Harnessing Information Thermodynamics: Conversion of DNA Information into Mechanical Work in RNA Transcription and Nanopore Sequencing
Abstract
1. Introduction
2. Methods
2.1. Transcription System Model
2.2. Information Thermodynamics of Gaining Mutual Information
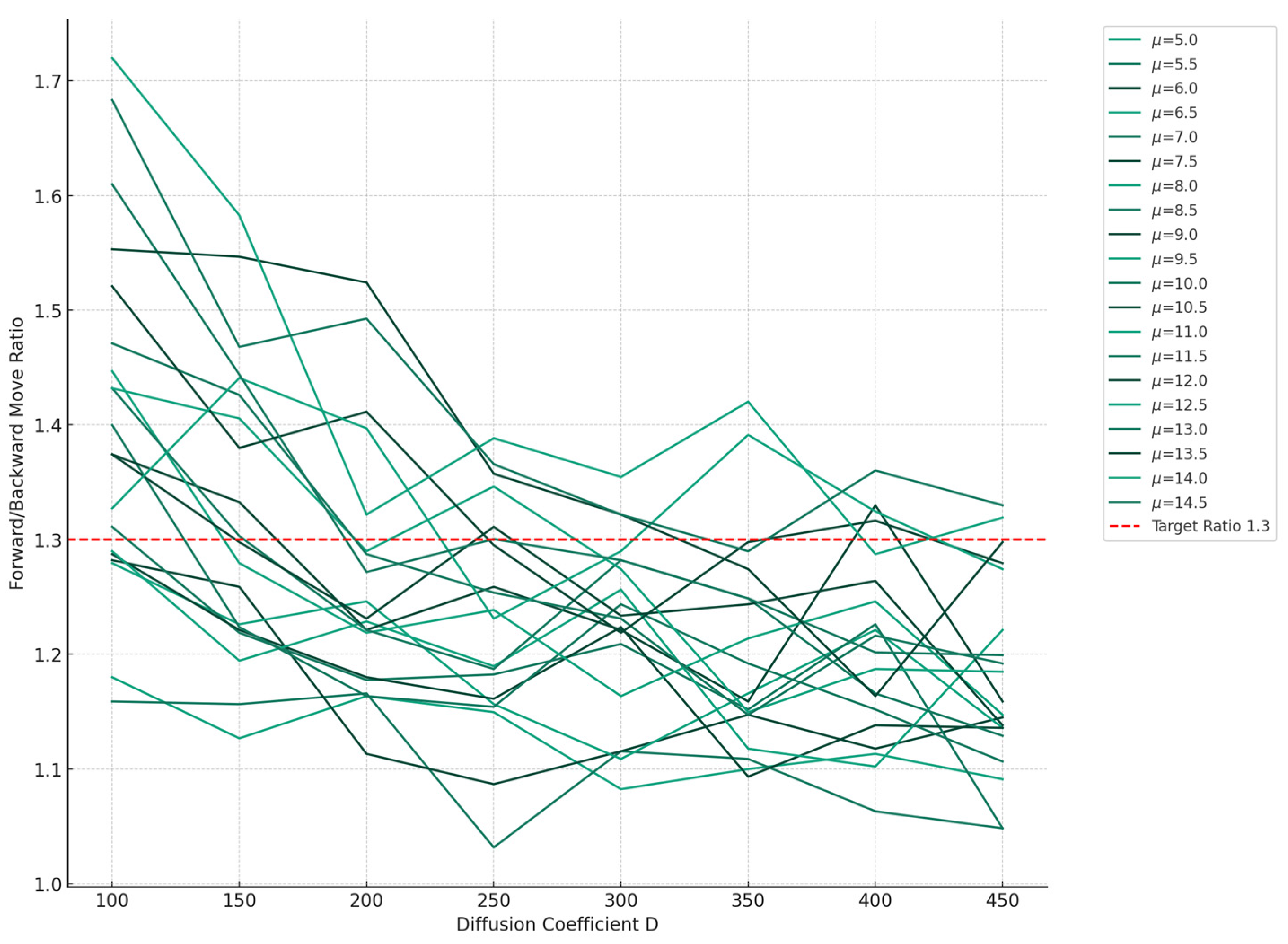
2.3. Free Energy Change during RNAP Translocation
2.4. A Proposal for the Information Thermodynamics of Nanopore Sequencing
3. Discussion
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| RNAP | RNA polymerase |
| m | Position of the dNTP in the DNA (1 ≤ m ≤ N) |
| L | The length of the template DNA, equal to Nd |
| P(x,t) | Probability that RNAP is located at any time, t, and position, x |
| Pf(x,t) | Probability distribution of RNAP moving forward at any time, t, and position, x |
| Pb(x,t) | Probability distribution of RNAP moving backward at any time, t, and position, x |
| D | The diffusion coefficient of RNAP movement |
| μ | Drift velocity of RNAP movement |
| Rf | Transitional probability in RNAP movement in the forward orientation |
| Rb | Transitional probability in RNAP movement in the backward orientation |
| Im | Mutual information gained by RNAP from dNTP at m |
| im | Stochastic mutual information gained by RNAP from dNTP at m |
| Stotm | Total entropy production in the entire system |
| stotm | Total stochastic entropy production in the entire system |
| Δs(m) | The entropy produced in the RNAP movement from m to m + 1 position per length of the template DNA, L |
| I′m | Mutual information feedback by dNTP at m |
| i′m | Stochastic mutual information feedback by dNTP at m |
| wext | Work done to RNAP at m by DNA |
| ΔGm | Gibbs’ free energy change before and after RNAP movement |
| ΔS | The entropy produced in the nanopore motor protein movement from m to m + 1 position |
| Δs | The entropy produced in the nanopore motor protein movement from m to m + 1 position per length of the template DNA, L |
| e | Electron charge |
| V | Voltage across the membrane in nanopore sequencing |
| ϕ | Electrochemical factors of the entropy |
| kB | Boltzmann constant |
| dNTP | Deoxyribonucleotide |
| dATP | Deoxyadenosine triphosphate |
| dCTP | Deoxycytidine triphosphate |
| dGTP | Deoxyguanosine triphosphate |
| dTTP | Deoxythymidine triphosphate |
| rNTP | Ribonucleoside Triphosphates |
| rATP | Ribonucleoside Triphosphate Adenine |
| rCTP | Ribonucleoside Triphosphate Cytidine |
| rGTP | Ribonucleoside Triphosphate Guanine |
| rUTP | Ribonucleoside Triphosphate Uracil |
References
- Berezhkovskii, A.M.; Bezrukov, S.M. Counting translocations of strongly repelling particles through single channels: Fluctuation theorem for membrane transport. Phys. Rev. Lett. 2008, 100, 038104. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, Y. Quantum Thermodynamic Uncertainty Relation for Continuous Measurement. Phys. Rev. Lett. 2020, 125, 050601. [Google Scholar] [CrossRef] [PubMed]
- Van Vu, T.; Hasegawa, Y. Geometrical Bounds of the Irreversibility in Markovian Systems. Phys. Rev. Lett. 2021, 126, 010601. [Google Scholar] [CrossRef] [PubMed]
- Sagawa, T.; Kikuchi, Y.; Inoue, Y.; Takahashi, H.; Muraoka, T.; Kinbara, K.; Ishijima, A.; Fukuoka, H. Single-cell E. coli response to an instantaneously applied chemotactic signal. Biophys. J. 2014, 107, 730–739. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; Sagawa, T. Maxwell’s demon in biochemical signal transduction with feedback loop. Nat. Commun. 2015, 6, 7498. [Google Scholar] [CrossRef] [PubMed]
- Andrieux, D.; Gaspard, P. Fluctuation theorems and the nonequilibrium thermodynamics of molecular motors. Phys. Rev. E. 2006, 74 Pt 1, 011906. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, K.; Ueno, H.; Iino, R.; Noji, H. Fluctuation theorem applied to F1-ATPase. Phys. Rev. Lett. 2010, 104, 218103. [Google Scholar] [CrossRef] [PubMed]
- Lau, A.W.; Lacoste, D.; Mallick, K. Nonequilibrium fluctuations and mechanochemical couplings of a molecular motor. Phys. Rev. Lett. 2007, 99, 158102. [Google Scholar] [CrossRef] [PubMed]
- McMillan, D.G.; Watanabe, R.; Ueno, H.; Cook, G.M.; Noji, H. Biophysical Characterization of a Thermoalkaliphilic Molecular Motor with a High Stepping Torque Gives Insight into Evolutionary ATP Synthase Adaptation. J. Biol. Chem. 2016, 291, 23965–23977. [Google Scholar] [CrossRef]
- Seifert, U. Stochastic thermodynamics of single enzymes and molecular motors. Eur. Phys. J. E Soft Matter 2011, 34, 26. [Google Scholar] [CrossRef]
- Crooks, G.E. Entropy production fluctuation theorem and the nonequilibrium work relation for free energy differences. Phys. Rev. E. 1999, 60, 2721–2726. [Google Scholar] [CrossRef]
- Gaspard, P. Fluctuation theorem for nonequilibrium reactions. J. Chem. Phys. 2004, 120, 8898–8905. [Google Scholar] [CrossRef] [PubMed]
- Sagawa, T.; Ueda, M. Fluctuation theorem with information exchange: Role of correlations in stochastic thermodynamics. Phys. Rev. Lett. 2012, 109, 180602. [Google Scholar] [CrossRef] [PubMed]
- Verley, G.; Lacoste, D. Fluctuation theorems and inequalities generalizing the second law of thermodynamics out of equilibrium. Phys. Rev. E. 2012, 86 Pt 1, 051127. [Google Scholar] [CrossRef] [PubMed]
- Minetti, C.A.; Remeta, D.P.; Miller, H.; Gelfand, C.A.; Plum, G.E.; Grollman, A.P.; Breslauer, K.J. The thermodynamics of template-directed DNA synthesis: Base insertion and extension enthalpies. Proc. Natl. Acad. Sci. USA 2003, 100, 14719–14724. [Google Scholar] [CrossRef]
- Dickson, K.S.; Burns, C.M.; Richardson, J.P. Determination of the free-energy change for repair of a DNA phosphodiester bond. J. Biol. Chem. 2000, 275, 15828–15831. [Google Scholar] [CrossRef] [PubMed]
- Thomen, P.; Lopez, P.J.; Bockelmann, U.; Guillerez, J.; Dreyfus, M.; Heslot, F. T7 RNA polymerase studied by force measurements varying cofactor concentration. Biophys. J. 2008, 95, 2423–2433. [Google Scholar] [CrossRef][Green Version]
- Wang, M.D.; Schnitzer, M.J.; Yin, H.; Landick, R.; Gelles, J.; Block, S.M. Force and velocity measured for single molecules of RNA polymerase. Science 1998, 282, 902–907. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Sousa, R. Translocation by T7 RNA polymerase: A sensitively poised Brownian ratchet. J. Mol. Biol. 2006, 358, 241–254. [Google Scholar] [CrossRef]
- Bai, L.; Fulbright, R.M.; Wang, M.D. Mechanochemical kinetics of transcription elongation. Phys. Rev. Lett. 2007, 98, 068103. [Google Scholar] [CrossRef]
- Tsuruyama, T. RNA polymerase is a unique Maxwell’s demon that converts its transcribing genetic information to free energy for its movement. Eur. Phys. J. Plus 2023, 138, 604. [Google Scholar] [CrossRef]
- Da, L.T.; Chao, E.; Shuai, Y.; Wu, S.; Su, X.D.; Yu, J. T7 RNA polymerase translocation is facilitated by a helix opening on the fingers domain that may also prevent backtracking. Nucleic Acids Res. 2017, 45, 7909–7921. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, Y.; Bollas, A.; Wang, Y.; Au, K.F. Nanopore sequencing technology, bioinformatics and applications. Nat. Biotechnol. 2021, 39, 1348–1365. [Google Scholar] [CrossRef]
- Deamer, D.W.; Branton, D. Characterization of nucleic acids by nanopore analysis. Acc. Chem. Res. 2002, 35, 817–825. [Google Scholar] [CrossRef] [PubMed]
- Szilard, L. On the decrease of entropy in a thermodynamic system by the intervention of intelligent beings. Behav. Sci. 1964, 9, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Sagawa, T.; Ueda, M. Second law of thermodynamics with discrete quantum feedback control. Phys. Rev. Lett. 2008, 100, 080403. [Google Scholar] [CrossRef]
- Mathé, J.; Visram, H.; Viasnoff, V.; Rabin, Y.; Meller, A. Nanopore unzipping of individual DNA hairpin molecules. Biophys. J. 2004, 87, 3205–3212. [Google Scholar] [CrossRef]
- Ling, X.S. DNA sequencing using nanopores and kinetic proofreading. Quant. Biol. 2020, 8, 187–194. [Google Scholar] [CrossRef]
- Angevine, C.E.; Robertson, J.W.F.; Dass, A.; Reiner, J.E. Laser-based temperature control to study the roles of entropy and enthalpy in polymer-nanopore interactions. Sci. Adv. 2021, 7, eabf5462. [Google Scholar] [CrossRef]
- Chen, Y.; Nie, F.; Xie, S.Q.; Zheng, Y.F.; Dai, Q.; Bray, T.; Wang, Y.X.; Xing, J.F.; Huang, Z.J.; Wang, D.P.; et al. Efficient assembly of nanopore reads via highly accurate and intact error correction. Nat. Commun. 2021, 12, 60. [Google Scholar] [CrossRef]
- Craig, J.M.; Laszlo, A.H.; Brinkerhoff, H.; Derrington, I.M.; Noakes, M.T.; Nova, I.C.; Tickman, B.I.; Doering, K.; de Leeuw, N.F.; Gundlach, J.H. Revealing dynamics of helicase translocation on single-stranded DNA using high-resolution nanopore tweezers. Proc. Natl. Acad. Sci. USA 2017, 114, 11932–11937. [Google Scholar] [CrossRef] [PubMed]
- Ben McNally, M.W.; Meller, A. Electro-mechanical unzipping of individual DNA molecules using synthetic sub-2 nm pores. Nano Lett. 2008, 8, 3418–3422. [Google Scholar] [CrossRef] [PubMed]
- Sagawa, T.; Ueda, M. Nonequilibrium thermodynamics of feedback control. Phys. Rev. E. 2012, 85 Pt 1, 021104. [Google Scholar] [CrossRef] [PubMed]
- Szilárd, L. Über die Entropieverminderung in einem thermodynamischen System bei Eingriffen intelligenter Wesen. Z. für Phys. 1929, 53, 840–856. [Google Scholar] [CrossRef]
- Hoang, T.M.; Pan, R.; Ahn, J.; Bang, J.; Quan, H.T.; Li, T. Experimental Test of the Differential Fluctuation Theorem and a Generalized Jarzynski Equality for Arbitrary Initial States. Phys. Rev. Lett. 2018, 120, 080602. [Google Scholar] [CrossRef]
- Toyabe, S.; Sagawa, T.; Ueda, M.; Muneyuki, E.; Sano, M. Experimental demonstration of information-to-energy conversion and validation of the generalized Jarzynski equality. Nat. Phys. 2010, 6, 988–992. [Google Scholar] [CrossRef]
- Sagawa, T.; Ueda, M. Minimal energy cost for thermodynamic information processing: Measurement and information erasure. Phys. Rev. Lett. 2009, 102, 250602. [Google Scholar] [CrossRef]
- Sagawa, T.; Ueda, M. Generalized Jarzynski equality under nonequilibrium feedback control. Phys. Rev. Lett. 2010, 104, 090602. [Google Scholar] [CrossRef]
- Hasegawa, Y.; Van Vu, T. Fluctuation Theorem Uncertainty Relation. Phys. Rev. Lett. 2019, 123, 110602. [Google Scholar] [CrossRef]
- Van Vu, T.; Vo, V.T.; Hasegawa, Y. Entropy production estimation with optimal current. Phys. Rev. E 2020, 101, 042138. [Google Scholar] [CrossRef]



| G | A | T(U) | C | Total | |
|---|---|---|---|---|---|
| Probability; P(m) | 0.22 | 0.38 | 0.16 | 0.25 | 1.00 |
| −P(m) log P(m) | 0.33 | 0.37 | 0.29 | 0.35 | 1.34 |
| −log P(m) | 1.51 | 0.96 | 1.83 | 1.38 | 1.42 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsuruyama, T. Harnessing Information Thermodynamics: Conversion of DNA Information into Mechanical Work in RNA Transcription and Nanopore Sequencing. Entropy 2024, 26, 324. https://doi.org/10.3390/e26040324
Tsuruyama T. Harnessing Information Thermodynamics: Conversion of DNA Information into Mechanical Work in RNA Transcription and Nanopore Sequencing. Entropy. 2024; 26(4):324. https://doi.org/10.3390/e26040324
Chicago/Turabian StyleTsuruyama, Tatsuaki. 2024. "Harnessing Information Thermodynamics: Conversion of DNA Information into Mechanical Work in RNA Transcription and Nanopore Sequencing" Entropy 26, no. 4: 324. https://doi.org/10.3390/e26040324
APA StyleTsuruyama, T. (2024). Harnessing Information Thermodynamics: Conversion of DNA Information into Mechanical Work in RNA Transcription and Nanopore Sequencing. Entropy, 26(4), 324. https://doi.org/10.3390/e26040324






