Tick-Tock Consider the Clock: The Influence of Circadian and External Cycles on Time of Day Variation in the Human Metabolome—A Review
Abstract
:1. Introduction
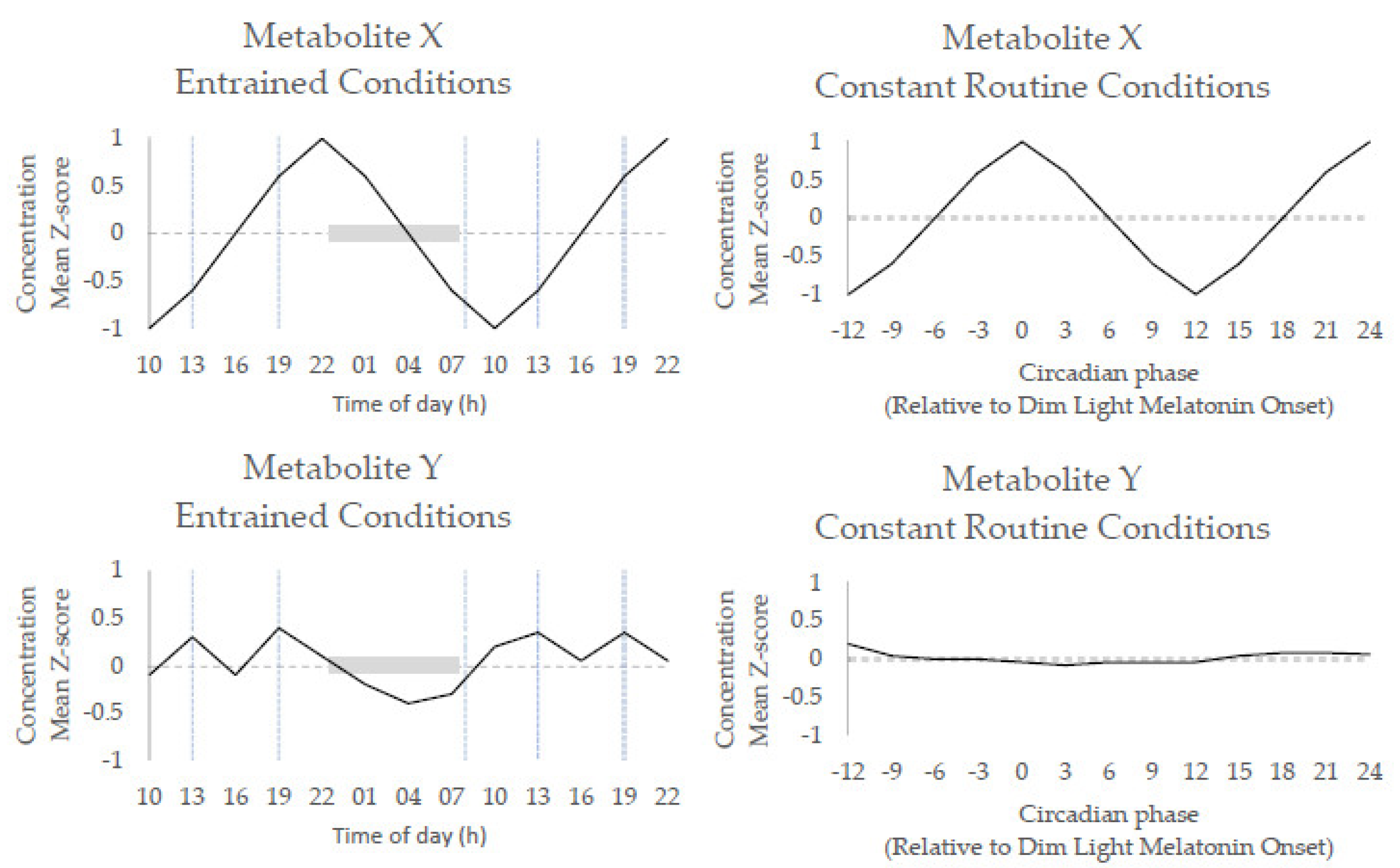
1.1. Key Concepts of Circadian Biology
- Establish the tissues in which time of day variation of metabolites have been observed, or failed to be observed, and the extent to which the metabolome is influenced.
- Establish the source(s) for this observed daily variation and, if applicable, which metabolite classes are most susceptible.
- Consider the implications of circadian/diurnal variation and the timing of sample collection on biomarker discovery and how this may undermine their potential clinical application.
1.2. Literature Search—Parameters and Outcomes
- The literature details original research, i.e., no derivative work such as reviews
- The research studied human participants over a time course
- Employed any metabolomics platform to analyse samples collected across the time course.
2. Literature Search—Results and Commentary
2.1. Blood
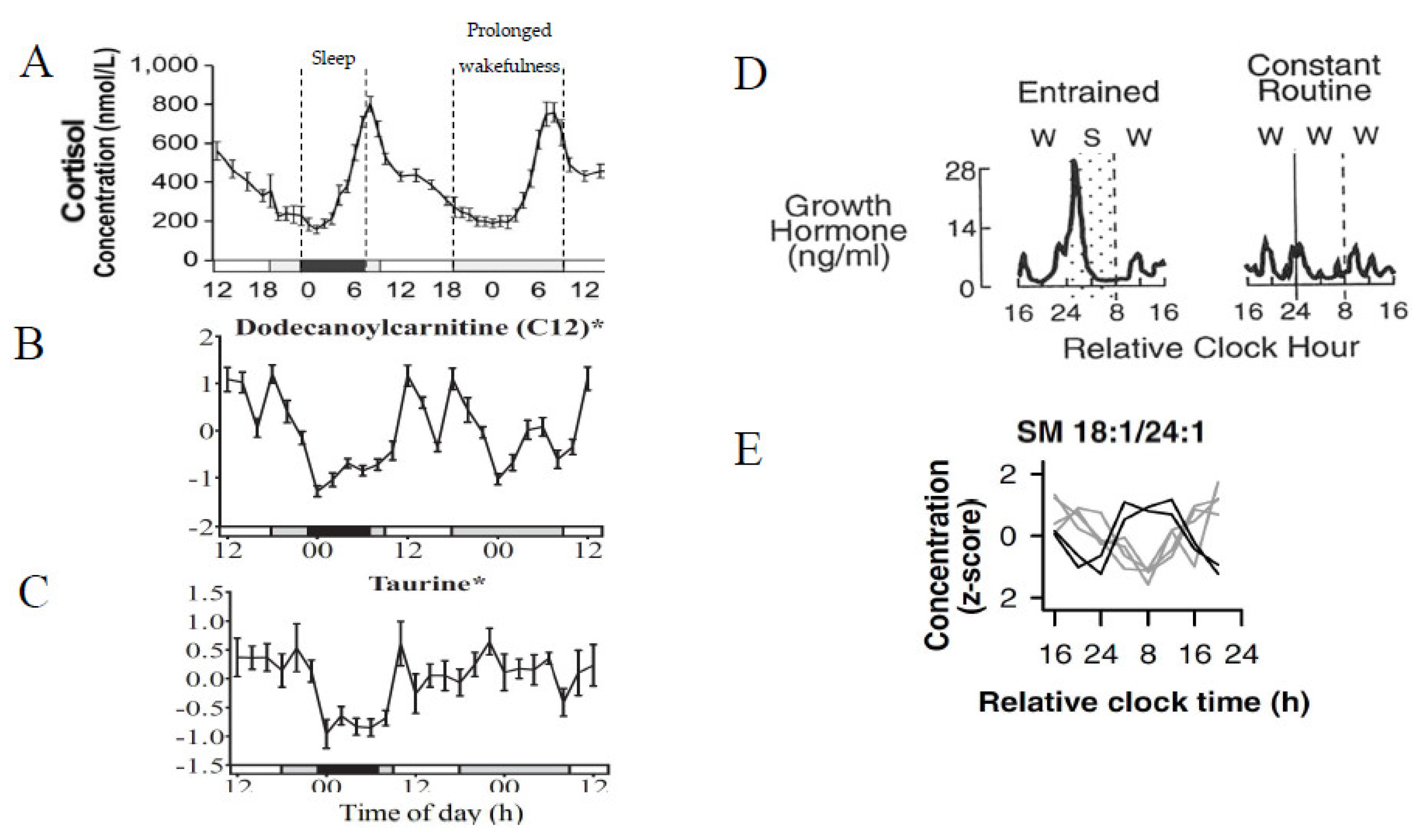
2.1.1. Circadian Variation
2.1.2. Sleep Deprivation and Prolonged Wakefulness
2.1.3. Shift Work
2.1.4. 24 h Diurnal Rhythms
2.1.5. Health Status
2.1.6. Diet Composition
2.1.7. Morning vs. Evening Studies
2.2. Urine
2.2.1. Sleep Deprivation and Prolonged Wakefulness
2.2.2. Shift Work
2.2.3. Creatinine
2.3. Saliva
2.3.1. Circadian Variation
2.3.2. Morning vs. Evening Studies
2.4. Breath
2.4.1. Morning vs. Evening Studies
2.4.2. 24 h Diurnal Rhythms
2.5. Skeletal Muscle
2.5.1. Diet Composition
2.5.2. 24 h Diurnal Rhythms
3. Discussion
3.1. Key Findings
- The number of studies investigating time of day variation of the human metabolome, to date, is small (n = 29).
- Endogenous metabolite rhythms, regulated by the circadian timing system, have been observed via constant routine studies in blood and saliva.
- Diurnal 24 h metabolite rhythms potentially evoked by external cues, either environmental (e.g., light/dark cycle) or behavioural (e.g., sleep/wake; feeding/fasting), have been observed in blood, urine, saliva, breath, and skeletal muscle.
- Acute changes in external cues, e.g., sleep/wake, feeding/fasting, activity/rest cycles and shift work, result in acute alterations to metabolite rhythms (timing and amplitude) that can persist after cessation of the change.
- Metabolite rhythms (timing and amplitude) may be sex dependent although sex has not been regularly investigated with regard to differences in 24 h metabolite rhythms.
- Specific physiological phenotypes and healthy vs. diseased state are shown to result in unique diurnal rhythms alongside the expected metabolite profiles of each phenotype.
- Lipids, in particular glycerophospholipids, and amino acids are the most frequently observed rhythmic metabolite classes. Lipid rhythms have shown the most variation between individuals with differences in phase (timing).
- Lipid rhythms may feature class-dependent temporal separation based upon carbon chain length and degree of saturation.
- A subset of metabolites are repeatedly reported as undergoing significant time of day variation across studies. A total of 35 putatively identified metabolites having been observed in at least five studies (Table 7) out of a total of 400 putatively identified across all studies.
3.2. Potential Consequences Resulting from Time of Day Variation
3.3. Proposed Updates to Minimum Reporting Guidelines in Human Metabolomics Studies
3.4. Investigating Metabolite Rhythms—The Next Steps
3.5. Summary
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dunn, W.B.; Broadhurst, D.I.; Atherton, H.J.; Goodacre, R.; Griffin, J. Systems level studies of mammalian metabolomes: The roles of mass spectrometry and nuclear magnetic resonance spectroscopy. Chem. Soc. Rev. 2011, 40, 387–426. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Sun, H.; Yan, G.; Wang, P.; Wang, X. Review article Metabolomics for Biomarker Discovery: Moving to the Clinic. BioMed. Res. Int. 2015, 2015, 354671. [Google Scholar] [CrossRef]
- López-López, Á.; López-Gonzálvez, Á.; Barker-Tejeda, T.C.; Barbas, C. A review of validated biomarkers obtained through metabolomics. Expert Rev. Mol. Diagn. 2018, 18, 557–575. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Sánchez, B.; Priego-Capote, F.; de Castro, M.L. Metabolomics analysis I. Selection of biological samples and practical aspects preceding sample preparation. TrAC Trends Anal. Chem. 2010, 29, 111–119. [Google Scholar] [CrossRef]
- Zang, X.; Monge, M.E.; Fernández, F.M. Mass spectrometry-based non-targeted metabolic profiling for disease detection: Recent developments. TrAC Trends Anal. Chem. 2019, 118, 158–169. [Google Scholar] [CrossRef]
- León, Z.; García-Cañaveras, J.C.; Donato, M.T.; Lahoz, A. Mammalian cell metabolomics: Experimental design and sample preparation. Electrophoresis 2013, 34, 2762–2775. [Google Scholar] [CrossRef]
- Khamis, M.M.; Adamko, D.J.; El-Aneed, A. Mass spectrometric based approaches in urine metabolomics and biomarker discovery. Mass Spectrom. Rev. 2017, 36, 115–134. [Google Scholar] [CrossRef]
- Hernandes, V.V.; Barbas, C.; Dudzik, D. A review of blood sample handling and pre-processing for metabolomics studies. Electrophoresis 2017, 38, 2232–2241. [Google Scholar] [CrossRef]
- Zelena, E.; Dunn, W.; Broadhurst, D.; Francis-McIntyre, S.; Carroll, K.M.; Begley, P.; O’Hagan, S.; Knowles, J.D.; Halsall, A.; Wilson, I.D.; et al. Development of a Robust and Repeatable UPLC−MS Method for the Long-Term Metabolomic Study of Human Serum. Anal. Chem. 2009, 81, 1357–1364. [Google Scholar] [CrossRef]
- Considine, E.C. The Search for Clinically Useful Biomarkers of Complex Disease: A Data Analysis Perspective. Metabolites 2019, 9, 126. [Google Scholar] [CrossRef] [Green Version]
- Di Guida, R.; Engel, J.; Allwood, J.W.; Weber, R.J.M.; Jones, M.R.; Sommer, U.; Viant, M.R.; Dunn, W.B. Non-targeted UHPLC-MS metabolomic data processing methods: A comparative investigation of normalisation, missing value imputation, transformation and scaling. Metabolomics 2016, 12, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Vuckovic, D. Current trends and challenges in sample preparation for global metabolomics using liquid chromatography–Mass spectrometry. Anal. Bioanal. Chem. 2012, 403, 1523–1548. [Google Scholar] [CrossRef]
- Ioannidis, J.P.A. Why Most Discovered True Associations Are Inflated. Epidemiology 2008, 19, 640–648. [Google Scholar] [CrossRef] [Green Version]
- Ioannidis, J.P.A. Effect of Formal Statistical Significance on the Credibility of Observational Associations. Am. J. Epidemiol. 2008, 168, 374–383. [Google Scholar] [CrossRef]
- Rosenbaum, P.R. Replicating Effects and Biases. Am. Stat. 2001, 55, 223–227. [Google Scholar] [CrossRef]
- Eshima, J.; Davis, T.J.; Bean, H.D.; Fricks, J.; Smith, B.S. A Metabolomic Approach for Predicting Diurnal Changes in Cortisol. Metabolites 2020, 10, 194. [Google Scholar] [CrossRef]
- Griffiths, W.J.; Koal, T.; Wang, Y.; Kohl, M.; Enot, D.P.; Deigner, H.-P. Targeted Metabolomics for Biomarker Discovery. Angew. Chem. Int. Ed. 2010, 49, 5426–5445. [Google Scholar] [CrossRef]
- Mastrangelo, A.; Ferrarini, A.; Rey-Stolle, F.; García, A.; Barbas, C. From sample treatment to biomarker discovery: A tutorial for untargeted metabolomics based on GC-(EI)-Q-MS. Anal. Chim. Acta 2015, 900, 21–35. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolomics by Gas Chromatography–Mass Spectrometry: Combined Targeted and Untargeted Profiling. Curr. Protoc. Mol. Biol. 2016, 114, 30.4.1–30.4.32. [Google Scholar] [CrossRef]
- Gertsman, I.; Barshop, B.A. Promises and pitfalls of untargeted metabolomics. J. Inherit. Metab. Dis. 2018, 41, 355–366. [Google Scholar] [CrossRef]
- Pezzatti, J.; Boccard, J.; Codesido, S.; Gagnebin, Y.; Joshi, A.; Picard, D.; González-Ruiz, V.; Rudaz, S. Implementation of liquid chromatography–high resolution mass spectrometry methods for untargeted metabolomic analyses of biological samples: A tutorial. Anal. Chim. Acta 2020, 1105, 28–44. [Google Scholar] [CrossRef] [PubMed]
- Manzi, M.; Riquelme, G.; Zabalegui, N.; Monge, M.E. Improving diagnosis of genitourinary cancers: Biomarker discovery strategies through mass spectrometry-based metabolomics. J. Pharm. Biomed. Anal. 2020, 178, 112905. [Google Scholar] [CrossRef] [PubMed]
- Dettmer, K.; Aronov, P.A.; Hammock, B.D. Mass spectrometry-based metabolomics. Mass Spectrom. Rev. 2006, 26, 51–78. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, M.; Carvalho, M.; Bastos, M.D.L.; De Pinho, P.G. Metabolomics Analysis for Biomarker Discovery: Advances and Challenges. Curr. Med. Chem. 2013, 20, 257–271. [Google Scholar] [CrossRef] [PubMed]
- Dudzik, D.; Barbas-Bernardos, C.; García, A.; Barbas, C. Quality assurance procedures for mass spectrometry untargeted metabolomics. a review. J. Pharm. Biomed. Anal. 2018, 147, 149–173. [Google Scholar] [CrossRef]
- Takahashi, J.S. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 2017, 18, 164–179. [Google Scholar] [CrossRef] [Green Version]
- Bunning, E. The Physiological Clock: Circadian Rhythms and Biological Chronometry, 3rd ed.; English University Press: London, UK, 1973. [Google Scholar]
- Patton, A.P.; Hastings, M.H. The suprachiasmatic nucleus. Curr. Biol. 2018, 28, R816–R822. [Google Scholar] [CrossRef] [Green Version]
- Koronowski, K.B.; Kinouchi, K.; Welz, P.-S.; Smith, J.; Zinna, V.M.; Shi, J.; Samad, M.; Chen, S.; Magnan, C.N.; Kinchen, J.M.; et al. Defining the Independence of the Liver Circadian Clock. Cell 2019, 177, 1448–1462.e141. [Google Scholar] [CrossRef]
- Huang, W.; Ramsey, K.M.; Marcheva, B.; Bass, J. Circadian rhythms, sleep, and metabolism. J. Clin. Investig. 2011, 121, 2133–2141. [Google Scholar] [CrossRef]
- Dyar, K.A.; Lutter, D.; Artati, A.; Ceglia, N.J.; Liu, Y.; Armenta, D.; Jastroch, M.; Schneider, S.; de Mateo, S.; Cervantes, M.; et al. Atlas of Circadian Metabolism Reveals System-wide Coordination and Communication between Clocks. Cell 2018, 174, 1571–1585. [Google Scholar] [CrossRef] [Green Version]
- Minami, Y.; Kasukawa, T.; Kakazu, Y.; Iigo, M.; Sugimoto, M.; Ikeda, S.; Yasui, A.; van der Horst, G.T.J.; Soga, T.; Ueda, H.R. Measurement of internal body time by blood metabolomics. Proc. Natl. Acad. Sci. USA 2009, 106, 9890–9895. [Google Scholar] [CrossRef] [Green Version]
- Lundell, L.S.; Parr, E.B.; Devlin, B.L.; Ingerslev, L.R.; Altıntaş, A.; Sato, S.; Sassone-Corsi, P.; Barrès, R.; Zierath, J.R.; Hawley, J.A. Time-restricted feeding alters lipid and amino acid metabolite rhythmicity without perturbing clock gene expression. Nat. Commun. 2020, 11, 1–11. [Google Scholar] [CrossRef]
- Zhang, R.; Lahens, N.; Balance, H.; Hughes, M.; Hogenesch, J. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc. Natl. Acad. Sci. USA 2014, 111, 16219–16224. [Google Scholar] [CrossRef] [Green Version]
- Masri, S.; Sassone-Corsi, P. The emerging link between cancer, metabolism, and circadian rhythms. Nat. Med. 2018, 24, 1795–1803. [Google Scholar] [CrossRef]
- Münch, M.; Bromundt, V. Light and chronobiology: Implications for health and disease. Dialogues Clin. Neurosci. 2012, 14, 448–453. [Google Scholar] [CrossRef]
- Logan, R.W.; McClung, C.A. Rhythms of life: Circadian disruption and brain disorders across the lifespan. Nat. Rev. Neurosci. 2018, 20, 49–65. [Google Scholar] [CrossRef]
- McHill, A.W.; Wright, K.P., Jr. Role of sleep and circadian disruption on energy expenditure and in metabolic predisposition to human obesity and metabolic disease. Obes. Rev. 2017, 18, 15–24. [Google Scholar] [CrossRef]
- Münch, M.; Wirz-Justice, A.; Brown, S.A.; Kantermann, T.; Martiny, K.; Stefani, O.; Vetter, C.; Wright, J.K.P.; Wulff, K.; Skene, D.J. The Role of Daylight for Humans: Gaps in Current Knowledge. Clocks Sleep 2020, 2, 8. [Google Scholar] [CrossRef] [Green Version]
- Zimmet, P.; Alberti, K.G.M.M.; Stern, N.; Bilu, C.; El-Osta, A.; Einat, H.; Kronfeld-Schor, N. The Circadian Syndrome: Is the Metabolic Syndrome and much more! J. Intern. Med. 2019, 286, 181–191. [Google Scholar] [CrossRef]
- Duffy, J.F.; Dijk, D.-J. Getting Through to Circadian Oscillators: Why Use Constant Routines? J. Biol. Rhythm. 2002, 17, 4–13. [Google Scholar] [CrossRef]
- Aschoff, J. Freerunning and Entrained Circadian Rhythms; Biological Rhythms; Springer: Boston, MA, USA, 1981; pp. 81–93. [Google Scholar] [CrossRef]
- Roenneberg, T.; Daan, S.; Merrow, M. The Art of Entrainment. J. Biol. Rhythm. 2003, 18, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Merrow, M.; Spoelstra, K.; Roenneberg, T. The circadian cycle: Daily rhythms from behaviour to genes. EMBO Rep. 2005, 6, 930–935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duarte, L.; Menna-Barreto, L.; Miguel, M.; Louzada, F.; Araújo, J.; Alam, M.; Areas, R.; Pedrazzoli, M. Chronotype ontogeny related to gender. Braz. J. Med Biol. Res. 2014, 47, 316–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischer, D.; Lombardi, D.A.; Marucci-Wellman, H.; Roenneberg, T. Chronotypes in the US—Influence of age and sex. PLoS ONE 2017, 12, e0178782. [Google Scholar] [CrossRef] [Green Version]
- Nowak, R.; McMillen, I.C.; Redman, J.; Short, R.V. The Correlation between Serum and Salivary Melatonin Concentrations and Urinary 6-Hydroxymelatonin Sulphate Excretion Rates: Two Non-Invasive Techniques for Monitoring Human Circadian Rhythmicity. Clin. Endocrinol. 1987, 27, 445–452. [Google Scholar] [CrossRef]
- Arendt, J. Melatonin and Human Rhythms. Chrono. Int. 2006, 23, 21–37. [Google Scholar] [CrossRef]
- Klerman, E.B.; Gershengorn, H.B.; Duffy, J.F.; Kronauer, R.E. Comparisons of the Variability of Three Markers of the Human Circadian Pacemaker. J. Biol. Rhythm. 2002, 17, 181–193. [Google Scholar] [CrossRef]
- Honma, A.; Revell, V.L.; Gunn, P.J.; Davies, S.K.; Middleton, B.; Raynaud, F.I.; Skene, D.J. Effect of acute total sleep deprivation on plasma melatonin, cortisol and metabolite rhythms in females. Eur. J. Neurosci. 2019, 51, 366–378. [Google Scholar] [CrossRef] [Green Version]
- Davies, S.K.; Ang, J.E.; Revell, V.L.; Holmes, B.; Mann, A.; Robertson, F.P.; Cui, N.; Middleton, B.; Ackermann, K.; Kayser, M.; et al. Effect of sleep deprivation on the human metabolome. Proc. Natl. Acad. Sci. USA 2014, 111, 10761–10766. [Google Scholar] [CrossRef] [Green Version]
- Czeisler, C.A.; Klerman, E.B. Circadian and sleep-dependent regulation of hormone release in humans. Recent Prog. Horm. Res. 1999, 54, 97–130. [Google Scholar]
- Chua, E.C.-P.; Shui, G.; Lee, I.T.-G.; Lau, P.; Tan, L.-C.; Yeo, S.-C.; Lam, B.D.; Bulchand, S.; Summers, S.A.; Puvanendran, K.; et al. Extensive diversity in circadian regulation of plasma lipids and evidence for different circadian metabolic phenotypes in humans. Proc. Natl. Acad. Sci. USA 2013, 110, 14468–14473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, Y.; Kim, S.B.; Wang, B.; Blanco, R.A.; Le, N.-A.; Wu, S.; Accardi, C.J.; Alexander, R.W.; Ziegler, T.R.; Jones, D.P. Individual variation in macronutrient regulation measured by proton magnetic resonance spectroscopy of human plasma. Am. J. Physiol. Integr. Comp. Physiol. 2009, 297, R202–R209. [Google Scholar] [CrossRef] [Green Version]
- Ang, J.E.; Revell, V.; Anuska, M.; Mäntele, S.; Otway, D.T.; Johnston, J.D.; Thumser, A.E.; Skene, D.J.; Raynaud, F. Identification of Human Plasma Metabolites Exhibiting Time-of-Day Variation Using an Untargeted Liquid Chromatography–Mass Spectrometry Metabolomic Approach. Chrono. Int. 2012, 29, 868–881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dallmann, R.; Viola, A.U.; Tarokh, L.; Cajochen, C.; Brown, S.A. The human circadian metabolome. Proc. Natl. Acad. Sci. USA 2012, 109, 2625–2629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasukawa, T.; Sugimoto, M.; Hida, A.; Minami, Y.; Mori, M.; Honma, S.; Honma, K.-I.; Mishima, K.; Soga, T.; Ueda, H.R. Human blood metabolite timetable indicates internal body time. Proc. Natl. Acad. Sci. USA 2012, 109, 15036–15041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, K.; Mall, C.; Taylor, S.L.; Hitchcock, S.; Zhang, C.; Wettersten, H.I.; Daniel Jones, A.; Chapman, A.; Weiss, R.H. Mealtime, temporal, and daily variability of the human urinary and plasma metabolomes in a tightly controlled environment. PLoS ONE 2014, 9, e86223. [Google Scholar] [CrossRef]
- Chua, E.C.-P.; Shui, G.; Cazenave-Gassiot, A.; Wenk, M.R.; Gooley, J.J. Changes in Plasma Lipids during Exposure to Total Sleep Deprivation. Sleep 2015, 38, 1683–1691. [Google Scholar] [CrossRef] [Green Version]
- Skarke, C.; Lahens, N.F.; Rhoades, S.D.; Campbell, A.; Bittinger, K.; Bailey, A.; Hoffmann, C.; Olson, R.S.; Chen, L.; Yang, G.; et al. A Pilot Characterization of the Human Chronobiome. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef]
- Isherwood, C.M.; Van Der Veen, D.R.; Johnston, J.D.; Skene, D.J. Twenty-four-hour rhythmicity of circulating metabolites: Effect of body mass and type 2 diabetes. FASEB J. 2017, 31, 5557–5567. [Google Scholar] [CrossRef] [Green Version]
- Gehrman, P.; Sengupta, A.; Harders, E.; Ubeydullah, E.; Pack, A.I.; Weljie, A. Altered diurnal states in insomnia reflect peripheral hyperarousal and metabolic desynchrony: A preliminary study. Sleep 2018, 41, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Sato, S.; Parr, E.B.; Devlin, B.L.; Hawley, J.A.; Sassone-Corsi, P. Human metabolomics reveal daily variations under nutritional challenges specific to serum and skeletal muscle. Mol. Metab. 2018, 16, 1–11. [Google Scholar] [CrossRef]
- Skene, D.J.; Skornyakov, E.; Chowdhury, N.R.; Gajula, R.P.; Middleton, B.; Satterfield, B.C.; Porter, K.I.; Van Dongen, H.P.A.; Gaddameedhi, S. Separation of circadian- and behavior-driven metabolite rhythms in humans provides a window on peripheral oscillators and metabolism. Proc. Natl. Acad. Sci. USA 2018, 115, 7825–7830. [Google Scholar] [CrossRef] [Green Version]
- Grant, L.K.; Ftouni, S.; Nijagal, B.; De Souza, D.P.; Tull, D.; McConville, M.J.; Rajaratnam, S.M.W.; Lockley, S.W.; Anderson, C. Circadian and wake-dependent changes in human plasma polar metabolites during prolonged wakefulness: A preliminary analysis. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Gu, F.; Klerman, E.B.; Kim, S.; Moore, S.; Yu, K.; Albert, P.S.; Caporaso, N.E. Diurnal variation of metabolites in three individual participants. Chrono. Int. 2018, 36, 332–342. [Google Scholar] [CrossRef]
- Kervezee, L.; Cermakian, N.; Boivin, D.B. Individual metabolomic signatures of circadian misalignment during simulated night shifts in humans. PLoS Biol. 2019, 17, e3000303. [Google Scholar] [CrossRef] [Green Version]
- Lusczek, E.R.; Parsons, L.S.; Elder, J.; Harvey, S.B.; Skube, M.; Muratore, S.; Beilman, G.; Cornelissen-Guillaume, G. Metabolomics Pilot Study Identifies Desynchronization of 24-H Rhythms and Distinct Intra-patient Variability Patterns in Critical Illness: A Preliminary Report. Front. Neurol. 2020, 11, 533915. [Google Scholar] [CrossRef]
- Talasniemi, J.P.; Pennanen, S.; Savolainen, H.; Niskanen, L.; Liesivuori, J. Analytical investigation: Assay of d-lactate in diabetic plasma and urine. Clin. Biochem. 2008, 41, 1099–1103. [Google Scholar] [CrossRef]
- Acosta, S.; Nilsson, T. Current status on plasma biomarkers for acute mesenteric ischemia. J. Thromb. Thrombolysis 2011, 33, 355–361. [Google Scholar] [CrossRef]
- Abbassi-Ghadi, N.; Kumar, S.; Huang, J.; Goldin, R.; Takats, Z.; Hanna, G. Metabolomic profiling of oesophago-gastric cancer: A systematic review. Eur. J. Cancer 2013, 49, 3625–3637. [Google Scholar] [CrossRef]
- Ueland, P.M.; Ulvik, A.; Rios-Avila, L.; Midttun, Ø.; Gregory, J.F. Direct and Functional Biomarkers of Vitamin B6 Status. Annu. Rev. Nutr. 2015, 35, 33–70. [Google Scholar] [CrossRef]
- Macrez, R.; Stys, P.K.; Vivien, D.; Lipton, A.S.; Docagne, F. Mechanisms of glutamate toxicity in multiple sclerosis: Biomarker and therapeutic opportunities. Lancet Neurol. 2016, 15, 1089–1102. [Google Scholar] [CrossRef]
- Boenzi, S.; Diodato, D. Biomarkers for mitochondrial energy metabolism diseases. Essays Biochem. 2018, 62, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Peoc’h, K.; Corcos, O. Biomarkers for acute mesenteric ischemia diagnosis: State of the art and perspectives. Ann. Biol. Clin. 2019, 77, 415–421. [Google Scholar] [CrossRef]
- Takahashi, J.S.; Hong, H.-K.; Ko, C.H.; McDearmon, E.L. The genetics of mammalian circadian order and disorder: Implications for physiology and disease. Nat. Rev. Genet. 2008, 9, 764–775. [Google Scholar] [CrossRef]
- Watson, N.F.; Buchwald, D.; Harden, K.P. A Twin Study of Genetic Influences on Diurnal Preference and Risk for Alcohol Use Outcomes. J. Clin. Sleep Med. 2013, 9, 1333–1339. [Google Scholar] [CrossRef]
- Koskenvuo, M.; Hublin, C.; Partinen, M.; Heikkilä, K.; Kaprio, J. Heritability of diurnal type: A nationwide study of 8753 adult twin pairs. J. Sleep Res. 2007, 16, 156–162. [Google Scholar] [CrossRef]
- Barclay, N.L.; Eley, T.C.; Buysse, D.J.; Archer, S.N.; Gregory, A.M. Diurnal preference and sleep quality: Same genes? A study of young adult twins. Chronobiol. Int. 2010, 27, 278–296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Von Schantz, M.; Taporoski, T.P.; Horimoto, A.R.V.R.; Duarte, N.E.; Vallada, H.; Krieger, J.E.; Pedrazzoli, M.; Negrão, A.B.; Pereira, A.C. Distribution and heritability of diurnal preference (chronotype) in a rural Brazilian family-based cohort, the Baependi study. Sci. Rep. 2015, 5, 9214. [Google Scholar] [CrossRef] [Green Version]
- Kelly, G. Body temperature variability (Part 1): A review of the history of body temperature and its variability due to site selection, biological rhythms, fitness, and aging. Altern. Med. Rev. 2006, 11, 278–293. [Google Scholar] [PubMed]
- Kelly, G.S. Body temperature variability (Part 2): Masking influences of body temperature variability and a review of body temperature variability in disease. Altern. Med. Rev. 2007, 12, 49–62. [Google Scholar]
- Jerjes, W.K.; Cleare, A.J.; Peters, T.J.; Taylor, N.F. Circadian rhythm of urinary steroid metabolites. Ann. Clin. Biochem. Int. J. Lab. Med. 2006, 43, 287–294. [Google Scholar] [CrossRef]
- Walsh, M.C.; Brennan, L.; Malthouse, J.P.G.; Roche, H.M.; Gibney, M.J. Effect of acute dietary standardization on the urinary, plasma, and salivary metabolomic profiles of healthy humans. Am. J. Clin. Nutr. 2006, 84, 531–539. [Google Scholar] [CrossRef] [Green Version]
- Slupsky, C.M.; Rankin, K.N.; Wagner, J.; Fu, H.; Chang, D.; Weljie, A.M.; Saude, E.J.; Lix, B.; Adamko, D.J.; Shah, S.; et al. Investigations of the effects of gender, diurnal variation, and age in human urinary metabolomic profiles. Anal. Chem. 2007, 79, 6995–7004. [Google Scholar] [CrossRef]
- Giskeødegård, G.F.; Davies, S.K.; Revell, V.L.; Keun, H.; Skene, D.J. Diurnal rhythms in the human urine metabolome during sleep and total sleep deprivation. Sci. Rep. 2015, 5, 14843. [Google Scholar] [CrossRef] [Green Version]
- Papantoniou, K.; Pozo, O.J.; Espinosa, A.; Marcos, J.; Castaño-Vinyals, G.; Basagaña, X.; Pagès, E.J.; Mirabent, J.; Martín, J.; Faro, P.S.; et al. Increased and Mistimed Sex Hormone Production in Night Shift Workers. Cancer Epidemiol. Biomark. Prev. 2015, 24, 854–863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Firsov, D.; Bonny, O. Circadian rhythms and the kidney. Nat. Rev. Nephrol. 2018, 14, 626–635. [Google Scholar] [CrossRef]
- Cook, T.; Ma, Y.; Gamagedara, S. Evaluation of statistical techniques to normalize mass spectrometry-based urinary metabolomics data. J. Pharm. Biomed. Anal. 2020, 177, 112854. [Google Scholar] [CrossRef]
- Dame, Z.; Aziat, F.; Mandal, R.; Krishnamurthy, R.; Bouatra, S.; Borzouie, S.; Guo, A.C.; Sajed, T.; Deng, L.; Lin, H.; et al. The human saliva metabolome. Metabolomics 2015, 11, 1864–1883. [Google Scholar] [CrossRef]
- Sinues, P.M.-L.; Kohler, M.; Zenobi, R. Monitoring Diurnal Changes in Exhaled Human Breath. Anal. Chem. 2012, 85, 369–373. [Google Scholar] [CrossRef]
- Sinues, P.M.-L.; Tarokh, L.; Li, X.; Kohler, M.; Brown, S.A.; Zenobi, R.; Dallmann, R. Circadian Variation of the Human Metabolome Captured by Real-Time Breath Analysis. PLoS ONE 2014, 9, e114422. [Google Scholar] [CrossRef]
- Wilkinson, M.; Maidstone, R.; Loudon, A.; Blaikley, J.; White, I.R.; Singh, D.; Ray, D.W.; Goodacre, R.; Fowler, S.J.; Durrington, H.J. Circadian rhythm of exhaled biomarkers in health and asthma. Eur. Respir. J. 2019, 54, 1901068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loizides-Mangold, U.; Perrin, L.; Vandereycken, B.; Betts, J.A.; Walhin, J.-P.; Templeman, I.; Chanon, S.; Weger, B.D.; Durand, C.; Robert, M.; et al. Lipidomics reveals diurnal lipid oscillations in human skeletal muscle persisting in cellular myotubes cultured in vitro. Proc. Natl. Acad. Sci. USA 2017, 114, E8565–E8574. [Google Scholar] [CrossRef] [Green Version]
- Held, N.M.; Wefers, J.; van Weeghel, M.; Daemen, S.; Hansen, J.; Vaz, F.M.; van Moorsel, D.; Hesselink, M.K.; Houtkooper, R.H.; Schrauwen, P. Skeletal muscle in healthy humans exhibits a day-night rhythm in lipid metabolism. Mol. Metab. 2020, 37, 100989. [Google Scholar] [CrossRef] [PubMed]
- Danlos, F.-X.; Grajeda-Iglesias, C.; Durand, S.; Sauvat, A.; Roumier, M.; Cantin, D.; Colomba, E.; Rohmer, J.; Pommeret, F.; Baciarello, G.; et al. Metabolomic analyses of COVID-19 patients unravel stage-dependent and prognostic biomarkers. Cell Death Dis. 2021, 12. [Google Scholar] [CrossRef]
- Leitner, M.; Fragner, L.; Danner, S.; Holeschofsky, N.; Leitner, K.; Tischler, S.; Doerfler, H.; Bachmann, G.; Sun, X.; Jaeger, W.; et al. Combined Metabolomic Analysis of Plasma and Urine Reveals AHBA, Tryptophan and Serotonin Metabolism as Potential Risk Factors in Gestational Diabetes Mellitus (GDM). Front. Mol. Biosci. 2017, 4, 84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morton, A.J.; Middleton, B.; Rudiger, S.; Bawden, C.S.; Kuchel, T.R.; Skene, D.J. Increased plasma melatonin in presymptomatic Huntington disease sheep (ovis aries): Compensatory neuroprotection in a neurodegenerative disease? J. Pineal Res. 2019, 68. [Google Scholar] [CrossRef]
- Roenneberg, T.; Winnebeck, E.C.; Klerman, E.B. Daylight Saving Time and Artificial Time Zones—A Battle Between Biological and Social Times. Front. Physiol. 2019, 10, 944. [Google Scholar] [CrossRef] [Green Version]
- Dallmann, R.; Brown, S.A.; Gachon, F. Chronopharmacology: New Insights and Therapeutic Implications. Annu. Rev. Pharmacol. Toxicol. 2014, 54, 339–361. [Google Scholar] [CrossRef] [Green Version]
- Yin, P.; Lehmann, R.; Xu, G. Effects of pre-analytical processes on blood samples used in metabolomics studies. Anal. Bioanal. Chem. 2015, 407, 4879–4892. [Google Scholar] [CrossRef] [Green Version]
- Fiehn, O.; Robertson, D.; Griffin, J.; Van Der Werf, M.; Nikolau, B.; Morrison, N.; Sumner, L.W.; Goodacre, R.; Hardy, N.W.; Taylor, C.; et al. The metabolomics standards initiative (MSI). Metabolomics 2007, 3, 175–178. [Google Scholar] [CrossRef]
- Griffin, J.L.; Nicholls, A.W.; Daykin, C.A.; Heald, S.; Keun, H.C.; Schuppe-Koistinen, I.; Griffiths, J.R.; Cheng, L.L.; Rocca-Serra, P.; Rubtsov, D.V.; et al. Standard reporting requirements for biological samples in metabolomics experiments: Mammalian/in vivo experiments. Metabolomics 2007, 3, 179–188. [Google Scholar] [CrossRef] [Green Version]
- Van Der Werf, M.J.; Takors, R.; Smedsgaard, J.; Nielsen, J.; Ferenci, T.; Portais, J.C.; Wittmann, C.; Hooks, M.; Tomassini, A.; Oldiges, M.; et al. Standard reporting requirements for biological samples in metabolomics experiments: Microbial and in vitro biology experiments. Metabolomics 2007, 3, 189–194. [Google Scholar] [CrossRef] [Green Version]
- Goodacre, R.; Broadhurst, D.; Smilde, A.K.; Kristal, B.S.; Baker, J.D.; Beger, R.; Bessant, C.; Connor, S.; Capuani, G.; Craig, A.; et al. Proposed minimum reporting standards for data analysis in metabolomics. Metabolomics 2007, 3, 231–241. [Google Scholar] [CrossRef]
- Balsalobre, A.; Damiola, F.; Schibler, U. A Serum Shock Induces Circadian Gene Expression in Mammalian Tissue Culture Cells. Cell 1998, 93, 929–937. [Google Scholar] [CrossRef] [Green Version]
- Ndikung, J.; Storm, D.; Violet, N.; Kramer, A.; Schönfelder, G.; Ertych, N.; Oelgeschläger, M. Restoring circadian synchrony in vitro facilitates physiological responses to environmental chemicals. Environ. Int. 2020, 134, 105265. [Google Scholar] [CrossRef]
- Ouyang, Y.; Andersson, C.R.; Kondo, T.; Golden, S.S.; Johnson, C.H. Resonating circadian clocks enhance fitness in cyanobacteria. Proc. Natl. Acad. Sci. USA 1998, 95, 8660–8664. [Google Scholar] [CrossRef] [Green Version]
- Rust, M.J.; Golden, S.S.; O’Shea, E.K. Light-Driven Changes in Energy Metabolism Directly Entrain the Cyanobacterial Circadian Oscillator. Science 2011, 331, 220–223. [Google Scholar] [CrossRef] [Green Version]
- De Almeida, E.A.; Di Mascio, P.; Harumi, T.; Spence, D.W.; Moscovitch, A.; Hardeland, R.; Cardinali, D.P.; Brown, G.M.; Pandi-Perumal, S.R. Measurement of melatonin in body fluids: Standards, protocols and procedures. Child’s Nerv. Syst. 2011, 27, 879–891. [Google Scholar] [CrossRef] [Green Version]
- Skene, D.J.; Middleton, B.; Fraser, C.K.; Pennings, J.L.A.; Kuchel, T.R.; Rudiger, S.R.; Bawden, C.S.; Morton, A.J. Metabolic profiling of presymptomatic Huntington’s disease sheep reveals novel biomarkers. Sci. Rep. 2017, 7, 43030. [Google Scholar] [CrossRef] [Green Version]
- Cornelissen, G. Cosinor-based rhythmometry. Theor. Biol. Med Model. 2014, 11, 16. [Google Scholar] [CrossRef] [Green Version]
- Wu, G.; Anafi, R.C.; Hughes, M.E.; Kornacker, K.; HogenEsch, J.B. MetaCycle: An integrated R package to evaluate periodicity in large scale data. Bioinformatics 2016, 32, 3351–3353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhake, R.; Russell, G.M.; Kershaw, Y.; Stevens, K.; Zaccardi, F.; Warburton, V.E.C.; Linthorst, A.C.E.; Lightman, S.L. Continuous Free Cortisol Profiles in Healthy Men. J. Clin. Endocrinol. Metab. 2019, 105. [Google Scholar] [CrossRef]
- Santos, H.D.L.; Bennett, K.P.; Hurley, J.M. MOSAIC: A joint modeling methodology for combined circadian and non-circadian analysis of multi-omics data. Bioinformatics 2021, 37, 767–774. [Google Scholar] [CrossRef] [PubMed]
- Thaben, P.F.; Westermark, P.O. Detecting Rhythms in Time Series with RAIN. J. Biol. Rhythm. 2014, 29, 391–400. [Google Scholar] [CrossRef] [Green Version]
- Santos, H.D.L.; Collins, E.J.; Mann, C.; Sagan, A.W.; Jankowski, M.S.; Bennett, K.P.; Hurley, J.M. ECHO: An application for detection and analysis of oscillators identifies metabolic regulation on genome-wide circadian output. Bioinformatics 2020, 36, 773–781. [Google Scholar] [CrossRef]
- Thaben, P.F.; Westermark, P.O. Differential rhythmicity: Detecting altered rhythmicity in biological data. Bioinformatics 2016, 32, 2800–2808. [Google Scholar] [CrossRef]
- Doherty, C.J.; Kay, S.A. Circadian Control of Global Gene Expression Patterns. Annu. Rev. Genet. 2010, 44, 419–444. [Google Scholar] [CrossRef] [Green Version]
- Pelikan, A.; Herzel, H.; Kramer, A. Studies overestimate the extent of circadian rhythm repogramming in response to dietary and genetic changes. BioRxiv 2020. [Google Scholar] [CrossRef]
- Mure, L.S.; Hiep, D.L.; Benegiamo, G.; Chang, M.W.; Rios, L.; Jilani, N.; Ngotho, M.; Kariuki, T.; Dkhissi-Benyahya, O.; Copper, H.; et al. Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science 2018, 359. [Google Scholar] [CrossRef] [Green Version]
| Search Terms | Database/Search Engine | ‘Hits’ | Relevant Papers (Based on Abstract) | Met Inclusion Criteria * | |
|---|---|---|---|---|---|
| Circadian Studies | Diurnal Studies | ||||
| “Human(s)” “Circadian Rhythm OR Circadian Clocks” “Metabolomics OR Metabolome” ** | PubMed (NCBI) | 70 | 133 | 6 | 19 |
| Web of Science | 52 | ||||
| “Metabolomic” “Circadian” “Rhythm” “Human” “Chronobiology” | Google Scholar | 212 | |||
| N/A | Further manual searches | 13 | |||
| “Human(s)” “Diurnal Variation OR Diurnal”, “Metabolome OR Metabolomics” *** | PubMed (NCBI) | 19 | 123 (Majority duplicates of prior search) | 3 | 16 |
| Web of Science | 28 | ||||
| “Metabolomic” “Metabolome” “Diurnal” “Rhythm” “Human” “Chronobiology” | Google Scholar | 92 | |||
| N/A | Further manual searches | 0 | |||
| Author(s) | Assay/Platform | Time Course Details | Study Setting/Conditions | Cohort Details | Rhythmic/Gradient Metabolites/ Features Observed | Rhythmic/Gradient Classes Primarily Observed |
|---|---|---|---|---|---|---|
| Park et al., (2009) [54] | Untargeted 1H NMR | Diurnal variation 24 h, 1 h intervals between samples | ‘Inpatient’ Standardised meals. Consistent light/dark cycle | N = 10, 5 males Age 22–83 BMI 18.5–32.6 | 34 | Amino acids Lipids (unidentified) |
| Ang et al., (2012) [55] | Untargeted UPLC/Q-TOF MS (Reversed Phase) | Diurnal variation (25 h, 3 h intervals between samples) | ‘Inpatient’ 17:8 wake/sleep, light/dark cycle. Hourly isocaloric meals Semi-recumbent position | N = 8 All male Age 53.6 ± 6.0 BMI 23.2 ± 1.4 | 203 features (19%) 34 metabolites | Amino acids Acylcarnitines LysoPEs LysoPCs |
| Dallmann et al., (2012) [56] | Untargeted GC-MS LC-MS (Reversed Phase) | Circadian variation (constant routine 40 h, 4 h intervals between samples) | ‘Inpatient’ Standard constant routine parameters (see [41]) | N = 10 (split into 2 equal groups, within which samples were pooled for each 4 h interval) All male Age 57.8 ± 1.0 & 61.0 ± 0.6 BMI 26.6 ± 0.6 & 25.1 ± 0.5 | 41 (15%) | Amino acids Glycerophospholipids Acylcarnitines Steroid hormones |
| Kasukawa et al., (2012) [57] | Untargeted LC-TOF MS (Reversed Phase) | Circadian variation (forced desynchrony 28 h, bookended by constant routine protocols (38 h each, 2 h intervals between samples) | ‘Inpatient’ Standard constant routine parameters (with the exception of meals every 2 h (see [41]) Controlled light/dark cycles, temperature during forced desynchrony | N = 6 All male Aged 20–23 | 312 features (7%) | Amino acids Steroid hormones |
| Chua et al., (2013) [53] | Targeted Lipidomics LC-MS/MS (Reversed Phase) | Circadian variation (constant routine 37 h, 4 h intervals between samples at 5 h onwards of constant routine) | ‘Inpatient’ Standard constant routine parameters (see [41]) | N = 20 All male Age 24.4 ± 1.8 3 ‘Overweight’ 17 ‘Healthy | 35 (13.3%) | Glycerolipids Glycerophospholipids |
| Davies et al., (2014) [51] | Untargeted UPLC/Q-TOF MS/MS and targeted FIA-MS UPLC-MS/MS (Reversed Phase) | Diurnal variation (24 h). 24 h wake/sleep cycle vs. 24 h prolonged wakefulness, 2 h intervals between samples 48 h | ‘Inpatient’ Standardised meals and mealtimes. Controlled light/dark cycle and activity/posture | N = 12 All male Age 23 ± 5, BMI 24.5 ± 2.3 | 109 (63.7%) sleep/wake 88 (51.5%) sleep deprivation 78 (45%) during both conditions | Amino acids Acylcarnitines LysoPCs Phosphatidylcholines Sphingolipids Fatty acids |
| Kim et al., (2014) [58] | Untargeted LC—TOF MS (Reversed Phase) | Diurnal variation Sampling 1, 3, 7, 9, 11, 14 h post-wake, first sample fasted. | ‘Inpatient’ Standardised meals and mealtimes | N = 26 14 males Age 33 ±10.9 BMI 24.3 ±3.3 | 11 (9%) | LysoPCs Phosphatidylinositol |
| Chua et al., (2015) [59] | Targeted Lipidomics LC-MS/MS (Reversed Phase) | Circadian variation (constant routine 37 h, 4 h intervals between samples at 5 h onwards of constant routine) | ‘Inpatient’ Standard constant routine parameters (see [41]} | N = 20 All male Age 23 ± 5 BMI 24.5 ± 2.3 | 4 (1.5%) decreased during sleep deprivation 21 (5.5%) increased during sleep deprivation | Sphingomyelins TAGs Phosphatidylcholines Phosphatidylinositol |
| Skarke et al., (2017) [60] | Targeted LC-MS/MS (HILIC) | Diurnal variation am vs. pm (48 h, 5 samples 12 h apart) | ‘Outpatient’ | N = 6 All male Age 32.3 ± 3.6 BMI 25.2 ± 3.4 | 9 (5.4%) | |
| Isherwood et al., (2017) [61] | Targeted FIA-MS UPLC-MS/MS (Reversed Phase) | Diurnal variation (24 h—2 h intervals between samples) | ‘Inpatient’ Controlled sleep/wake, light/dark cycle, and posture Hourly isocaloric meals | N = 23 All male BMI/Age Lean group 23.2 ± 1.4/53.6 ± 6.0 OW/OB 29.8 ± 2.3/51.0 ± 7.7 T2DM group 31 ± 1.6/57.3 ± 4.8 | 50/130 (38.5%) total 35—lean 39—OW/OB 20—T2DM | Amino acids Phosphatidylcholines LysoPCs Acylcarnitines |
| Gehrman et al., (2018) [62] | Targeted 1H NMR | Diurnal variation (48 h—2 h intervals between samples) | ‘Inpatient’ Habitual sleep/wake cycle Hourly isocaloric meals | N = 30 20 male and 10 females (split equally into 2 groups) BMI < 29 Healthy Age 35.0 ± 7.5 Insomnia Age 37 ± 7.9 | 24 (total) 11 common to both groups 6 unique to healthy 7 unique to insomnia | Amino acids |
| Sato et al., (2018) [63] | Untargeted UHPLC-MS/MS GC-MS | Diurnal variation am vs. pm | ‘Outpatient’ Standardised meals and mealtimes | N = 8 All male Age 30–45 BMI 27–32.5 | 532, 130, 349 features (50%, 12%, 33%) time of day, diet, time of day diet interaction, respectively. After HFD 13% features lost daily variation, 17% gained new daily variation After HCD 7% features lost daily variation 14% gained new daily variation | Amino acids Fatty acyls Glycerolipids Glycerophospholipids Sphingolipids Carbohydrates Xenobiotics |
| Skene et al., (2018) [64] | Targeted FIA-MS UPLC-MS/MS (Reversed Phase) | Circadian variation (constant routine 24 h, 11 samples at 1–3 h intervals) Day shift vs. night shift (simulation)) circadian vs. behavioural control | ‘Inpatient’ Standard constant routine parameters (see [41]) During baseline & shift work—controlled sleep/wake, light/dark cycle, temperature. Standardised meals and mealtimes | Night shift: N = 7 6 males Age 27.6 ± 3.2 BMI 25.6 ± 3.3 Day shift: N = 7, 4 males Age 24.0 ± 2.2 BMI 25.9 ±3.4 | 65 (49.2%) across both shift patterns, 27 (20.5%) common to both | Amino acids LysoPCs Phosphatdylcholines Acylcarnitines Glycerophospholipids Sphingolipids |
| Grant et al., (2019) [65] | Untargeted & Targeted LC-QTOF/MS (HILIC) | Circadian variation (24 h) Circadian- vs. wake-dependent changes | ‘Inpatient’ Standard constant routine parameters (see [41]) | N = 13 9 males Age 25.0 ± 4.3 BMI 22.0 ± 2.1 | Targeted: Group level 28/99 (28.3%) (rhythmic, rhythmic & linear) 4/99 (4%) linear Untargeted: Group level 361 (22%) rhythmic features 8% linear features Individual level 14% rhythmic profiles 4% linear profile | Amino acids Organic acids |
| Gu et al., (2019) [66] | Untargeted UHPLC-MS (Reversed phase) & GC-MS/MS | Diurnal variation (26–48 h) (48 h time course for N = 2, 26 h for N = 1 participants), | ‘Inpatient’ Standardised meals and mealtimes Habitual sleep time (10 h sleep) | N = 3 2 males Age 20–31 BMI 18 < 29.9 | 100/663 (15.1%) rhythmic in at least 1 individual 26/663 (3.9%) rhythmic in at least 2 individuals. | Amino acids DAGs Lysolipids Phospholipids Steroid lipids |
| Kervezee et al., (2019) [67] | Targeted DI-MS LC-MS/MS (Reversed phase) | Diurnal variation (24 h—2 h intervals between samples) Baseline vs. forced misalignment post-simulated shift work | ‘Inpatient’ Controlled sleep/wake, light/dark cycle and hourly isocaloric meals during sampling periods | N = 9 8 males Age 22.6 ± 3.4 BMI 21.3 (19.6–23) | 51 (39.2%) baseline 53 (40.8%) night shift 32 (24.6%) both, 24 phase shifted, 27 (21%) significantly changed post-night shift | Amino acids Fatty acids Organic acids Lysophospholipids PCs |
| Honma et al., (2020) [50] | Targeted FIA-MS UPLC-MS/MS (Reversed Phase) | Diurnal variation (70 h, 2 h intervals between samples) 16:8 wake/sleep cycle > 40 h prolonged wakefulness > 8 h recovery sleep | ‘Inpatient’ Standardised meals and mealtimes. Controlled light/dark cycle and activity/posture | N = 12 All female Age 25 ± 4 BMI 24.9 ± 3.6 | Total 97/130, 58 (44.6%) common for all conditions. Baseline 78 (60%) 8 unique. Sleep deprivation 76 (58.5%) 5 unique Recovery sleep 80 (61.5%) 5 unique | Glycerophospholipids Sphingolipids Amino acids Biogenic amines Acylcarnitines |
| Lusczek et al., (2020) [68] | Untargeted UHPLC/MS (Reversed Phase) | Diurnal variation (24 h—4 h intervals between samples) | ‘Inpatient’ Self-selected light/dark, feeding/fasting, sleep/wake cycle for healthy participants | Healthy cohort N = 5 2 males, Age 45–72 BMI 22.4–33.3 ICU cohort N = 5 2 males Age 43–66 BMI 31.0–57.3 | 10 (16.7%) in healthy 0 in ICU | Amino acids Acyl carnitines LysoPEs |
| Author(s) | Assay/Platform | Time Course Details | Study Setting/Conditions | Cohort Details | Rhythmic/Gradient Metabolites/Features Observed | Rhythmic/Gradient Classes Primarily Observed |
|---|---|---|---|---|---|---|
| Jerjes et al., (2006) [83] | Targeted GC-MS | Diurnal variation (24 h—3 h intervals between samples) | N = 20 10 males Age 32 ± 5.4 BMI 23.5 ± 2 | 9 | Androgens Cortisol metabolites | |
| Walsh et al., (2006) [84] | Untargeted 1H NMR | Diurnal variation am vs. pm | ‘Outpatient’ Standardised meals | N = 60 30 males Age 19–69 | 1 | |
| Slupsky et al., (2007) [85] | Targeted 1H NMR | Diurnal variation am vs. pm | ‘Outpatient’ | N = 30 23 females Age 24.7 ± 2.7 BMI 22.7 ± 0.97 | 6 | |
| Kim et al., (2014) [58] | Untargeted LC—TOF MS (Reversed Phase) | Diurnal variation Sampling 1, 3, 7, 9, 11, 14 h post-wake, first sample fasted. | ‘Inpatient’ Standardised meals and mealtimes | N = 26 14 males Age 33 ± 10.9 BMI 24.3 ± 3.3 | 135 (46%) | Glycerophospholipids LysoPCs Phosphatidylinositol |
| Giskeødegård et al., (2015) [86] | Untargeted 1H NMR | Diurnal variation (48 h) Samples at 2–4 h intervals when awake, 8 h overnight | ‘Inpatient’ Standardised meals and mealtimes. Controlled light/dark cycle and activity/posture | N = 15 All male Age 23.7 ± 5.4 | 5 (15.6%)—sleep/wake cycle 7 (22%) during 24 h wakefulness During sleep deprivation 8 increased, 8 decreased | Amino acids Fatty acids |
| Papantoniou et al., (2015) [87] | Targeted GC-MS | Diurnal variation (24 h) | ‘Outpatient’ Day vs. night shift workers | N = 117 63 males Age 22–64 BMI 22.6–30.6 | 5 (31.3%) significantly different in premenopausal day vs. night workers | Progestagens Androgens |
| Authors | Assay/Platform | Time Course Details | Study Setting/Conditions | Cohort Details | Rhythmic/Gradient Metabolites/Features Observed | Rhythmic/Gradient Classes Primarily Observed |
|---|---|---|---|---|---|---|
| Walsh et al., (2006) [84] | Untargeted 1H NMR | Diurnal variation am vs. pm | ‘Outpatient’ Standardised meals | N = 60 30 males Age 19–69 | 1 | No gradient metabolite classes identified |
| Dallmann et al., (2012) [56] | Untargeted GC-MS LC-MS (Reversed Phase) | Circadian variation (constant routine 40 h, 4 h intervals between samples) | ‘Inpatient’ Standard constant routine parameters (see [41]) | N = 10 (split into 2 equal groups within which samples were pooled for each 4 h interval) All male Age 57.8 ± 1.0 & 61.0 ± 0.6 BMI 26.6 ± 0.6 & 25.1 ± 0.5 | 29 (15%) | Amino acids |
| Dame et al., (2015) [90] | Untargeted 1H NMR | Diurnal variation sampling at prebreakfast vs. 2 h post-breakfast vs. 2 h post-lunch | N = 16 8 males & females Age (24–42) (only N = 2 took part in observation of diurnal variation) | 8 (10.5%) | Amino acids | |
| Skarke et al., (2017) [60] | Targeted LC-MS/MS (HILIC) | Diurnal variation am vs. pm (48 h, 5 samples 12 h apart) | ‘Outpatient’ | N = 6 All male Age 32.3 ± 3.6 BMI 25.2 ± 3.4 | 14 (5.6%) | Amino acids |
| Authors | Assay/Platform | Time Course Details | Study Setting/Conditions | Cohort Details | Rhythmic/Gradient Metabolites/Features Observed | Rhythmic/Gradient Classes Primarily Observed |
|---|---|---|---|---|---|---|
| Sinues et al., (2012) [91] | Untargeted SESI-MS | Diurnal variation (4 time periods) 8:00–11:00, 11:00–13:00, 13:00–15:00, 15:00–18:00 | ‘Outpatient’ | N = 12 7 males | Diurnal changes observed but number of rhythmic features not reported | No metabolites structurally identified |
| Sinues et al., (2014) [92] | Untargeted SESI-MS | Diurnal variation (24 h, 1 h intervals, 5–7 repeats per sample) | ‘Inpatient’ Controlled laboratory conditions: hourly isocaloric meals, constant wakefulness, consistent light conditions | N = 3 2 males Age 33–38 | 40 (36%) of features (49% in N = 1) | No metabolites structurally identified |
| Wilkinson et al., (2019) [93] | Untargeted GC-MS | Diurnal variation (24 h—4 time points: 16:00, 22:00, 04:00, 10:00) | Standardised meals and feeding schedule. Maintained habitual bedtime | Healthy N = 10 7 males Age 27.5–49.3 BMI 23.4–30.5 Asthma N = 9 7 male Age 26.0–49.5 BMI 22.3–27.2 | Combined dataset 5/102 (4.9%) metabolites Asthma 3/102 (~2.9%) metabolites, 1 of which is unique to this group in addition to rhyth-micity of exhaled nitric oxide fraction Healthy 2/102 (~2%) metabolites rhythmic and unique to this group | Volatile organic compounds |
| Authors | Performed Assay | Time Course Details | Study Setting/Conditions | Cohort Details | Rhythmic/Gradient Metabolites/Features Observed | Rhythmic/Gradient Classes Primarily Observed |
|---|---|---|---|---|---|---|
| Loizides-Mangold et al., (2017) [94] | Targeted (Lipidomics) LC-MS | Diurnal variation (24 h—4 h intervals between samples) | ‘Inpatient’ Controlled sleep/wake, light/dark cycle, temperature. Isocaloric meals | N = 10, 9 males Age 29.9 ± 9.8 BMI 24.1 ± 2.7 | 106 of 1058 metabolites (10%) | TAGs, PCs, Pes PIs, PSs, CLs Cers, GlcCers, SMs |
| Sato et al., (2018) [63] | Untargeted UHPLC-MS/MS GC-MS | Diurnal variation am vs. pm | ‘Outpatient’ Standardised meals and mealtimes | N = 8, All male Age 30–45 BMI 27–32.5 | 163 & 19 of 625 features (26% & 3%) as a result of time of day & diet, respectively | Amino acids Fatty acyls Glycerolipids Glycerophospholipids Sphingolipids Carbohydrates Xenobiotics |
| Held et al., (2020) [95] | Semi-targeted Lipidomics UPLC/HRMS (reversed & normal phase) | Diurnal variation (24 h—5 h intervals between samples) | ‘Inpatient’ Controlled sleep/wake, light/dark cycle. Standardised meals and mealtimes | N = 12, All male Age 22.2 ± 2.3 BMI 22.4 ± 2.0 | 126 of 971 (13%) | Glycerophospholipids TAGs Sphingolipids DAGs Sterol Lipids |
| Rank | Putative Identification of Rhythmic/Gradient Metabolites | InChIKey | Number of Studies Significant Changes were Observed in |
|---|---|---|---|
| 1 | Proline | ONIBWKKTOPOVIA-BYPYZUCNSA-N | 11 |
| 2 | Leucine | ROHFNLRQFUQHCH-YFKPBYRVSA-N | 10 |
| 3 | PC(32:0) | - | 10 |
| 4 | Phenylalanine | COLNVLDHVKWLRT-QMMMGPOBSA-N | 9 |
| 5 | Ornithine | 9 | |
| 6 | Tyrosine | OUYCCCASQSFEME-QMMMGPOBSA-N | 9 |
| 7 | Glutamic acid | WHUUTDBJXJRKMK-VKHMYHEASA-N | 8 |
| 8 | Isoleucine | AGPKZVBTJJNPAG-WHFBIAKZSA-N | 8 |
| 9 | LysoPC(18:2) and/or LysoPE (18:2) | - | 8 |
| 10 | PC(34:3) | - | 8 |
| 11 | Citrulline | RHGKLRLOHDJJDR-BYPYZUCNSA-N | 7 |
| 12 | Taurine | XOAAWQZATWQOTB-UHFFFAOYSA-N | 7 |
| 13 | Tryptophan | QIVBCDIJIAJPQS-VIFPVBQESA-N | 7 |
| 14 | Valine | KZSNJWFQEVHDMF-BYPYZUCNSA-N | 7 |
| 15 | LysoPC(18:1) | - | 6 |
| 16 | LysoPC(16:0) | - | 6 |
| 17 | Aminoadipic acid | OYIFNHCXNCRBQI-BYPYZUCNSA-N | 6 |
| 18 | Citric acid | KRKNYBCHXYNGOX-UHFFFAOYSA-N | 6 |
| 19 | Cortisone | MFYSYFVPBJMHGN-ZPOLXVRWSA-N | 6 |
| 20 | Creatinine | DDRJAANPRJIHGJ-UHFFFAOYSA-N | 6 |
| 21 | Glycine | DHMQDGOQFOQNFH-UHFFFAOYSA-N | 6 |
| 22 | Kynurenine | YGPSJZOEDVAXAB-UHFFFAOYSA-N | 6 |
| 23 | PC C36:2 | - | 6 |
| 24 | Alanine | QNAYBMKLOCPYGJ-REOHCLBHSA-N | 5 |
| 25 | Cortisol | JYGXADMDTFJGBT-VWUMJDOOSA-N | 5 |
| 26 | Lysine | KDXKERNSBIXSRK-YFKPBYRVSA-N | 5 |
| 27 | LysoPC(17:0) | - | 5 |
| 28 | PC C34:1 | - | 5 |
| 29 | PC C34:2 | - | 5 |
| 30 | PC(32:1) | - | 5 |
| 31 | Pregnenolone sulfate | DIJBBUIOWGGQOP-OZIWPBGVSA-N | 5 |
| 32 | Sarcosine | FSYKKLYZXJSNPZ-UHFFFAOYSA-N | 5 |
| 33 | SM(20:2) | - | 5 |
| 34 | Threonine | AYFVYJQAPQTCCC-GBXIJSLDSA-N | 5 |
| 35 | Trimethylamine N-oxide (TMAO) | UYPYRKYUKCHHIB-UHFFFAOYSA-N | 5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hancox, T.P.M.; Skene, D.J.; Dallmann, R.; Dunn, W.B. Tick-Tock Consider the Clock: The Influence of Circadian and External Cycles on Time of Day Variation in the Human Metabolome—A Review. Metabolites 2021, 11, 328. https://doi.org/10.3390/metabo11050328
Hancox TPM, Skene DJ, Dallmann R, Dunn WB. Tick-Tock Consider the Clock: The Influence of Circadian and External Cycles on Time of Day Variation in the Human Metabolome—A Review. Metabolites. 2021; 11(5):328. https://doi.org/10.3390/metabo11050328
Chicago/Turabian StyleHancox, Thomas P. M., Debra J. Skene, Robert Dallmann, and Warwick B. Dunn. 2021. "Tick-Tock Consider the Clock: The Influence of Circadian and External Cycles on Time of Day Variation in the Human Metabolome—A Review" Metabolites 11, no. 5: 328. https://doi.org/10.3390/metabo11050328
APA StyleHancox, T. P. M., Skene, D. J., Dallmann, R., & Dunn, W. B. (2021). Tick-Tock Consider the Clock: The Influence of Circadian and External Cycles on Time of Day Variation in the Human Metabolome—A Review. Metabolites, 11(5), 328. https://doi.org/10.3390/metabo11050328







