Assessment of Evolutionary Relationships for Prioritization of Myxobacteria for Natural Product Discovery
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Isolation of Environmental Myxobacteria
2.3. Genomic DNA Isolation, Sequencing, Assembly, and Annotation
2.4. Comparative Genomic Studies
2.5. BIG-SCAPE Analysis
3. Results
3.1. Comparative Genomics and Taxonomic Assessment of Archangium Primigenium, Chondrococcus Macrosporus, and Environmental Isolates
3.2. Biosynthetic Potential and Genus Level Correlations
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Awal, R.P.; Garcia, R.; Gemperlein, K.; Wink, J.; Kunwar, B.; Parajuli, N.; Muller, R. Vitiosangium cumulatum gen. nov., sp. nov. and Vitiosangium subalbum sp. nov., soil myxobacteria, and emended descriptions of the genera Archangium and Angiococcus, and of the family Cystobacteraceae. Int. J. Syst. Evol. Microbiol. 2017, 67, 1422–1430. [Google Scholar] [CrossRef]
- Awal, R.P.; Garcia, R.; Muller, R. Racemicystis crocea gen. nov., sp. nov., a soil myxobacterium in the family Polyangiaceae. Int. J. Syst. Evol. Microbiol. 2016, 66, 2389–2395. [Google Scholar] [CrossRef] [Green Version]
- Chambers, J.; Sparks, N.; Sydney, N.; Livingstone, P.G.; Cookson, A.R.; Whitworth, D.E. Comparative Genomics and Pan-Genomics of the Myxococcaceae, including a Description of Five Novel Species: Myxococcus eversor sp. nov., Myxococcus llanfairpwllgwyngyllgogerychwyrndrobwllllantysiliogogogochensis sp. nov., Myxococcus vastator sp. nov., Pyxidicoccus caerfyrddinensis sp. nov., and Pyxidicoccus trucidator sp. nov. Genome Biol. Evol. 2020, 12, 2289–2302. [Google Scholar] [CrossRef]
- Garcia, R.; Gemperlein, K.; Muller, R. Minicystis rosea gen. nov., sp. nov., a polyunsaturated fatty acid-rich and steroid-producing soil myxobacterium. Int. J. Syst. Evol. Microbiol. 2014, 64, 3733–3742. [Google Scholar] [CrossRef]
- Garcia, R.; Muller, R. Simulacricoccus ruber gen. nov., sp. nov., a microaerotolerant, non-fruiting, myxospore-forming soil myxobacterium and emended description of the family Myxococcaceae. Int. J. Syst. Evol. Microbiol. 2018, 68, 3101–3110. [Google Scholar] [CrossRef]
- Garcia, R.; Stadler, M.; Gemperlein, K.; Muller, R. Aetherobacter fasciculatus gen. nov., sp. nov. and Aetherobacter rufus sp. nov., novel myxobacteria with promising biotechnological applications. Int. J. Syst. Evol. Microbiol. 2016, 66, 928–938. [Google Scholar] [CrossRef] [PubMed]
- Iizuka, T.; Jojima, Y.; Hayakawa, A.; Fujii, T.; Yamanaka, S.; Fudou, R. Pseudenhygromyxa salsuginis gen. nov., sp. nov., a myxobacterium isolated from an estuarine marsh. Int. J. Syst. Evol. Microbiol. 2013, 63, 1360–1369. [Google Scholar] [CrossRef]
- Livingstone, P.G.; Ingleby, O.; Girdwood, S.; Cookson, A.R.; Morphew, R.M.; Whitworth, D.E. Predatory Organisms with Untapped Biosynthetic Potential: Descriptions of Novel Corallococcus Species C. aberystwythensis sp. nov., C. carmarthensis sp. nov., C. exercitus sp. nov., C. interemptor sp. nov., C. llansteffanensis sp. nov., C. praedator sp. nov., C. sicarius sp. nov., and C. terminator sp. nov. Appl. Environ. Microbiol. 2020, 86. [Google Scholar] [CrossRef]
- Mohr, K.I.; Garcia, R.O.; Gerth, K.; Irschik, H.; Muller, R. Sandaracinus amylolyticus gen. nov., sp. nov., a starch-degrading soil myxobacterium, and description of Sandaracinaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2012, 62, 1191–1198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohr, K.I.; Moradi, A.; Glaeser, S.P.; Kampfer, P.; Gemperlein, K.; Nubel, U.; Schumann, P.; Muller, R.; Wink, J. Nannocystis konarekensis sp. nov., a novel myxobacterium from an Iranian desert. Int. J. Syst. Evol. Microbiol. 2018, 68, 721–729. [Google Scholar] [CrossRef]
- Mohr, K.I.; Wolf, C.; Nubel, U.; Szafranska, A.K.; Steglich, M.; Hennessen, F.; Gemperlein, K.; Kampfer, P.; Martin, K.; Muller, R.; et al. A polyphasic approach leads to seven new species of the cellulose-decomposing genus Sorangium, Sorangium ambruticinum sp. nov., Sorangium arenae sp. nov., Sorangium bulgaricum sp. nov., Sorangium dawidii sp. nov., Sorangium kenyense sp. nov., Sorangium orientale sp. nov. and Sorangium reichenbachii sp. nov. Int. J. Syst. Evol. Microbiol. 2018, 68, 3576–3586. [Google Scholar] [CrossRef] [PubMed]
- Moradi, A.; Ebrahimipour, G.H.; Mohr, K.I.; Kampfer, P.; Glaeser, S.P.; Hennessen, F.; Gemperlein, K.; Awal, R.P.; Wolf, C.; Muller, R.; et al. Racemicystis persica sp. nov., a myxobacterium from soil. Int. J. Syst. Evol. Microbiol. 2017, 67, 472–478. [Google Scholar] [CrossRef]
- Sood, S.; Awal, R.P.; Wink, J.; Mohr, K.I.; Rohde, M.; Stadler, M.; Kampfer, P.; Glaeser, S.P.; Schumann, P.; Garcia, R.; et al. Aggregicoccus edonensis gen. nov., sp. nov., an unusually aggregating myxobacterium isolated from a soil sample. Int. J. Syst. Evol. Microbiol. 2015, 65, 745–753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, E.; Muramatsu, H.; Nagai, K. Vulgatibacter incomptus gen. nov., sp. nov. and Labilithrix luteola gen. nov., sp. nov., two myxobacteria isolated from soil in Yakushima Island, and the description of Vulgatibacteraceae fam. nov., Labilitrichaceae fam. nov. and Anaeromyxobacteraceae fam. nov. Int. J. Syst. Evol. Microbiol. 2014, 64, 3360–3368. [Google Scholar] [CrossRef]
- Bader, C.D.; Panter, F.; Muller, R. In depth natural product discovery–Myxobacterial strains that provided multiple secondary metabolites. Biotechnol. Adv. 2020, 39, 107480. [Google Scholar] [CrossRef] [PubMed]
- Bretl, D.J.; Kirby, J.R. Molecular Mechanisms of Signaling in Myxococcus xanthus Development. J. Mol. Biol. 2016, 428, 3805–3830. [Google Scholar] [CrossRef] [PubMed]
- Mercier, R.; Mignot, T. Regulations governing the multicellular lifestyle of Myxococcus xanthus. Curr. Opin. Microbiol. 2016, 34, 104–110. [Google Scholar] [CrossRef]
- Mohr, K.I. Diversity of Myxobacteria-We Only See the Tip of the Iceberg. Microorganisms 2018, 6, 84. [Google Scholar] [CrossRef] [Green Version]
- Pathak, D.T.; Wei, X.; Wall, D. Myxobacterial tools for social interactions. Res. Microbiol. 2012, 163, 579–591. [Google Scholar] [CrossRef] [Green Version]
- Petters, S.; Gross, V.; Sollinger, A.; Pichler, M.; Reinhard, A.; Bengtsson, M.M.; Urich, T. The soil microbial food web revisited: Predatory myxobacteria as keystone taxa? ISME J. 2021. [Google Scholar] [CrossRef]
- Sah, G.P.; Wall, D. Kin recognition and outer membrane exchange (OME) in myxobacteria. Curr. Opin. Microbiol. 2020, 56, 81–88. [Google Scholar] [CrossRef]
- Thiery, S.; Kaimer, C. The Predation Strategy of Myxococcus xanthus. Front. Microbiol. 2020, 11, 2. [Google Scholar] [CrossRef]
- Whitworth, D.E. Genome-wide analysis of myxobacterial two-component systems: Genome relatedness and evolutionary changes. BMC Genom. 2015, 16, 780. [Google Scholar] [CrossRef] [Green Version]
- Baltz, R.H. Molecular beacons to identify gifted microbes for genome mining. J. Antibiot. 2017, 70, 639–646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baltz, R.H. Gifted microbes for genome mining and natural product discovery. J. Ind. Microbiol. Biotechnol. 2017, 44, 573–588. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, J.; Fayad, A.A.; Muller, R. Natural products from myxobacteria: Novel metabolites and bioactivities. Nat. Prod. Rep. 2017, 34, 135–160. [Google Scholar] [CrossRef] [PubMed]
- Landwehr, W.; Wolf, C.; Wink, J. Actinobacteria and Myxobacteria-Two of the Most Important Bacterial Resources for Novel Antibiotics. Curr. Top. Microbiol. Immunol. 2016, 398, 273–302. [Google Scholar] [CrossRef] [PubMed]
- Weissman, K.J.; Muller, R. Myxobacterial secondary metabolites: Bioactivities and modes-of-action. Nat. Prod. Rep. 2010, 27, 1276–1295. [Google Scholar] [CrossRef]
- Wenzel, S.C.; Muller, R. Myxobacteria--‘microbial factories’ for the production of bioactive secondary metabolites. Mol. Biosyst. 2009, 5, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, T.; Krug, D.; Bozkurt, N.; Duddela, S.; Jansen, R.; Garcia, R.; Gerth, K.; Steinmetz, H.; Muller, R. Correlating chemical diversity with taxonomic distance for discovery of natural products in myxobacteria. Nat. Commun. 2018, 9, 803. [Google Scholar] [CrossRef]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.W.; De Meyer, S.; et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Chun, J.; Rainey, F.A. Integrating genomics into the taxonomy and systematics of the Bacteria and Archaea. Int. J. Syst. Evol. Microbiol. 2014, 64, 316–324. [Google Scholar] [CrossRef] [PubMed]
- Garcia, R.; Gerth, K.; Stadler, M.; Dogma, I.J., Jr.; Muller, R. Expanded phylogeny of myxobacteria and evidence for cultivation of the ‘unculturables’. Mol. Phylogenet. Evol. 2010, 57, 878–887. [Google Scholar] [CrossRef]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.; Oh, H.S.; Park, S.C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Livingstone, P.G.; Morphew, R.M.; Whitworth, D.E. Genome Sequencing and Pan-Genome Analysis of 23 Corallococcus spp. Strains Reveal Unexpected Diversity, With Particular Plasticity of Predatory Gene Sets. Front. Microbiol. 2018, 9, 3187. [Google Scholar] [CrossRef] [Green Version]
- Sangal, V.; Goodfellow, M.; Jones, A.L.; Schwalbe, E.C.; Blom, J.; Hoskisson, P.A.; Sutcliffe, I.C. Next-generation systematics: An innovative approach to resolve the structure of complex prokaryotic taxa. Sci. Rep. 2016, 6, 38392. [Google Scholar] [CrossRef] [Green Version]
- Shimkets, L.; Woese, C.R. A phylogenetic analysis of the myxobacteria: Basis for their classification. Proc. Natl. Acad. Sci. USA 1992, 89, 9459–9463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sproer, C.; Reichenbach, H.; Stackebrandt, E. The correlation between morphological and phylogenetic classification of myxobacteria. Int. J. Syst. Evol. Microbiol. 1999, 49 Pt 3, 1255–1262. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Pauker, O.; Steiner, U.; Schumann, P.; Straubler, B.; Heibei, S.; Lang, E. Taxonomic characterization of members of the genus Corallococcus: Molecular divergence versus phenotypic coherency. Syst. Appl. Microbiol. 2007, 30, 109–118. [Google Scholar] [CrossRef]
- Bouhired, S.; Rupp, O.; Blom, J.; Schaberle, T.F.; Schiefer, A.; Kehraus, S.; Pfarr, K.; Goesmann, A.; Hoerauf, A.; Konig, G. Complete Genome Sequence of the Corallopyronin A-Producing Myxobacterium Corallococcus coralloides B035. Microbiol. Resour. Announc. 2019, 8, e00050-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huntley, S.; Zhang, Y.; Treuner-Lange, A.; Kneip, S.; Sensen, C.W.; Sogaard-Andersen, L. Complete genome sequence of the fruiting myxobacterium Corallococcus coralloides DSM 2259. J. Bacteriol. 2012, 194, 3012–3013. [Google Scholar] [CrossRef] [Green Version]
- McDonald, J.C. Studies on the genus Archangium (Myxobacterales). II. The effect of temperature and carbohydrates on some physiological processes. Mycologia 1967, 59, 1059–1068. [Google Scholar] [CrossRef] [PubMed]
- McDonald, J.C. Studies on the genus Archangium (Myxobacterales) I. Morphology. Mycologia 1965, 57, 737–747. [Google Scholar] [CrossRef]
- Shimkets, L.J.; Dworkin, M.; Reichenbach, H. The Myxobacteria. In The Prokaryotes; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006. [Google Scholar] [CrossRef]
- McCurdy, H.D. Studies on the Taxonomy of the Myxobacterales. Int. J. Syst. Bacteriol. 1971, 21, 50–54. [Google Scholar] [CrossRef]
- Starr, M.P.; Skerman, V.B. Bacterial diversity: The natural history of selected morphologically unusual bacteria. Annu. Rev. Microbiol. 1965, 19, 407–454. [Google Scholar] [CrossRef] [PubMed]
- Sly, L.I. Taxonomic Note: V. B. D. Skerman (1921–1993), a Reforming Force in Bacterial Systematics and Nomenclature. Int. J. Syst. Evol. Microbiol. 1995, 45, 412–413. [Google Scholar] [CrossRef]
- Karwowski, J.P.; Sunga, G.N.; Kadam, S.; McAlpine, J.B. A method for the selective isolation of Myxococcus directly from soil. J. Ind. Microbiol. 1996, 16, 230–236. [Google Scholar] [CrossRef]
- Lang, E.; Stackebrandt, E. Emended descriptions of the genera Myxococcus and Corallococcus, typification of the species Myxococcus stipitatus and Myxococcus macrosporus and a proposal that they be represented by neotype strains. Request for an Opinion. Int. J. Syst. Evol. Microbiol. 2009, 59, 2122–2128. [Google Scholar] [CrossRef]
- Dawid, W. Biology and global distribution of myxobacteria in soils. FEMS Microbiol. Rev. 2000, 24, 403–427. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [Green Version]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [Green Version]
- Meier-Kolthoff, J.P.; Goker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Ouk Kim, Y.; Park, S.C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-R, L.M.; Konstantinidis, K.T. The enveomics collection: A toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ 2016, 4, e1900v1. [Google Scholar]
- Medema, M.H.; Blin, K.; Cimermancic, P.; de Jager, V.; Zakrzewski, P.; Fischbach, M.A.; Weber, T.; Takano, E.; Breitling, R. antiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011, 39, W339–W346. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kautsar, S.A.; Blin, K.; Shaw, S.; Navarro-Munoz, J.C.; Terlouw, B.R.; van der Hooft, J.J.J.; van Santen, J.A.; Tracanna, V.; Suarez Duran, H.G.; Pascal Andreu, V.; et al. MIBiG 2.0: A repository for biosynthetic gene clusters of known function. Nucleic Acids Res. 2020, 48, D454–D458. [Google Scholar] [CrossRef] [Green Version]
- Navarro-Munoz, J.C.; Selem-Mojica, N.; Mullowney, M.W.; Kautsar, S.A.; Tryon, J.H.; Parkinson, E.I.; De Los Santos, E.L.C.; Yeong, M.; Cruz-Morales, P.; Abubucker, S.; et al. A computational framework to explore large-scale biosynthetic diversity. Nat. Chem. Biol. 2020, 16, 60–68. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rossello-Mora, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bentley, R.; Meganathan, R. Geosmin and methylisoborneol biosynthesis in streptomycetes. Evidence for an isoprenoid pathway and its absence in non-differentiating isolates. FEBS Lett. 1981, 125, 220–222. [Google Scholar] [CrossRef] [Green Version]
- Dickschat, J.S.; Bode, H.B.; Mahmud, T.; Muller, R.; Schulz, S. A novel type of geosmin biosynthesis in myxobacteria. J. Org. Chem. 2005, 70, 5174–5182. [Google Scholar] [CrossRef]
- Bhat, S.; Ahrendt, T.; Dauth, C.; Bode, H.B.; Shimkets, L.J. Two lipid signals guide fruiting body development of Myxococcus xanthus. mBio 2014, 5, e00939-13. [Google Scholar] [CrossRef] [Green Version]
- Lorenzen, W.; Bozhuyuk, K.A.; Cortina, N.S.; Bode, H.B. A comprehensive insight into the lipid composition of Myxococcus xanthus by UPLC-ESI-MS. J. Lipid Res. 2014, 55, 2620–2633. [Google Scholar] [CrossRef] [Green Version]
- Botella, J.A.; Murillo, F.J.; Ruiz-Vazquez, R. A cluster of structural and regulatory genes for light-induced carotenogenesis in Myxococcus xanthus. Eur. J. Biochem. 1995, 233, 238–248. [Google Scholar] [CrossRef]
- Cervantes, M.; Murillo, F.J. Role for vitamin B(12) in light induction of gene expression in the bacterium Myxococcus xanthus. J. Bacteriol. 2002, 184, 2215–2224. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Rubio, J.J.; Elias-Arnanz, M.; Padmanabhan, S.; Murillo, F.J. A repressor-antirepressor pair links two loci controlling light-induced carotenogenesis in Myxococcus xanthus. J. Biol. Chem. 2002, 277, 7262–7270. [Google Scholar] [CrossRef] [Green Version]
- Perez-Marin, M.C.; Padmanabhan, S.; Polanco, M.C.; Murillo, F.J.; Elias-Arnanz, M. Vitamin B12 partners the CarH repressor to downregulate a photoinducible promoter in Myxococcus xanthus. Mol. Microbiol. 2008, 67, 804–819. [Google Scholar] [CrossRef] [PubMed]
- Gaitatzis, N.; Kunze, B.; Muller, R. In vitro reconstitution of the myxochelin biosynthetic machinery of Stigmatella aurantiaca Sg a15: Biochemical characterization of a reductive release mechanism from nonribosomal peptide synthetases. Proc. Natl. Acad. Sci. USA 2001, 98, 11136–11141. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Weissman, K.J.; Muller, R. Myxochelin biosynthesis: Direct evidence for two- and four-electron reduction of a carrier protein-bound thioester. J. Am. Chem. Soc. 2008, 130, 7554–7555. [Google Scholar] [CrossRef]
- Cortina, N.S.; Krug, D.; Plaza, A.; Revermann, O.; Muller, R. Myxoprincomide: A natural product from Myxococcus xanthus discovered by comprehensive analysis of the secondary metabolome. Angew. Chem. Int. Ed. 2012, 51, 811–816. [Google Scholar] [CrossRef] [PubMed]
- Sasse, F.; Steinmetz, H.; Hofle, G.; Reichenbach, H. Rhizopodin, a new compound from Myxococcus stipitatus (myxobacteria) causes formation of rhizopodia-like structures in animal cell cultures. Production, isolation, physico-chemical and biological properties. J. Antibiot. 1993, 46, 741–748. [Google Scholar] [CrossRef] [PubMed]
- Pistorius, D.; Muller, R. Discovery of the rhizopodin biosynthetic gene cluster in Stigmatella aurantiaca Sg a15 by genome mining. Chembiochem 2012, 13, 416–426. [Google Scholar] [CrossRef]
- Park, S.; Hyun, H.; Lee, J.S.; Cho, K. Identification of the Phenalamide Biosynthetic Gene Cluster in Myxococcus stipitatus DSM 14675. J. Microbiol. Biotechnol. 2016, 26, 1636–1642. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kautsar, S.A.; Medema, M.H.; Weber, T. The antiSMASH database version 3: Increased taxonomic coverage and new query features for modular enzymes. Nucleic Acids Res. 2021, 49, D639–D643. [Google Scholar] [CrossRef]
- Meiser, P.; Weissman, K.J.; Bode, H.B.; Krug, D.; Dickschat, J.S.; Sandmann, A.; Muller, R. DKxanthene biosynthesis--understanding the basis for diversity-oriented synthesis in myxobacterial secondary metabolism. Chem. Biol. 2008, 15, 771–781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panter, F.; Krug, D.; Muller, R. Novel Methoxymethacrylate Natural Products Uncovered by Statistics-Based Mining of the Myxococcus fulvus Secondary Metabolome. ACS Chem. Biol. 2019, 14, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Momen, A.Z.; Hoshino, T. Biosynthesis of violacein: Intact incorporation of the tryptophan molecule on the oxindole side, with intramolecular rearrangement of the indole ring on the 5-hydroxyindole side. Biosci. Biotechnol. Biochem. 2000, 64, 539–549. [Google Scholar] [CrossRef]
- Brady, S.F.; Chao, C.J.; Handelsman, J.; Clardy, J. Cloning and heterologous expression of a natural product biosynthetic gene cluster from eDNA. Org. Lett. 2001, 3, 1981–1984. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, T. Violacein and related tryptophan metabolites produced by Chromobacterium violaceum: Biosynthetic mechanism and pathway for construction of violacein core. Appl. Microbiol. Biotechnol. 2011, 91, 1463–1475. [Google Scholar] [CrossRef]
- Erol, O.; Schaberle, T.F.; Schmitz, A.; Rachid, S.; Gurgui, C.; El Omari, M.; Lohr, F.; Kehraus, S.; Piel, J.; Muller, R.; et al. Biosynthesis of the myxobacterial antibiotic corallopyronin A. Chembiochem 2010, 11, 1253–1265. [Google Scholar] [CrossRef] [PubMed]
- Pogorevc, D.; Panter, F.; Schillinger, C.; Jansen, R.; Wenzel, S.C.; Muller, R. Production optimization and biosynthesis revision of corallopyronin A, a potent anti-filarial antibiotic. Metab. Eng. 2019, 55, 201–211. [Google Scholar] [CrossRef]
- Weinig, S.; Hecht, H.J.; Mahmud, T.; Muller, R. Melithiazol biosynthesis: Further insights into myxobacterial PKS/NRPS systems and evidence for a new subclass of methyl transferases. Chem. Biol. 2003, 10, 939–952. [Google Scholar] [CrossRef] [Green Version]
- Schieferdecker, S.; Konig, S.; Weigel, C.; Dahse, H.M.; Werz, O.; Nett, M. Structure and biosynthetic assembly of gulmirecins, macrolide antibiotics from the predatory bacterium Pyxidicoccus fallax. Chemistry 2014, 20, 15933–15940. [Google Scholar] [CrossRef] [PubMed]
- Surup, F.; Viehrig, K.; Mohr, K.I.; Herrmann, J.; Jansen, R.; Muller, R. Disciformycins A and B: 12-membered macrolide glycoside antibiotics from the myxobacterium Pyxidicoccus fallax active against multiresistant staphylococci. Angew. Chem. Int. Ed. 2014, 53, 13588–13591. [Google Scholar] [CrossRef]
- Panter, F.; Krug, D.; Baumann, S.; Muller, R. Self-resistance guided genome mining uncovers new topoisomerase inhibitors from myxobacteria. Chem. Sci. 2018, 9, 4898–4908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gregory, K.; Salvador, L.A.; Akbar, S.; Adaikpoh, B.I.; Stevens, D.C. Survey of Biosynthetic Gene Clusters from Sequenced Myxobacteria Reveals Unexplored Biosynthetic Potential. Microorganisms 2019, 7, 781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
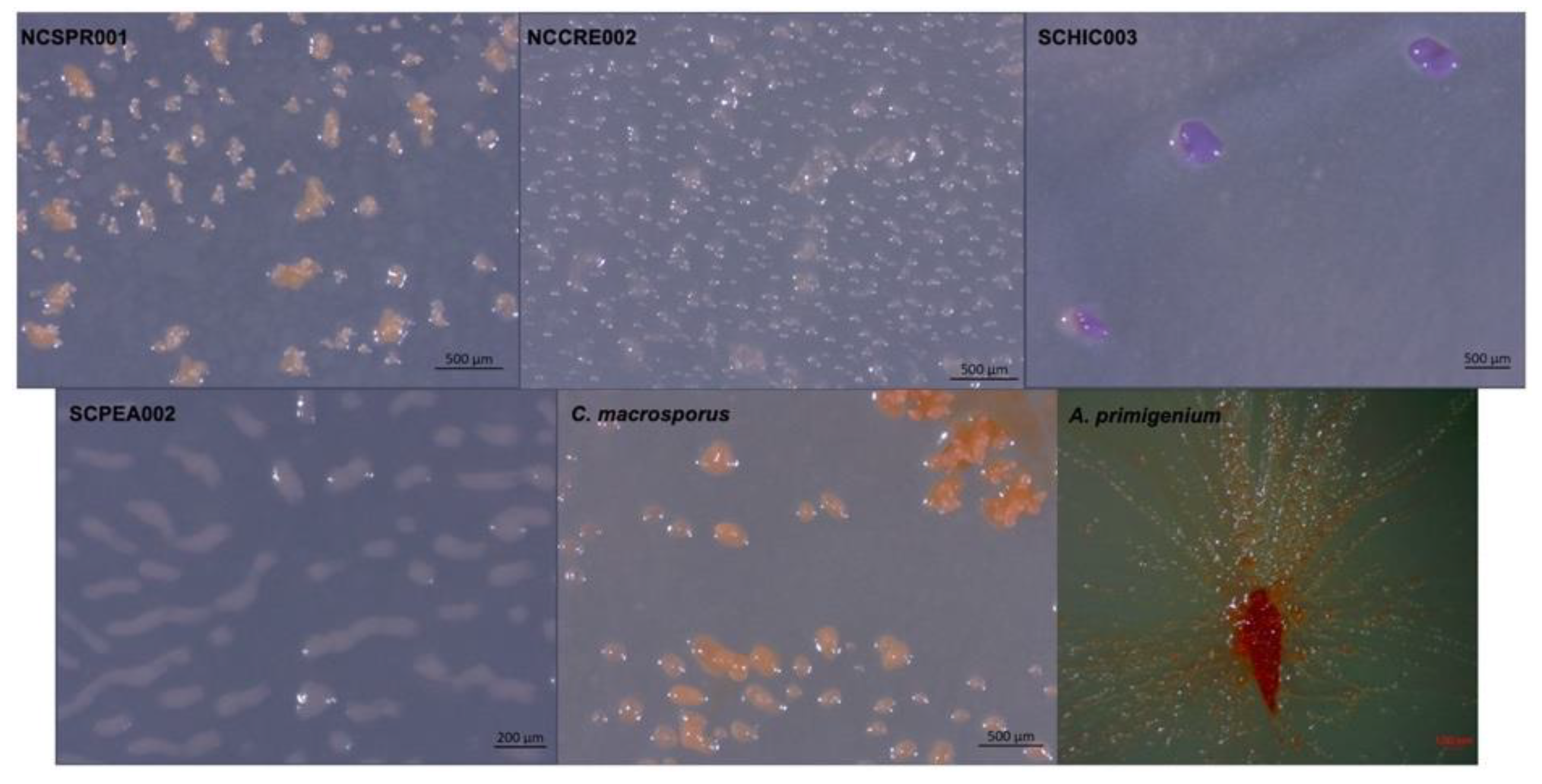

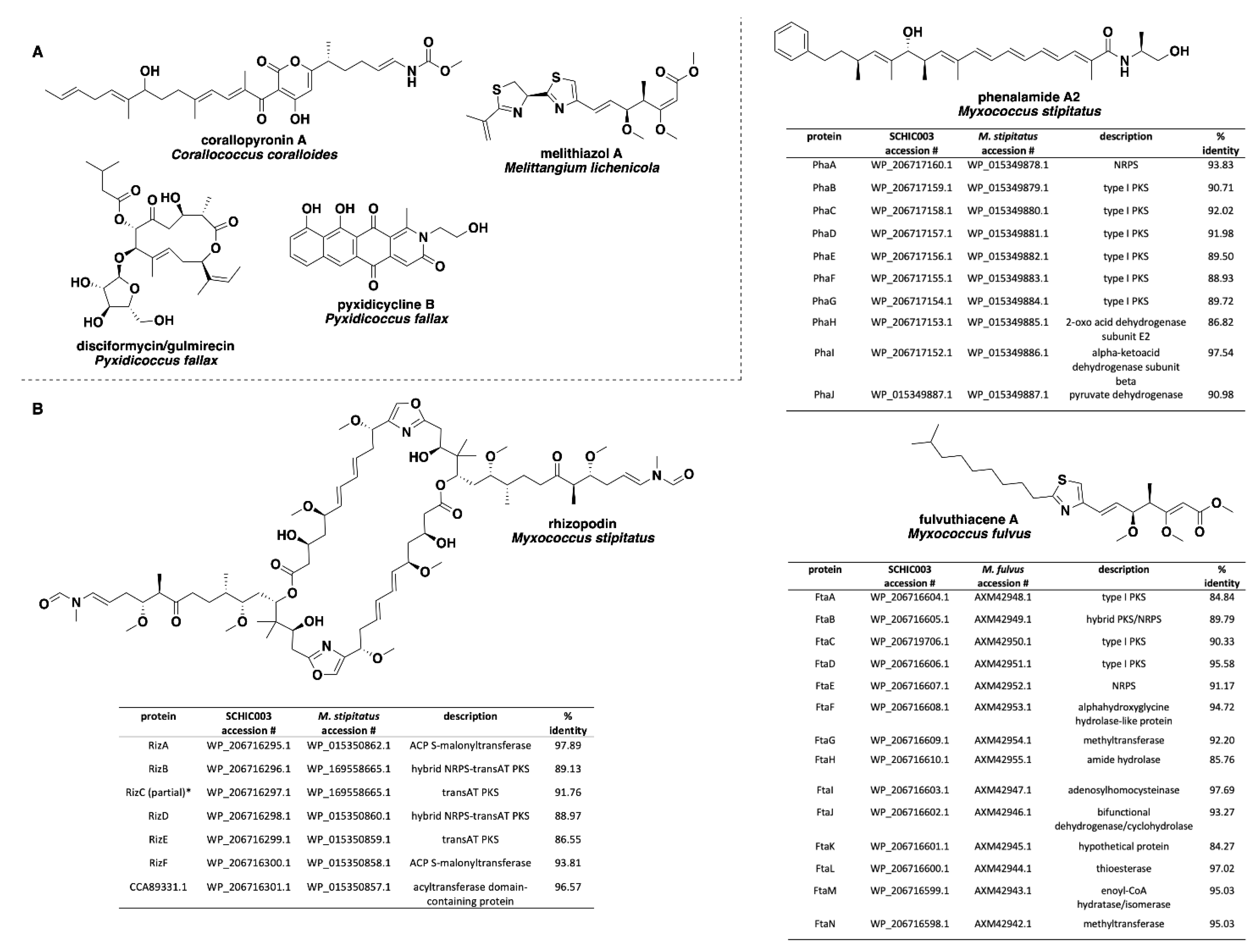

| Species | Size (bp) | CDS | GC% | N50 | L50 | Contigs | Coverage |
|---|---|---|---|---|---|---|---|
| A. primigenium | 9,491,554 | 7873 | 70.7% | 9,468,833 | 1 | 3 | 441x |
| C. macrosporus | 9,811,739 | 7977 | 70.4% | 1,094,727 | 2 | 17 | 300x |
| NCSPR001 | 9,785,177 | 8033 | 70.1% | 9,343,940 | 1 | 3 | 312x |
| NCCRE002 | 10,538,407 | 8589 | 69.7% | 3,024,381 | 2 | 8 | 479x |
| SCPEA002 | 13,211,253 | 10,588 | 69.6% | N/A | 1 | 1 | 144x |
| SCHIC003 | 10,367,529 | 8339 | 68.6% | N/A | 1 | 1 | 301x |
| Species | 16s rRNA | dDDH | ANI |
|---|---|---|---|
| M. boletus DSM 14713T | 98.89% | 29.5 | 86.1% |
| C. fuscus DSM 52655 | 98.7% | 24.5 | 83.29% |
| A. gephyra DSM 2261T | 97.72% | 23.2 | 81.41% |
| S. aurantiaca DW43-1 | 96.06% | 20 | 78.9% |
| M. macrosporus DSM 14697T | 96.63% | 19.8 | 78.85% |
| NCCRE002 | NCSPR001 | Chondrococcus macrosporus | Corallococcus interemptor T | Corallococcus terminator T | Corallococcus sicarius T | Corallococcus praedator T | Corallococcus macrosporus HW1 | Corallococcus llansteffanensis T | Corallococcus exiguus T | Corallococcus exercitus T | Corallococcus coralloides T | Corallococcus carmarthensis T | Corallococcus aberystwythensis T | Corallococcus Z5C101001 | Corallococcus ZKHCc1_1396 | Corallococcus CA053C | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NCCRE002 | 100% | dDDH: 51 | 41 | 44 | 29 | 29 | 30 | 21 | 30 | 66 | 43 | 54 | 43 | 43 | 34 | 29 | 30 |
| ANI: 94 | 91 | 92 | 86 | 86 | 86 | 81 | 87 | 96 | 91 | 94 | 92 | 91 | 88 | 86 | 86 | ||
| NCSPR001 | 99.8 | 100% | dDDH: 41 | 45 | 29 | 30 | 30 | 21 | 30 | 51 | 43 | 54 | 43 | 43 | 34 | 29 | 30 |
| ANI: 91 | 92 | 86 | 87 | 87 | 81 | 87 | 93 | 92 | 94 | 91 | 91 | 88 | 87 | 87 | |||
| Chondrococcus macrosporus | 99.15 | 99.22 | 100% | dDDH: 42 | 30 | 30 | 30 | 21 | 31 | 41 | 51 | 42 | 43 | 44 | 34 | 30 | 31 |
| ANI: 91 | 86 | 87 | 87 | 81 | 87 | 91 | 94 | 91 | 92 | 92 | 89 | 87 | 87 | ||||
| Corallococcus interemptor T | 99.8 | 99.87 | 99.35 | 100% | dDDH: 29 | 30 | 30 | 21 | 30 | 44 | 42 | 46 | 42 | 42 | 34 | 30 | 30 |
| ANI: 86 | 87 | 87 | 81 | 87 | 92 | 91 | 92 | 91 | 91 | 88 | 87 | 87 | |||||
| Corallococcus terminator T | 99.03 | 98.96 | 99.09 | 98.96 | 100% | dDDH: 35 | 49 | 21 | 35 | 29 | 30 | 29 | 30 | 30 | 31 | 42 | 34 |
| ANI: 89 | 93 | 81 | 89 | 86 | 87 | 86 | 87 | 87 | 87 | 91 | 89 | ||||||
| Corallococcus sicarius T | 98.89 | 98.83 | 98.98 | 98.83 | 99.61 | 100% | dDDH: 35 | 21 | 50 | 30 | 31 | 30 | 31 | 31 | 31 | 43 | 50 |
| ANI: 89 | 81 | 93 | 86 | 87 | 87 | 87 | 87 | 87 | 89 | 93 | |||||||
| Corallococcus praedator T | 99.03 | 98.96 | 99.09 | 98.96 | 100 | 99.61 | 100% | dDDH: 21 | 36 | 30 | 31 | 30 | 31 | 31 | 32 | 43 | 35 |
| ANI: 81 | 89 | 86 | 87 | 87 | 87 | 87 | 87 | 92 | 89 | ||||||||
| Corallococcus macrosporus HW1 | 97.73 | 97.66 | 98.37 | 97.79 | 97.72 | 97.72 | 97.72 | 100% | dDDH: 22 | 21 | 21 | 21 | 21 | 21 | 21 | 21 | 21 |
| ANI: 81 | 81 | 81 | 81 | 81 | 81 | 81 | 81 | 81 | |||||||||
| Corallococcus llansteffanensis T | 98.83 | 98.76 | 98.89 | 98.76 | 99.54 | 99.93 | 99.54 | 97.73 | 100% | dDDH: 30 | 32 | 31 | 32 | 31 | 33 | 36 | 54 |
| ANI: 87 | 88 | 87 | 87 | 87 | 88 | 89 | 94 | ||||||||||
| Corallococcus exiguus T | 99.93 | 99.87 | 99.22 | 99.87 | 99.09 | 98.96 | 99.09 | 97.79 | 98.89 | 100% | dDDH: 43 | 54 | 44 | 43 | 34 | 30 | 30 |
| ANI: 91 | 94 | 92 | 91 | 88 | 86 | 86 | |||||||||||
| Corallococcus exercitus T | 99.02 | 99.09 | 99.87 | 99.22 | 99.22 | 99.09 | 99.22 | 98.37 | 99.02 | 99.09 | 100% | dDDH: 44 | 48 | 47 | 36 | 31 | 32 |
| ANI: 92 | 93 | 93 | 89 | 87 | 87 | ||||||||||||
| Corallococcus coralloides T | 99.67 | 99.61 | 99.09 | 99.74 | 98.83 | 98.7 | 98.83 | 97.66 | 98.63 | 99.74 | 98.96 | 100% | dDDH: 44 | 44 | 34 | 30 | 30 |
| ANI: 92 | 92 | 88 | 87 | 87 | |||||||||||||
| Corallococcus carmarthensis T | 99.22 | 99.28 | 99.93 | 99.28 | 99.15 | 99.02 | 99.15 | 98.31 | 98.96 | 99.28 | 99.8 | 99.02 | 100% | dDDH: 48 | 36 | 31 | 31 |
| ANI: 93 | 89 | 87 | 87 | ||||||||||||||
| Corallococcus aberystwythensis T | 99.35 | 99.15 | 99.8 | 99.15 | 99.15 | 99.02 | 99.15 | 98.31 | 98.96 | 99.26 | 99.67 | 99.02 | 99.87 | 100% | dDDH: 35 | 31 | 31 |
| ANI: 89 | 87 | 87 | |||||||||||||||
| Corallococcus Z5C101001 | 98.76 | 98.83 | 99.48 | 98.83 | 99.22 | 99.22 | 99.22 | 98.11 | 99.15 | 98.83 | 99.61 | 98.57 | 99.54 | 99.41 | 100% | dDDH: 32 | 32 |
| ANI: 88 | 88 | ||||||||||||||||
| Corallococcus ZKHCc1_1396 | 98.7 | 98.89 | 98.89 | 98.76 | 99.67 | 99.35 | 99.67 | 97.4 | 99.28 | 98.76 | 99.02 | 98.5 | 98.96 | 98.83 | 99.09 | 100% | dDDH: 35 |
| ANI: 88 | |||||||||||||||||
| Corallococcus CA053C | 98.57 | 98.5 | 98.7 | 98.5 | 99.54 | 99.67 | 99.54 | 97.59 | 99.61 | 98.63 | 98.83 | 98.37 | 98.76 | 98.76 | 99.22 | 99.28 | 100% |
| SCPEA002 | SCHIC003 | M. fulvus 124B02 | P. fallax T | M. stipitatus T | M. hansupus | M. eversor T | M. llanfair T | M. vastator T | M. virescens T | P. trucidator T | P. caerfyrddinensis T | M. xanthus DK1622 | M. macrosporus T | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SCPEA002 | 100% | dDDH: 22 | 23 | 28 | 22 | 23 | 23 | 28 | 25 | 24 | 29 | 34 | 24 | 24 |
| ANI: 82 | 82 | 85 | 82 | 83 | 82 | 82 | 83 | 83 | 86 | 88 | 83 | 83 | ||
| SCHIC003 | 99.15% | 100% | dDDH: 26 | 23 | 49 | 22 | 27 | 27 | 23 | 22 | 22 | 22 | 22 | 22 |
| ANI: 84 | 82 | 93 | 81 | 85 | 85 | 82 | 82 | 82 | 82 | 81 | 82 | |||
| M. fulvus 124B02 | 99.61% | 99.41% | 100% | dDDH: 23 | 26 | 22 | 28 | 28 | 23 | 23 | 23 | 23 | 22 | 23 |
| ANI: 82 | 84 | 82 | 85 | 85 | 82 | 82 | 82 | 82 | 82 | 82 | ||||
| P. fallax T | 99.54% | 98.96% | 99.41% | 100% | dDDH: 23 | 24 | 23 | 23 | 25 | 25 | 30 | 29 | 25 | 25 |
| ANI: 82 | 83 | 82 | 82 | 84 | 83 | 86 | 86 | 83 | 84 | |||||
| M. stipitatus T | 99.15% | 100% | 99.41% | 98.96% | 100% | dDDH: 22 | 27 | 27 | 23 | 22 | 22 | 22 | 29 | 22 |
| ANI: 81 | 85 | 85 | 82 | 82 | 82 | 82 | 81 | 82 | ||||||
| M. hansupus | 98.89% | 98.44% | 98.89% | 98.57% | 98.44% | 100% | dDDH: 22 | 23 | 32 | 32 | 24 | 24 | 31 | 32 |
| ANI: 82 | 82 | 88 | 87 | 83 | 83 | 87 | 88 | |||||||
| M. eversor T | 98.83% | 98.24% | 98.83% | 98.50% | 98.24% | 99.15% | 100% | dDDH: 41 | 23 | 22 | 24 | 23 | 22 | 23 |
| ANI: 91 | 82 | 82 | 82 | 82 | 82 | 82 | ||||||||
| M. llanfair T | 98.76% | 98.31% | 98.89% | 98.44% | 98.31% | 99.09% | 99.93% | 100% | dDDH: 23 | 23 | 24 | 23 | 23 | 23 |
| ANI: 82 | 82 | 83 | 82 | 82 | 82 | |||||||||
| M. vastator T | 98.70% | 98.24% | 98.70% | 98.37% | 98.24% | 99.41% | 98.96% | 98.89% | 100% | dDDH: 52 | 25 | 25 | 52 | 41 |
| ANI: 94 | 84 | 84 | 94 | 91 | ||||||||||
| M. virescens T | 98.63% | 98.14% | 98.63% | 98.31% | 98.18% | 99.35% | 98.89% | 98.83% | 99.67% | 100% | dDDH: 25 | 24 | 73 | 40 |
| ANI: 83 | 83 | 97 | 90 | |||||||||||
| P. trucidator T | 99.09% | 98.37% | 98.70% | 99.02% | 98.37% | 98.89% | 98.83% | 98.76% | 98.70% | 98.63% | 100% | dDDH: 33 | 24 | 25 |
| ANI: 88 | 83 | 84 | ||||||||||||
| P. caerfyrddinensis T | 99.48% | 98.76% | 99.09% | 99.41% | 98.76% | 99.15% | 98.96% | 98.89% | 98.96% | 98.96% | 99.61% | 100% | dDDH: 24 | 25 |
| ANI: 83 | 83 | |||||||||||||
| M. xanthus DK1622 | 98.57% | 98.11% | 98.57% | 98.24% | 98.11% | 99.28% | 98.83% | 98.76% | 99.74% | 99.93% | 98.57% | 98.83% | 100% | dDDH: 40 |
| ANI: 90 | ||||||||||||||
| M. macrosporus T | 98.89% | 98.44% | 98.89% | 98.57% | 98.44% | 99.48% | 99.15% | 99.09% | 99.67% | 99.61% | 98.89% | 99.15% | 99.54% | 100% |
| Myxobacteria | # of Total BGCs | # and % of Singletons | # of Edges Formed with other BGCs | # of BGCs with 1 or 2 Edges | # of BGCs with 3 or More Edges |
|---|---|---|---|---|---|
| A. primigenium ATCC 29037 | 32 | 24 (75%) | 21 | 6 | 2 |
| C. macrosporus ATCC 29039 * | 42 | 9 (21.4%) | 228 | 4 | 29 |
| NCSPR001 | 32 | 1 (3.1%) | 248 | 7 | 24 |
| NCCRE002 * | 36 | 3 (16.7%) | 231 | 7 | 26 |
| SCPEA002 | 34 | 26 (76.5%) | 62 | 4 | 4 |
| SCHIC003 | 29 | 8 (27.6%) | 85 | 13 | 8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahearne, A.; Albataineh, H.; Dowd, S.E.; Stevens, D.C. Assessment of Evolutionary Relationships for Prioritization of Myxobacteria for Natural Product Discovery. Microorganisms 2021, 9, 1376. https://doi.org/10.3390/microorganisms9071376
Ahearne A, Albataineh H, Dowd SE, Stevens DC. Assessment of Evolutionary Relationships for Prioritization of Myxobacteria for Natural Product Discovery. Microorganisms. 2021; 9(7):1376. https://doi.org/10.3390/microorganisms9071376
Chicago/Turabian StyleAhearne, Andrew, Hanan Albataineh, Scot E. Dowd, and D. Cole Stevens. 2021. "Assessment of Evolutionary Relationships for Prioritization of Myxobacteria for Natural Product Discovery" Microorganisms 9, no. 7: 1376. https://doi.org/10.3390/microorganisms9071376
APA StyleAhearne, A., Albataineh, H., Dowd, S. E., & Stevens, D. C. (2021). Assessment of Evolutionary Relationships for Prioritization of Myxobacteria for Natural Product Discovery. Microorganisms, 9(7), 1376. https://doi.org/10.3390/microorganisms9071376






