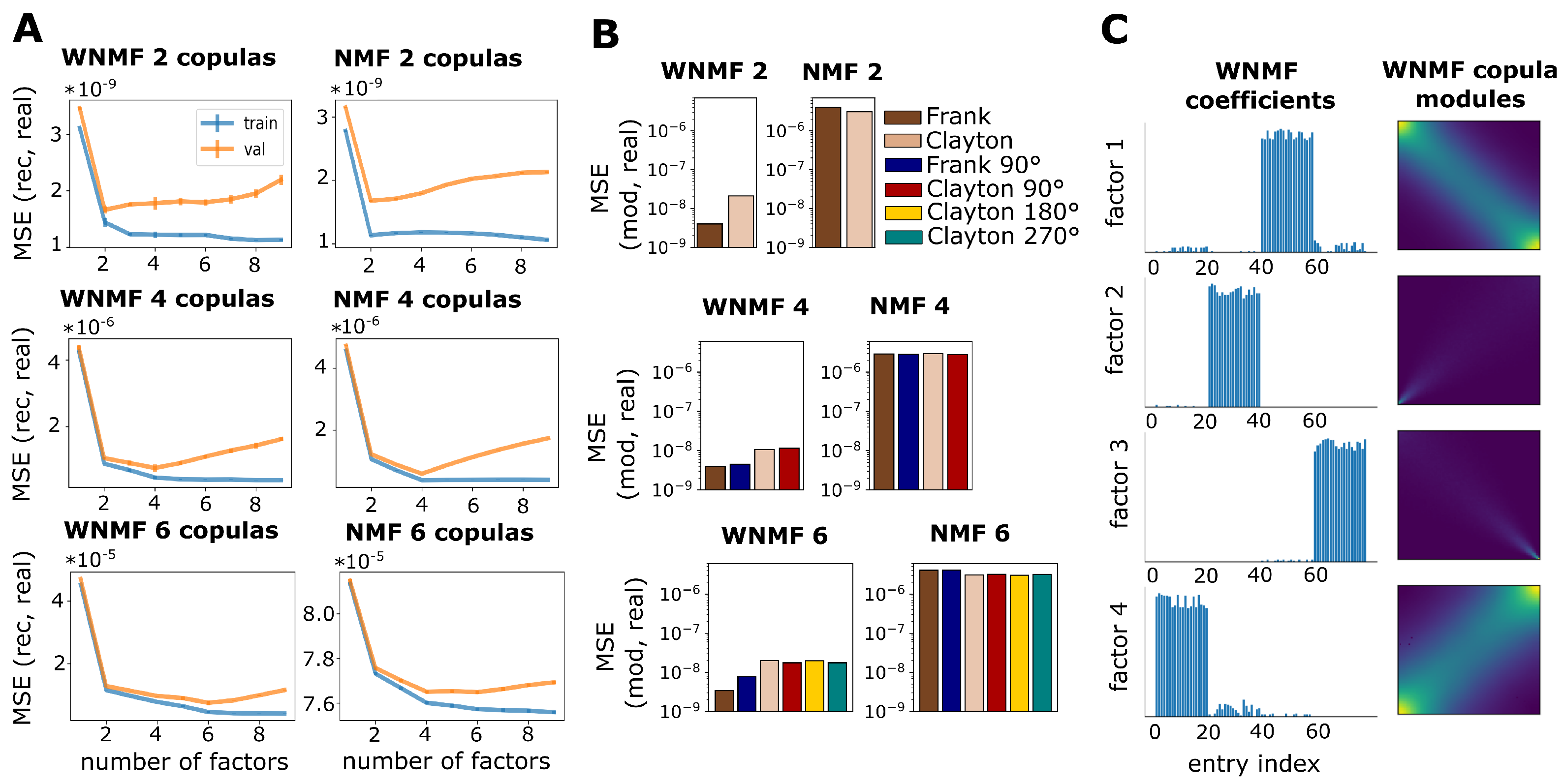
2.5. Weighted Non-Negative Matrix Factorization
2.6. Synthetic Data
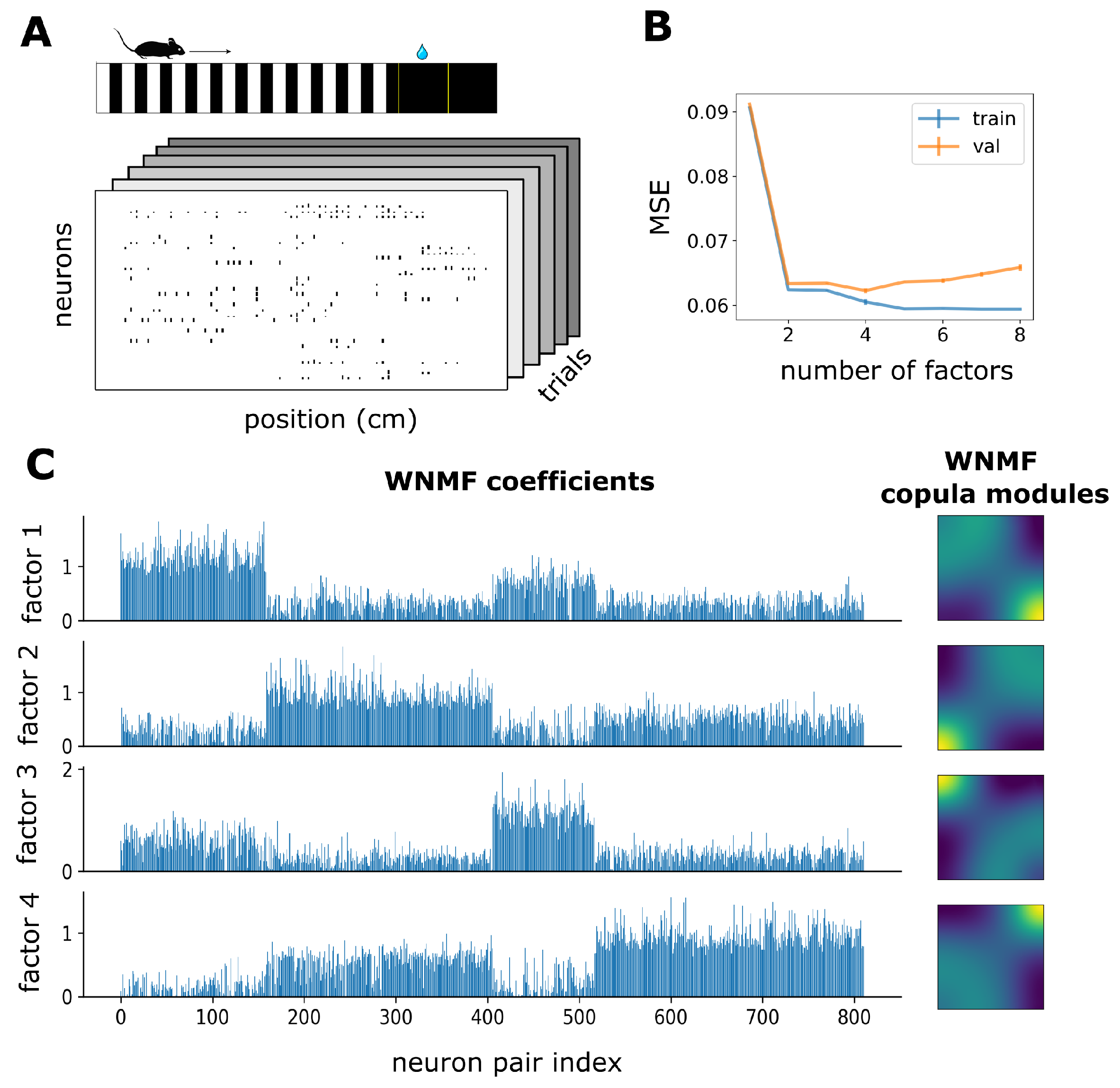
2.7. Experimental Data
3. Results
3.1. Validation on Synthetic Data
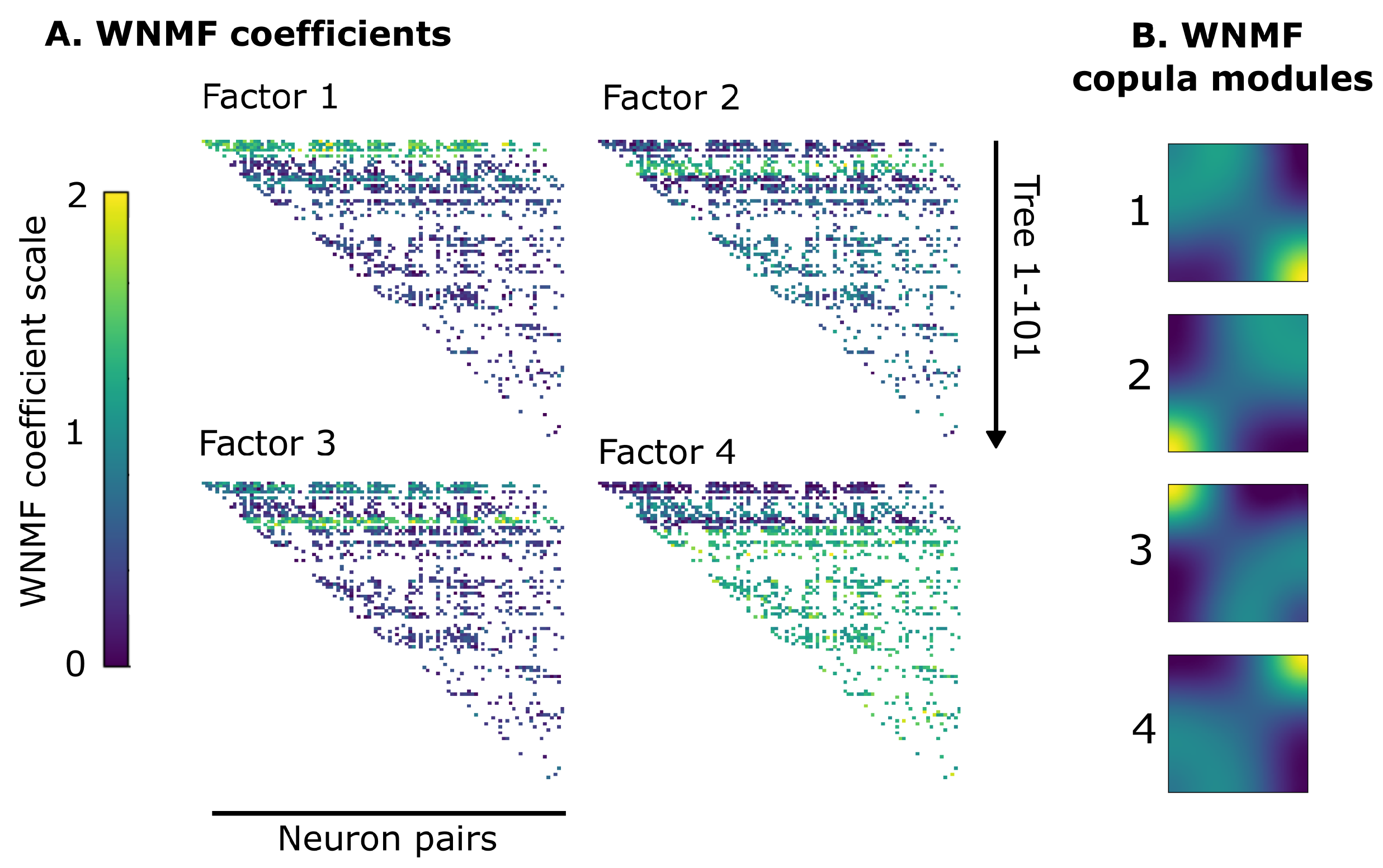
3.2. WNMF Identifies Shared Latent Structures of Neural Dependencies in Visual Cortex
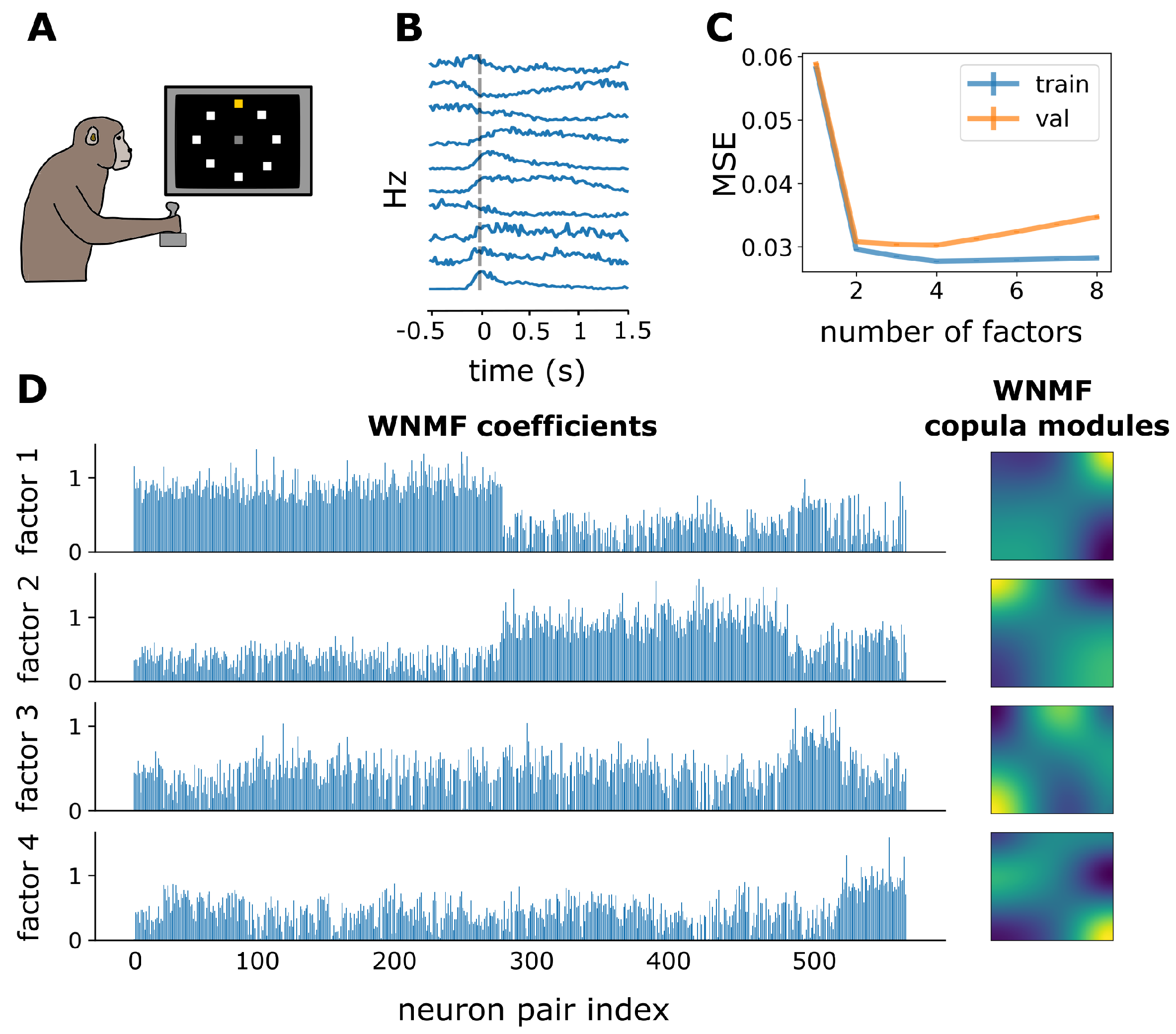
3.3. WNMF Identifies Shared Latent Structures of Neural Dependencies in Macaque Motor Cortex
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Saxena, S.; Cunningham, J.P. Towards the neural population doctrine. Curr. Opin. Neurobiol. 2019, 55, 103–111. [Google Scholar] [CrossRef]
- Vyas, S.; Golub, M.D.; Sussillo, D.; Shenoy, K.V. Computation through neural population dynamics. Annu. Rev. Neurosci. 2020, 43, 249–275. [Google Scholar] [CrossRef]
- Urai, A.E.; Doiron, B.; Leifer, A.M.; Churchland, A.K. Large-scale neural recordings call for new insights to link brain and behavior. Nat. Neurosci. 2022, 25, 11–19. [Google Scholar] [CrossRef]
- Chen, X.; Leischner, U.; Varga, Z.; Jia, H.; Deca, D.; Rochefort, N.L.; Konnerth, A. LOTOS-based two-photon calcium imaging of dendritic spines in vivo. Nat. Protoc. 2012, 7, 1818–1829. [Google Scholar] [CrossRef] [PubMed]
- Jun, J.J.; Steinmetz, N.A.; Siegle, J.H.; Denman, D.J.; Bauza, M.; Barbarits, B.; Lee, A.K.; Anastassiou, C.A.; Andrei, A.; Aydın, Ç.; et al. Fully integrated silicon probes for high-density recording of neural activity. Nature 2017, 551, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.C.K.; David, S.V.; Gallant, J.L. Complete functional characterization of sensory neurons by system identification. Annu. Rev. Neurosci. 2006, 29, 477–505. [Google Scholar] [CrossRef] [PubMed]
- Rolls, E.T.; Treves, A. The neuronal encoding of information in the brain. Prog. Neurobiol. 2011, 95, 448–490. [Google Scholar] [CrossRef]
- Kass, R.E.; Amari, S.I.; Arai, K.; Brown, E.N.; Diekman, C.O.; Diesmann, M.; Doiron, B.; Eden, U.T.; Fairhall, A.L.; Fiddyment, G.M.; et al. Computational neuroscience: Mathematical and statistical perspectives. Annu. Rev. Stat. Appl. 2018, 5, 183–214. [Google Scholar] [CrossRef]
- Hurwitz, C.; Kudryashova, N.; Onken, A.; Hennig, M.H. Building population models for large-scale neural recordings: Opportunities and pitfalls. Curr. Opin. Neurobiol. 2021, 70, 64–73. [Google Scholar] [CrossRef]
- Ohiorhenuan, I.E.; Mechler, F.; Purpura, K.P.; Schmid, A.M.; Hu, Q.; Victor, J.D. Sparse coding and high-order correlations in fine-scale cortical networks. Nature 2010, 466, 617–621. [Google Scholar] [CrossRef]
- Yu, S.; Yang, H.; Nakahara, H.; Santos, G.S.; Nikolić, D.; Plenz, D. Higher-order interactions characterized in cortical activity. J. Neurosci. 2011, 31, 17514–17526. [Google Scholar] [CrossRef] [PubMed]
- Shimazaki, H.; Amari, S.I.; Brown, E.N.; Grün, S. State-space analysis of time-varying higher-order spike correlation for multiple neural spike train data. PLoS Comput. Biol. 2012, 8, e1002385. [Google Scholar] [CrossRef] [PubMed]
- Montangie, L.; Montani, F. Higher-order correlations in common input shapes the output spiking activity of a neural population. Phys. A Stat. Mech. Appl. 2017, 471, 845–861. [Google Scholar] [CrossRef]
- Zohary, E.; Shadlen, M.N.; Newsome, W.T. Correlated neuronal discharge rate and its implications for psychophysical performance. Nature 1994, 370, 140–143. [Google Scholar] [CrossRef]
- Brown, E.N.; Kass, R.E.; Mitra, P.P. Multiple neural spike train data analysis: State-of-the-art and future challenges. Nat. Neurosci. 2004, 7, 456–461. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Bote, R.; Beck, J.; Kanitscheider, I.; Pitkow, X.; Latham, P.; Pouget, A. Information-limiting correlations. Nat. Neurosci. 2014, 17, 1410–1417. [Google Scholar] [CrossRef]
- Kohn, A.; Coen-Cagli, R.; Kanitscheider, I.; Pouget, A. Correlations and neuronal population information. Annu. Rev. Neurosci. 2016, 39, 237–256. [Google Scholar] [CrossRef]
- Panzeri, S.; Moroni, M.; Safaai, H.; Harvey, C.D. The structures and functions of correlations in neural population codes. Nat. Rev. Neurosci. 2022, 23, 551–567. [Google Scholar] [CrossRef]
- Onken, A.; Grünewälder, S.; Munk, M.H.; Obermayer, K. Analyzing short-term noise dependencies of spike-counts in macaque prefrontal cortex using copulas and the flashlight transformation. PLoS Comput. Biol. 2009, 5, e1000577. [Google Scholar] [CrossRef]
- Kudryashova, N.; Amvrosiadis, T.; Dupuy, N.; Rochefort, N.; Onken, A. Parametric Copula-GP model for analyzing multidimensional neuronal and behavioral relationships. PLoS Comput. Biol. 2022, 18, e1009799. [Google Scholar] [CrossRef]
- Pillow, J.W.; Shlens, J.; Paninski, L.; Sher, A.; Litke, A.M.; Chichilnisky, E.; Simoncelli, E.P. Spatio-temporal correlations and visual signalling in a complete neuronal population. Nature 2008, 454, 995–999. [Google Scholar] [CrossRef] [PubMed]
- Michel, M.M.; Jacobs, R.A. The costs of ignoring high-order correlations in populations of model neurons. Neural Comput. 2006, 18, 660–682. [Google Scholar] [CrossRef] [PubMed]
- Jaworski, P.; Durante, F.; Härdle, W.K. Copulae in Mathematical and Quantitative Finance: Proceedings of the Workshop Held in Cracow, 10–11 July 2012; Springer: Berlin/Heidelberg, Germany, 2012; Volume 10, p. 11. [Google Scholar]
- Jenison, R.L.; Reale, R.A. The shape of neural dependence. Neural Comput. 2004, 16, 665–672. [Google Scholar] [CrossRef] [PubMed]
- Berkes, P.; Wood, F.; Pillow, J. Characterizing neural dependencies with copula models. Adv. Neural Inf. Process. Syst. 2008, 21, 129–136. [Google Scholar]
- Onken, A.; Grünewälder, S.; Munk, M.; Obermayer, K. Modeling short-term noise dependence of spike counts in macaque prefrontal cortex. Adv. Neural Inf. Process. Syst. 2008, 21, 85117. [Google Scholar]
- Sacerdote, L.; Tamborrino, M.; Zucca, C. Detecting dependencies between spike trains of pairs of neurons through copulas. Brain Res. 2012, 1434, 243–256. [Google Scholar] [CrossRef]
- Onken, A.; Panzeri, S. Mixed vine copulas as joint models of spike counts and local field potentials. Adv. Neural Inf. Process. Syst. 2016, 29, 910122. [Google Scholar]
- Faugeras, O.P. Inference for copula modeling of discrete data: A cautionary tale and some facts. Depend. Model. 2017, 5, 121–132. [Google Scholar] [CrossRef]
- Genest, C.; Nešlehová, J. A primer on copulas for count data. ASTIN Bull. J. IAA 2007, 37, 475–515. [Google Scholar] [CrossRef]
- Nagler, T. A generic approach to nonparametric function estimation with mixed data. Stat. Probab. Lett. 2018, 137, 326–330. [Google Scholar] [CrossRef]
- Aas, K.; Czado, C.; Frigessi, A.; Bakken, H. Pair-copula constructions of multiple dependence. Insur. Math. Econ. 2009, 44, 182–198. [Google Scholar] [CrossRef]
- Song, P.X.K.; Li, M.; Yuan, Y. Joint regression analysis of correlated data using Gaussian copulas. Biometrics 2009, 65, 60–68. [Google Scholar] [CrossRef] [PubMed]
- de Leon, A.R.; Wu, B. Copula-based regression models for a bivariate mixed discrete and continuous outcome. Stat. Med. 2011, 30, 175–185. [Google Scholar] [CrossRef]
- Smith, M.S.; Khaled, M.A. Estimation of copula models with discrete margins via Bayesian data augmentation. J. Am. Stat. Assoc. 2012, 107, 290–303. [Google Scholar] [CrossRef]
- Panagiotelis, A.; Czado, C.; Joe, H. Pair copula constructions for multivariate discrete data. J. Am. Stat. Assoc. 2012, 107, 1063–1072. [Google Scholar] [CrossRef]
- Racine, J.S. Mixed data kernel copulas. Empir. Econ. 2015, 48, 37–59. [Google Scholar] [CrossRef]
- Geenens, G.; Charpentier, A.; Paindaveine, D. Probit transformation for nonparametric kernel estimation of the copula density. Bernoulli 2017, 23, 1848–1873. [Google Scholar] [CrossRef]
- Schallhorn, N.; Kraus, D.; Nagler, T.; Czado, C. D-vine quantile regression with discrete variables. arXiv 2017, arXiv:1705.08310. [Google Scholar]
- Nagler, T.; Schellhase, C.; Czado, C. Nonparametric estimation of simplified vine copula models: Comparison of methods. Depend. Model. 2017, 5, 99–120. [Google Scholar] [CrossRef]
- Mitskopoulos, L.; Amvrosiadis, T.; Onken, A. Mixed vine copula flows for flexible modelling of neural dependencies. Front. Neurosci. 2022, 16, 1645. [Google Scholar] [CrossRef]
- Durkan, C.; Bekasov, A.; Murray, I.; Papamakarios, G. Neural spline flows. In Proceedings of the 33rd Conference on Neural Information Processing Systems (NeurIPS 2019), Vancouver, BC, Canada, 8–14 December 2019. [Google Scholar]
- Rezende, D.; Mohamed, S. Variational inference with normalizing flows. In Proceedings of the International Conference on Machine Learning, Lille, France, 6–11 July 2015; pp. 1530–1538. [Google Scholar]
- Lee, D.D.; Seung, H.S. Learning the parts of objects by non-negative matrix factorization. Nature 1999, 401, 788–791. [Google Scholar] [CrossRef] [PubMed]
- Russo, A.A.; Bittner, S.R.; Perkins, S.M.; Seely, J.S.; London, B.M.; Lara, A.H.; Miri, A.; Marshall, N.J.; Kohn, A.; Jessell, T.M.; et al. Motor cortex embeds muscle-like commands in an untangled population response. Neuron 2018, 97, 953–966. [Google Scholar] [CrossRef] [PubMed]
- Guillamet, D.; Vitria, J.; Schiele, B. Introducing a weighted non-negative matrix factorization for image classification. Pattern Recognit. Lett. 2003, 24, 2447–2454. [Google Scholar] [CrossRef]
- Zhou, Q.; Feng, Z.; Benetos, E. Adaptive noise reduction for sound event detection using subband-weighted NMF. Sensors 2019, 19, 3206. [Google Scholar] [CrossRef] [PubMed]
- Sklar, M. Fonctions de repartition an dimensions et leurs marges. Publ. Inst. Stat. Univ. Paris 1959, 8, 229–231. [Google Scholar]
- Bedford, T.; Cooke, R.M. Vines—A new graphical model for dependent random variables. Ann. Stat. 2002, 30, 1031–1068. [Google Scholar] [CrossRef]
- Czado, C. Analyzing Dependent Data with Vine Copulas; Lecture Notes in Statistics; Springer: Berlin/Heidelberg, Germany, 2019. [Google Scholar]
- Haff, I.H.; Aas, K.; Frigessi, A. On the simplified pair-copula construction—Simply useful or too simplistic? J. Multivar. Anal. 2010, 101, 1296–1310. [Google Scholar] [CrossRef]
- Bergstra, J.; Bengio, Y. Random search for hyper-parameter optimization. J. Mach. Learn. Res. 2012, 13, 281–305. [Google Scholar]
- Fasano, G.; Franceschini, A. A multidimensional version of the Kolmogorov—Smirnov test. Mon. Not. R. Astron. Soc. 1987, 225, 155–170. [Google Scholar] [CrossRef]
- Owen, A.B.; Perry, P.O. Bi-cross-validation of the SVD and the nonnegative matrix factorization. Ann. Appl. Stat. 2009, 3, 564–594. [Google Scholar] [CrossRef]
- Nelsen, R.B. An Introduction to Copulas; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2007. [Google Scholar]
- Henschke, J.U.; Dylda, E.; Katsanevaki, D.; Dupuy, N.; Currie, S.P.; Amvrosiadis, T.; Pakan, J.M.; Rochefort, N.L. Reward association enhances stimulus-specific representations in primary visual cortex. Curr. Biol. 2020, 30, 1866–1880. [Google Scholar] [CrossRef] [PubMed]
- Churchland, M.M.; Cunningham, J.P.; Kaufman, M.T.; Foster, J.D.; Nuyujukian, P.; Ryu, S.I.; Shenoy, K.V. Neural population dynamics during reaching. Nature 2012, 487, 51–56. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mitskopoulos, L.; Onken, A. Discovering Low-Dimensional Descriptions of Multineuronal Dependencies. Entropy 2023, 25, 1026. https://doi.org/10.3390/e25071026
Mitskopoulos L, Onken A. Discovering Low-Dimensional Descriptions of Multineuronal Dependencies. Entropy. 2023; 25(7):1026. https://doi.org/10.3390/e25071026
Chicago/Turabian StyleMitskopoulos, Lazaros, and Arno Onken. 2023. "Discovering Low-Dimensional Descriptions of Multineuronal Dependencies" Entropy 25, no. 7: 1026. https://doi.org/10.3390/e25071026
APA StyleMitskopoulos, L., & Onken, A. (2023). Discovering Low-Dimensional Descriptions of Multineuronal Dependencies. Entropy, 25(7), 1026. https://doi.org/10.3390/e25071026





