Abstract
The protection of grapevine biodiversity and the safeguarding of genetic variability are certainly primary and topical objectives for wine research, especially in territories historically devoted to viticulture. To assess the autochthonous germplasm of three different districts of Southern Umbria (Central Italy), the plant material of 70 grapevines retrieved from reforested land plots or old vineyards was collected, and their genetic identity was investigated using 13 microsatellite markers (SSR). The results revealed the presence of 39 unique genotypes, divided into 24 already-known cultivars and 15 never-reported SSR profiles. Most of the grapevine accessions were then vegetatively propagated and cultivated in a vineyard collection both to be protected from extinction and to be evaluated at the ampelographic level. Overall, this work emphasizes the need for recovering the threatened genetic variability that characterizes minor neglected grapevine cultivars or biotypes of Southern Umbria germplasm, and the requirement to revalue and exploit the more valuable genetic resources to enhance the local agri-food economy.
1. Introduction
Umbria is one of the smallest regions in the central part of the Italian Peninsula and is characterized by a landlocked landscape of rolling hills. Because of its pivotal position, Umbria always had important historical roles, often being a territory of disputes between the different neighboring domains, but above all, it was a key hub for major commercial routes and agricultural trades, in both north–south (from and to Rome) and east–west directions (between the Adriatic and Tyrrhenian Seas). Umbria has developed a well-established winemaking tradition that dates to the Etruscan period and persists to this day due to its favorable climate [1]. Until the recent past, wine production in this region was exclusively for family use and was primarily represented by mixed viticulture [2]. Throughout the 1950s, in fact, the most common training system still used was “viti maritate” (or “arbusta” according to Plinio il Vecchio), in which the vines are grown on other supporting trees (e.g., poplars) within a promiscuous cropping system [3]. Afterward, starting from the 1960s, there was a decisive change that oriented viticulture towards intensive monoculture and quality winemaking, thanks to the adoption of modern agronomic and oenological techniques [4].
Currently, the wine-growing acreage of Umbria is about 12,000 hectares [5] with two denomination of controlled and guaranteed origin (DOCG) and 13 denomination of controlled origin (DOC) wines, which are rather appreciated on the national and international markets. The main cultivated variety is Sangiovese (20% of the total area), followed by Trebbiano Toscano (12%), Grechetto (11%), Merlot (10%), Sagrantino (7%), and Cabernet Sauvignon (4%). Despite the limited availability of autochthonous cultivars in the official productive panorama, the Umbrian viticulture retains a huge biodiversity heritage [6]. As a matter of fact, several minor grapevine varieties (belonging to known genotypes or showing a unique fingerprinting) can easily be found in abandoned reforested land plots or in old vineyards, and some historical documents describe certain spontaneous vines varieties in wooded areas called “Viti vicciute” [7]. Moreover, there is evidence that several autochthonous varieties are still grown locally, although unnamed and neither identified nor described [8]. The real origins of the different grapevine varieties historically cultivated in Umbria are still unclear, and the presence of sparse wild vines (as Vitis vinifera L. subsp. sylvestris) is reported [9].
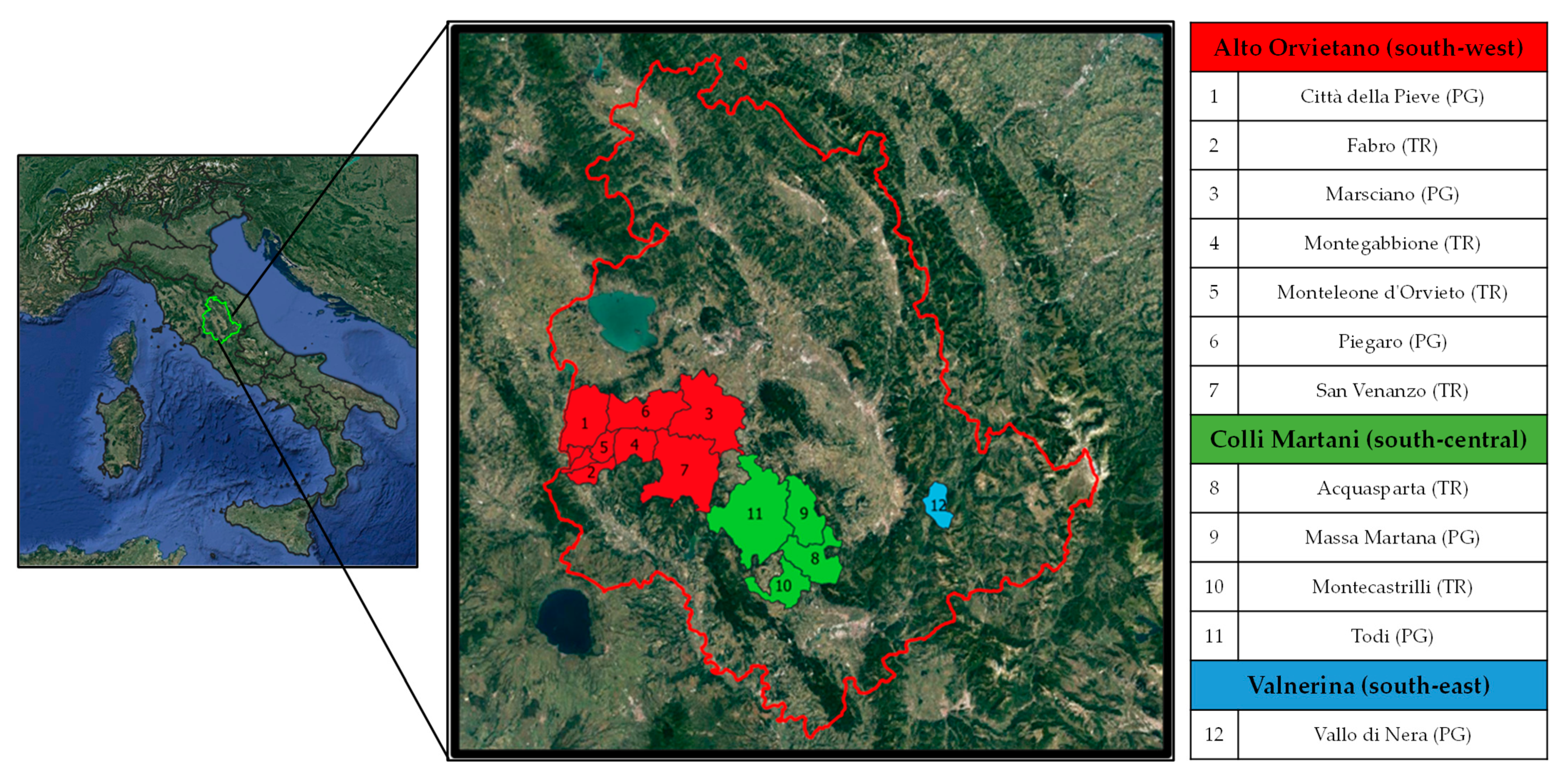
In the period 2013–2020, a research project funded by the Umbria Region was developed to protect the traditional germplasm of Umbria. Several inspections were carried out in 12 municipalities included in Alto Orvietano (southwest), Colli Martani (south-central), and Valnerina (southeast) territories of both the provinces of Perugia and Terni in Southern Umbria (Figure 1) to find vines of possible interest in private farms, in old and abandoned vineyards, or in wooded areas no longer devoted to agriculture. This work aimed to search, identify, rescue from genetic erosion, and revive some local grapevine varieties belonging to fragile and disadvantaged agricultural districts that are slowly recovering thanks to the economic boost of food and wine tourism [10].
Figure 1.
Geographical location and list of the 12 municipalities of Southern Umbria where the 70 grapevine samples included in the study were recovered. The color refers to the three territories considered: Alto Orvietano (in red), Colli Martani (in green), and Valnerina (in light blue). In brackets: abbreviation of the two provinces of Umbria, PG = Perugia; TR = Terni.
2. Results
2.1. Genetic Characterization of Grapevine Germplasm
The molecular analyses were performed on 70 grapevine samples collected in three different districts located in Southern Umbria (Alto Orvietano, Colli Martani, and Valnerina). Thirty-nine genotypes were found (Table 1), with 52 samples belonging to 24 known grapevine varieties and 18 samples showing 15 never-reported SSR profiles (Table 2).

Table 1.
List of the 70 grapevine samples grouped by their genotype. Original locations (municipality and province), berry color, true-to-type prime name, SSR profile ID number, and Vitis International Variety Catalogue [11] code are reported. In brackets: Italian common synonyms of some grapevine cultivars, according to the Italian Catalogue of Grapevine Varieties [12].

Table 2.
List of 39 unique genetic profiles obtained at 13 SSR loci.
2.2. Statistics for SSR Data
Statistics on the analyzed 13 SSRs are shown in Table 3. One hundred and fifteen alleles were detected, with a mean of 8.8 alleles per locus and a mean number of effective alleles of 4.9. The mean Ho and mean He were 0.831 and 0.789, respectively.

Table 3.
Statistics on the 13 SSR markers analyzed.
2.3. Genetic Similarity
The dendrogram of genetic similarity is shown in Figure 2. Cutting the dendrogram around nine on the X scale, nine groups were obtained. The main group (A) encompassing 20 genotypes (around half of the total genotypes found) includes varieties grown in the nearby Marche Region, such as Sgranarella and Famoso Marchigiano, and predominant Italian varieties such as Sangiovese, Trebbiano Toscano, and Montonico Bianco. Two pairs of varieties are grouped in B, i.e., Malvasia Bianca Lunga and Primitivo, likely of Balkan origin [14]; another two cultivars are in C, i.e., Muscat Rouge de Madère (also called Moscato Violetto) and Csaba Gyoengye (Perla di Csaba), both with muscat flavor and related to Muscat à Petits Grains Blancs as offspring and grandchild, respectively. Lambrusca di Alessandria is the only cultivar in group D. Agostenga groups with Unknown 2 in E. Interestingly, Verdicchio Bianco clusters with three unknown genotypes (Unknown 13, Unknown 3, and Unknown 14) (F); looking at SSR data, the three unknown genotypes share at least one allele per locus with Verdicchio Bianco, therefore they could be parent–offspring (PO) related. Likewise, Pignoletto could be PO-related with Unknown 11 in group G.
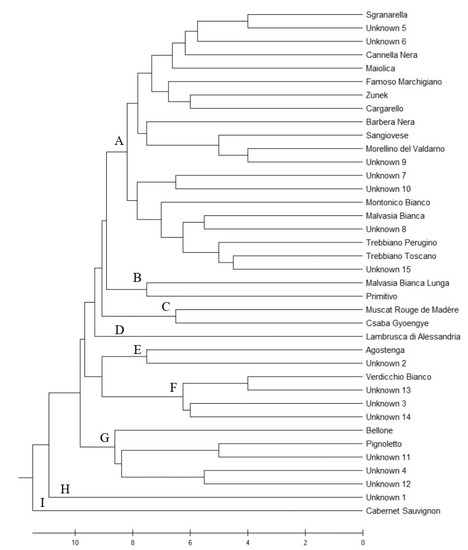
Figure 2.
Dendrogram of genetic similarity of 38 genotypes with 12 SSR markers. The optimal tree was inferred using the UPGMA method [15] and MEGA X software [16]. The tree is drawn to scale, with branch lengths in the same units as those of the genetic distances used to infer the tree. The genetic distances were computed using GenAlEx software.
Even if collected in a small area, unknown genotypes were shown to be overall highly different and did not group together in the tree. Interestingly, Unknown 1 (H) and Cabernet Sauvignon (I) seem to be outsiders.
2.4. Vineyard Collection Establishment and Ampelographic Descriptions
After genetic identification, a first group of 38 samples was vegetatively multiplied by grafting a wood branch on the same rootstock (1103 Paulsen); an experimental vineyard of Southern Umbria germplasm was started in 2015 with the rooted cuttings obtained; in the following years, additional accessions were added to the collection. Currently, the repository hosts 53 grapevine accessions out of the 70 vines considered in this study (Table 1, sample names in bold). In particular, they correspond to 29 different genotypes and, among them, 17 are already known varieties, while 12 are unknown genotypes.
For 38 accessions with already adult vines (over three years old, grape-producing plants), the ampelographic descriptions were performed following the recommended OIV —International Organisation of Vine and Wine methodology, based on the primary descriptor priority list [13]. These 38 accessions (marked with asterisks in Table 1) belong to 20 different genotypes, eight never described before, and 12 already known cultivars. Among the known cultivars, eight are already included in the Italian Catalogue of Grapevine Varieties [12] (i.e., Verdicchio Bianco, Trebbiano Toscano, Sangiovese, Montonico Bianco, Malvasia Bianca Lunga, Cargarello/Canaiolo Bianco, Lambrusca di Alessandria, and Primitivo) and four are not yet enrolled (i.e., Trebbiano Perugino, Famoso Marchigiano, Cannella Nera, and Zunek).
Ampelographic data of the eight unknown genotypes (11 accessions) are presented in Table 4, while those of the 12 already known varieties (27 accessions) are shown in Supplementary file S1. In addition to the 14 characters included in the primary descriptor priority list, we also detected the character OIV 151, related to the sexual organs of the flowers. As reported in Table 4, Casevecchie 2 (Unknown 01) has male flowers with fully developed stamens and reduced gynoecium; consequently, this accession produces very loose bunches with few fully developed berries.

Table 4.
Ampelographic descriptions of 11 grapevine accessions belonging to eight different unknown genotypes present in the vineyard germplasm collection at Castello di Montegiove Estate, municipality of Montegabbione (TR). The set of 14 standardized OIV primary descriptor (PDs) plus OIV 151, grouped according to each target vine organ (young shoot: OIV 001, 004; shoot: OIV 016; young leaf: OIV 051; mature leaf: OIV 067, 068, 070, 076, 079, 081-2, 084, 087; berry: OIV 223, 225; flower: OIV 151), are shown for each SSR profile.
3. Discussion
To explore the biodiversity of autochthonous grapevine from Southern Umbria, samples from 70 vines were collected in marginal vineyards not subjected to renewal in the last decades or in wooded areas where they were found as isolated relic plants. Genotyping was applied using a set of 13 SSR markers encompassing the nine SSR markers recommended by the European project GrapeGen06 [17]. The addition of VMC6E1, VMC6F1, VMC6G1 [18], and VMCNG4b9 [19] is a routine methodological approach of our laboratory that increases the resolution of the results and offers additional information to grapevine genotyping [20]. The screening by the internationally recognized microsatellite markers method permitted accurate and univocal identification of 39 different genetic profiles (24 known varieties and 15 unknown genotypes), giving an objective varietal classification [21]. The SSR profiles obtained also allowed the comparison of the Umbrian grapevine germplasm with literature data and molecular databases, searching for synonyms in neighboring countries.
Among the 70 grapevine samples retrieved, Verdicchio Bianco was the most common genotype (nine samples); this variety is rather widespread in Central and Northeastern Italy; in particular, it is one of the most prized white grapes of Marche [22], a region of the Adriatic coast bordering Umbria. Verdicchio Bianco has some officially recognized synonyms [23]: Trebbiano di Soave (appellation mainly used in several provinces of Veneto region), Verdello (the synonym used in Umbria), Verduschia or Duropersico (in Tuscany), and Trebbiano Verde (in Latium). Trebbiano Toscano (or Ugni Blanc) was also commonly recovered, as well as Sangiovese (8 and 7 samples, respectively); they are certainly the major white and red grape cultivar, and historically ubiquitous in the viticultural areas under investigation [24]. Many other recovered genotypes demonstrate the eclectic nature of the viticultural heritage of southern Umbria, which originates from the strategic importance of these territories in the trade routes of the past.
Trebbiano Perugino is a not a very common variety and is often confused with Trebbiano Toscano. The Trebbiano Perugino origin is unknown, but it is counted in ampelographic treatises as early as 1600 and then in the following centuries [25]. In the past, it was mainly present in the province of Perugia. Trebbiano Perugino has a remarkable vigor, and gives abundant and constant grape yields; it is characterized by elongated, large clusters and medium-small golden-yellow berries [25].
Montonico Bianco is an interesting, but now declining, cultivar, used both for wine and as a table grape. It is very resistant to late cold and has medium resistance of leaves and grapes to downy and powdery mildew [26]. Montonico Bianco is grown under a profusion of different names in the Italian Adriatic regions of Abruzzo, Marche, Apulia, but also in Tuscany, Umbria, and Calabria, suggesting a former wide distribution throughout Central and Southern Italy [27,28].
Famoso Marchigiano, different from Famoso from Emilia–Romagna, is a minor variety recently recovered in the Marche region [22]. Sgranarella is a white-berried variety also originating from Marche. It is mentioned in various ampelographic treaties of the past [29] and has recently been enrolled in the Italian Catalogue of Grapevine Varieties (2019) [12]. Both varieties are parent–offspring related with Crepolino/Visparola, one of the main founders of Italian grapevine germplasm [22].
Two “Cornacchione” samples from Valnerina showed an SSR profile different from the Cornacchione reported in the paper of [8] about the ancestral germplasm of Umbria and turned out to correspond to Cannella Nera [30], sometimes listed with the synonym Panfinone, a rare black-berried variety counted in the VIVC [11] and lacking any other bibliographic information.
Csaba Gyoengye (also called Perla di Csaba) is an early-ripening white grape variety, not surprisingly called by the keepers “Lugliolo”, namely “grapes that ripen in the month of July”. Csaba Gyoengye, a cross between Madeleine Angevine and Muscat Fleur d’Oranger [31], is widespread, especially in Northern Europe, and is traditionally grown as a table grape for fresh consumption; however, in Hungary, the country of origin, it is also used for winemaking [32]. Some enological trials on Csaba Gyoengye grapes were recently carried out in the territory of Colli Martani, giving interesting perspectives (unpublished data). Thus, based on these results, this cultivar was very recently approved by the Ministry of Agriculture for dual use in Italy, as well [12].
Villard Blanc is a Vitis interspecific crossing (Seibel 6468 X Seibel 6905 or Subéreux) of French origin, selected for its resistance to fungal diseases. In fact, it has the Rpv 3.1 gene in its genome (located in chromosome 18), which confers tolerance to downy mildew [33]. During the last century, direct–producer hybrids were commonly grown, trying to successfully obtain pest tolerance [34], and Villard Blanc was rather widely cultivated in some Italian regions (Veneto, for example). However, since the 1970s, its presence has dropped drastically, and the use of vines with non-Vitis vinifera DNA was even banned.
Originally from Slovenia, Zunek is known as Uva Sacra in Italy, where it is produced for table grapes [35]. Within this research, two different accessions from different wine-growing districts of southern Umbria (Alto Orvietano and Valnerina) turned out to be Zunek/Uva Sacra. The relic plants discovered were both capable of producing white grapes, in huge clusters with large and juicy berries, suitable for fresh consumption.
Morellino del Valdarno is a minor Tuscan cultivar native to the province of Arezzo which displayed a first-degree genetic relationship with Sangiovese [36] and showed interesting characteristics in monovarietal enological trials [37]. Since Morellino del Valdarno was also found in the territory under study, it can be considered a variety worth being monitored and safeguarded from the risk of genetic erosion.
Lambrusca di Alessandria is a cultivar native to Piedmont, as deducible from its name: the Alessandria province was indeed the largest growing area [38]. The name (as for Lambrusco) can be traced back to the Latin term Labruscae that designates wild grapevines and refers to the typical characteristics of the grapes, with a high content of tannins and anthocyanins. It was recently discovered that Lambrusca di Alessandria had a prominent genetic role as a parent, showing several presumed descendant cultivars [14,39]. Today it is considered as a variety with tiny commercial importance and is widespread exclusively in the oldest vineyards. To our knowledge, its presence in Central Italy is reported here for the first time.
Barbera is another grape variety from Piedmont that we retrieved in Umbria. It is a historical vine, already described in the very first treatises on ampelography; in the 1970s, it was the most common black-berried grape variety in Italy, probably due to its rusticity and ability to tolerate overseas parasites [40]. Currently, the cultivation area is much smaller than in the past and is limited to the suitable areas of Northwest Italy [40]. In the literature, the presence of Barbera and other “new vines introduced from outside that spread in a not indifferent way” is mentioned in the territory of Gubbio (PG), already at the end of the nineteenth century [41]; in Umbria, nowadays, Barbera is used for monovarietal vinification by a few local winegrowers only, for example in Valnerina.
Agostenga is an early-ripening cultivar (as can be guessed from the reference to the month of August in its name) also of Piedmont origin, but it is no longer present there. It is better called Prié Blanc or Blanc de Morgex in Aosta Valley (Northwest Italy), where it is cultivated almost exclusively today [29]. It grows at nearly prohibitive altitudes at the foot of Mont Blanc (900–1200 m above sea level), where there is no phylloxera pressure, therefore it does not require grafting on resistant rootstocks [42]. A few years ago, the synonymy with a geographically distant Spanish cultivar called Legiruela was established, as its SSR profile (34 loci) exhibited a perfect match with Agostenga/Prié Blanc [43].
Other varieties, quite common in Central Italy vineyards, were found: Malvasia Bianca Lunga and Malvasia Bianca, the first more and the second less widespread, respectively; Cargarello or Canaiolo Bianco/Drupeggio, an ancient minor variety part of the viticultural tradition of Tuscany and Umbria [44], currently poorly spread and poorly propagated in nurseries; Pignoletto, so-called in Emilia Romagna but grown as Grechetto Gentile in Umbria; Maiolica, a black-berried ancient cultivar grown in the near Marche region and in Tuscany (where it is given the synonym of Sanforte), offspring of the prolific Cascarello/Visparola and one of the parents of the Apulian Negroamaro [14]; Muscat Rouge de Madère or Moscato Violetto, a pleasant Muscat variety, sporadically present in Tuscany, offspring of Muscat à Petits Grains Blancs and Sciaccarello [14,45]; the Apulian Primitivo, an international black wine variety that probably originated in Montenegro, where it is mainly called Kratošija [46,47], and successfully grown in California as Zinfandel; Bellone, a white variety typical of Latium.
As common in germplasm assessment studies, one prominent “international grape variety” was found, Cabernet Sauvignon, which is grown worldwide thanks to its phenotypic plasticity and adaptability to the environment.
Concerning the 15 unknown genotypes, Rantola 32 accession recovered in San Venanzo (TR) was classified as Unknown 15; this genotype was already found in the province of Arezzo (Tuscany) in 2005 (unpublished data). As for Morellino del Valdarno, in this case we also tracked down, in South Umbria, some grapevines already present in Tuscan winegrowing areas, perhaps widespread vegetatively propagated cultivars in the past, now almost extinct. Casevecchie 2 (Unknown 01) showed the most divergent genotype among those here analyzed (except for Cabernet Sauvignon). The particularity of flowers with fully developed stamens and reduced gynoecium suggests a link with local Vitis sylvestris vines.
Alongside the genetic identification of the plant materials found, the propagation and the subsequent cultivation of most of the retrieved vines in a vineyard collection (as a repository) allowed us not only to safeguard several genotypes from extinction and to preserve the already known cultivars as a source of intravarietal diversity, but also to plan the evaluation of the grapes in terms of yields and quality.
4. Materials and Methods
4.1. Plant Materials
Young leaves of 70 vines were collected in situ in 12 different municipalities in the provinces of Perugia and Terni, covering a total area of about 1100 km2 (Figure 1), through various samplings that took place in the time frame of 2013–2019. The vines (mostly seedlings or not-grafted relic plants) were cataloged with a denomination given by local winegrowers or according to the place of discovery (Table 1). In any plant materials recovered, putative variety and grape characteristics (e.g., berry color) were neither already known nor assumed.
4.2. SSR Genotyping
Thirteen SSR markers were used for genotyping: the nine proposed as common grape markers for international use within the framework of the Grapegen06 European project (VVS2, VVMD5, VVMD7, VVMD25, VVMD27, VVMD28, VVMD32, VrZAG62, VrZAG79) [17], plus VMC6E1, VMC6F1, VMC6G1 [18], and VMCNG4b9 [19]. The SSR analyses were performed following the protocol detailed in [20]; two internationally required SSR markers (VVMD25 and VVMD32) [17] were also added and the set of 13 SSR markers were analyzed by two multiplex PCRs using fluorescent primers and an ABI3130xl genetic analyzer (Applied Biosystems, Foster City, CA, USA). The presence of PCR products was assessed by electrophoresis in a 1.5% agarose gel and quantified by comparison with a MassRuler DNA ladder mix (Thermo Fisher Scientific, Waltham, MA, USA). PCR products (0.5 μL) were mixed with 9.35 μL of formamide and 0.15 μL of the GeneScan™ 500 LIZ Size Standard (Life Tech, Carlsbad, CA, USA). Capillary electrophoresis was conducted in an ABI 3130xl Genetic Analyzer (Life Tech, CA, USA). Allele calling was performed with GeneMapper 5.0, using the 500 LIZ size standard as an internal ladder and a homemade bin set built with reference varieties. Allele sizes were recorded in bp (using the VIVC allele sizing [11]), and genotypes showing a single peak at a given locus were considered homozygous.
Identifications were performed by comparing the obtained SSR profiles with the CREA Viticulture and Enology molecular database (which currently contains about 5000 unique profiles, and is constantly updated), literature information, and the Vitis International Variety Catalogue [11]. In detail, Famoso Marchigiano, Morellino del Valdarno, Drupeggio, and Cannella Nera were identified referring to [22,30,36,44], respectively; Zunek, Csaba Gyongye, Villard Blanc, and Muscat Rouge de Madère were identified through the VIVC [11]; Trebbiano Perugino using the CREA-VE molecular database; all the remaining varieties using the molecular database of the Italian Catalogue of Grapevine Varieties [12].
4.3. Statistics on SSR Markers
Cervus 3.0 software [48] and GenAlEx 6.5 software [49] were used to determine the number of different alleles (No alleles), effective number of alleles (Ne alleles), observed (Ho), expected heterozygosity (He), polymorphic information content (PIC), Hardy–Weinberg equilibrium (HW), probability of null alleles (F (null)), and identity probability between two unrelated individuals and between two hypothetical full siblings (PID and PIDSIBS).
4.4. Dendrogram of Genetic Similarities
Genetic distances were computed using GenAlEx 6.5 software [49,50] and a dendrogram of genetic similarity was computed using 38 genotypes (Villard Blanc was excluded for being a hybrid) and 12 SSR markers (VMC6F1 was excluded due to missing data). The optimal tree was inferred using the UPGMA method [15] and MEGA X software [16].
4.5. Grapevine Germplasm Collection
An experimental germplasm collection of 2000 m2 was set up from 2015 until 2020 at Castello di Montegiove Estate in the municipality of Montegabbione (province of Terni, Umbria, Italy; 44°91′80″ N, 12°15′66″ E). The vines, grafted on 1103 Paulsen rootstock, were planted with an east–west orientation; planting distances were 0.90 m within rows and 2.80 m between rows. The vines were trained on upward vertical shoot positioned trellis, with spur cordon pruning and an average of 10 buds per vine. Over the years, the vineyard was conducted with homogeneous agronomic conditions, and pest management was scheduled with calendar sprays at 10-day intervals. For each of the 53 accessions actually present (sample names in bold in Table 1), there are at least 30 vines; Sangiovese and Trebbiano Toscano were added as reference cultivars for Central Italy grapevine germplasm.
4.6. Ampelographic Description
The ampelographic description of the main grapevine morphological traits was carried out in the experimental vineyard in Montegabbione (TR, Italy) during the vegetative season of 2019 on adult vines (over 3 years old), according to the set of 14 standardized OIV primary descriptor priority list [13]: young shoot (OIV 001, 004), shoot (OIV 016), young leaf (OIV 051), mature leaf (OIV 067, 068, 070, 076, 079, 081-2, 084, 087), berry (OIV 223, 225); as reputed interesting, the OIV 151 characteristic about flower sexual organs was added to the list.
5. Conclusions
The present study made it possible to explore a small part of the grapevine biodiversity of Southern Umbria by highlighting the hypothetical composition of the autochthonous germplasm that survived the phylloxera epidemics of the late 1800s and the widespread diffusion of the most common international varieties from the second half of the 20th century.
The 15 unknown genotypes recovered thanks to the present research work will soon be officially added to the VIVC [11], and the evaluations of their ampelographic, agronomic, and oenological characteristics are underway. Once enough data is collected, we plan to compare the available literature information about the old grapevine germplasm of Umbria to attribute the potential identity to any vines historically present in the districts under investigation.
The rediscovery and preservation of indigenous endangered grapevine cultivars or biotypes, especially in marginal areas such as the districts of Southern Umbria considered (Alto Orvietano, Colli Martani, and Valnerina), can give benefits to the local wine industry providing the basis for niche wines, an added value that can definitely help to relaunch the local economy in the territory of origin.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/plants10081539/s1, Supplementary file S1: Ampelographic descriptions of 27 grapevine samples belonging to twelve different known genotypes present in the vineyard germplasm collection at Castello di Montegiove Estate, municipality of Montegabbione (TR, Italy). Sangiovese and Trebbiano Toscano were added as reference cultivars. The tables contain the set of 14 standardized OIV primary descriptors plus OIV 151, grouped according to each target vine organ (young shoot: OIV 001, 004; shoot: OIV 016; young leaf: OIV 051; mature leaf: OIV 067, 068, 070, 076, 079, 081-2, 084, 087; berry: OIV 223, 225, flower: OIV 151) for each SSR profile.
Author Contributions
Conceptualization, P.S. and P.V.; methodology, P.S., M.C., A.Z., P.V. and D.M.; data curation, A.Z., A.C. and D.M.; validation, A.Z. and D.M.; formal analysis, A.Z., D.M. and M.C.; writing—original draft preparation, A.Z.; writing—review and editing, A.Z., M.C., P.S. and D.M.; visualization, A.Z. and M.C.; supervision, P.S. and M.C.; funding acquisition, P.S. and P.V.; project administration, P.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partially funded by Regione Umbria—Servizio sviluppo rurale e agricoltura sostenibile through the project “VIVIUMBRIA: Progetto di recupero e valorizzazione del germoplasma viticolo autoctono dell’Alto Orvietano—Città della Pieve”.
Data Availability Statement
The original data presented in this research work are stored in the databases of CREA—Research Centre for Viticulture and Enology.
Acknowledgments
The authors are grateful to the winemakers belonging to “Associazione Terre della Vicciuta” and Castello di Montegiove Estate for the essential role in the experimental vineyard management. Moreover, the authors thank the colleagues of CREA—Research Centre for Viticulture and Enology of Arezzo for their help in field surveys, sampling of plant materials, and laboratory analyses. This research was also supported by the Service for grapevine identification of CREA—Research Centre for Viticulture and Enology of Conegliano (TV).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Melelli, A.; Moretti, M. La fama del vino di Orvieto nel tempo—The Orvieto wine reputation over the time. In L’Orvieto. Studio per la Caratterizzazione Vitivinicola Dell’area a D.O.C. “Orvieto Classico” [The Orvieto. Study for the Wine Production Characterization of the “Orvieto Classico” D.O.C. Area]; Arusia: Città di Castello, Italy, 2004; pp. 13–30. [Google Scholar]
- Castagnoli, D. The wine-producing territory of Orvieto. J. Wine Res. 2013, 24, 253–263. [Google Scholar] [CrossRef]
- Desplanques, H. Campagne Umbre. Contributo allo Studio dei Paesaggi Rurali nell’Italia Centrale; Quattroemme: Perugia, Italy, 1975; ISBN 9788889398166. [Google Scholar]
- Unwin, T. Wine and the Vine: An Historical Geography of Viticulture and the Wine Trade; Routledge-Taylor & Francis Group: London, UK; New York, NY, USA, 1991; ISBN 0203013263. [Google Scholar]
- Gismondi, R. ISTAT Working Papers. Available online: www.istat.it/it/files/2021/01/IWP_8-2020.pdf (accessed on 27 June 2021).
- Baldeschi, G. I Vitigni e i Vini dell’Umbria. In Annuario Generale per la Viticoltura e l’Enologia-Anno II; Circolo Enofilo Italiano: Rome, Italy, 1893; pp. 26–38. [Google Scholar]
- Mattesini, E.; Ugoccioni, N. Vocabolario del Dialetto del Territorio Orvietano; Opera del Vocabolario Dialettale Umbro: Perugia, Italy, 1992. [Google Scholar]
- Panara, F.; Petoumenou, D.; Calderini, O.; Dini, F.; D’Onofrio, C.; Bedini, L.; Palliotti, A. Ampelographic and genetic characterisation of ancestral grapevine (Vitis vinifera L.) accessions present in the Umbria Region (Central Italy). J. Hortic. Sci. Biotechnol. 2013, 88, 525–530. [Google Scholar] [CrossRef]
- Anzani, R.; Failla, O.; Scienza, A.; Campostrini, F. Wild grapevine (Vitis vinifera var. silvestris) in Italy: Distribution, characteristics and germplasm preservation: 1989 report. Vitis 1990, 97–112. [Google Scholar] [CrossRef]
- Gregori, L. Soft Economy: Il Paesaggio del Vino in Umbria come Risorsa Geo-Turistica-Soft Economy: “The Wine Landscape” in the Umbria Region as a Geologic-Tourist Resource. Boll. AIC 2006, 185–202. [Google Scholar]
- Vitis International Variety Catalogue VIVC. Available online: https://www.vivc.de/ (accessed on 27 June 2021).
- Italian Catalogue of Grapevine Varieties-Catalogo Nazionale delle Varietà di Vite. Available online: http://catalogoviti.politicheagricole.it/catalogo.php (accessed on 27 June 2021).
- OIV-International Organisation of Vine and Wine. Second Edition of the OIV Descriptor List for Grape Varieties and Vitis Species. Available online: www.oiv.int/public/medias/2274/code-2e-edition-finale.pdf (accessed on 27 June 2021).
- D’Onofrio, C.; Tumino, G.; Gardiman, M.; Crespan, M.; Bignami, C.; de Palma, L.; Barbagallo, M.G.; Muganu, M.; Morcia, C.; Novello, V.; et al. Parentage Atlas of Italian Grapevine Varieties as Inferred From SNP Genotyping. Front. Plant Sci. 2021, 11, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Sneath, P.H.A.; Sokal, R.R. Numerical Taxonomy. The Principles and Practice of Numerical Classification; WH Freeman and Company: San Francisco, CA, USA, 1973; ISBN 0-7167-0697-0. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Maul, E.; Sudharma, K.N.; Kecke, S.; Marx, G.; Müller, C.; Audeguin, L.; Boselli, M.; Boursiquot, J.M.; Bucchetti, B.; Cabello, F.; et al. The European vitis database (www.eu-vitis.de)—A technical innovation through an online uploading and interactive modification system E. Vitis J. Grapevine Res. 2012, 51, 79–85. [Google Scholar] [CrossRef]
- Crespan, M. The parentage of Muscat of Hamburg. Vitis 2003, 42, 193–197. [Google Scholar] [CrossRef]
- Welter, L.J.; Göktürk-Baydar, N.; Akkurt, M.; Maul, E.; Eibach, R.; Töpfer, R.; Zyprian, E.M. Genetic mapping and localization of quantitative trait loci affecting fungal disease resistance and leaf morphology in grapevine (Vitis vinifera L). Mol. Breed. 2007, 20, 359–374. [Google Scholar] [CrossRef]
- Migliaro, D.; Morreale, G.; Gardiman, M.; Landolfo, S.; Crespan, M. Direct multiplex PCR for grapevine genotyping and varietal identification. Plant Genet. Resour. Characterisation Util. 2013, 11, 182–185. [Google Scholar] [CrossRef]
- Pelsy, F. Molecular and cellular mechanisms of diversity within grapevine varieties. Heredity (Edinb) 2010, 104, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Crespan, M.; Migliaro, D.; Larger, S.; Pindo, M.; Palmisano, M.; Manni, A. Grapevine (Vitis vinifera L.) varietal assortment and evolution in the Marche region (central Italy). OENO One 2021, 3, 17–37. [Google Scholar] [CrossRef]
- Crespan, M.; Armanni, A.; Da Rold, G.; De Nardi, B.; Gardiman, M.; Migliaro, D. ‘Verdello’, ‘Verdicchio’ and ‘Verduschia’: An example of integrated multidisciplinary study to clarify grapevine cultivar identity. Adv. Hortic. Sci. 2013, 26, 92–99. [Google Scholar] [CrossRef]
- Piñeiro, M.V. Storia Regionale della Vite e del Vino in Italia: Umbria; Volumnia: Perugia, Italy, 2012. [Google Scholar]
- Cosmo, I.; Bruni, B. Trebbiano Perugino. In Principali Vitigni da Vino Coltivati in Italia; Ministero dell’Agricoltura e delle Foreste: Rome, Italy, 1965. [Google Scholar]
- Cosmo, I. Montonico Bianco. In Principali Vitigni da Vino Coltivati in Italia; Ministero dell’Agricoltura e delle Foreste: Rome, Italy, 1965. [Google Scholar]
- Crespan, M.; Storchi, P.; Migliaro, D. Grapevine cultivar mantonico bianco is the second parent of the Sicilian Catarratto. Am. J. Enol. Vitic. 2017, 68, 258–262. [Google Scholar] [CrossRef]
- De Lorenzis, G.; Mercati, F.; Bergamini, C.; Cardone, M.F.; Lupini, A.; Mauceri, A.; Caputo, A.R.; Abbate, L.; Barbagallo, M.G.; Antonacci, D.; et al. SNP genotyping elucidates the genetic diversity of Magna Graecia grapevine germplasm and its historical origin and dissemination. BMC Plant Biol. 2019, 19, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Di Rovasenda, G. Saggio di Un’ampelografia Universale; Loescher: Torino, Italy, 1877. [Google Scholar]
- De Palma, L.; D’Onofrio, C.; De Michele, M.; Limosani, P.; Scalabrelli, G. Cannella Nera. Available online: http://vitisdb.it/varieties/show/1423 (accessed on 27 June 2021).
- Lacombe, T.; Boursiquot, J.M.; Laucou, V.; Di Vecchi-Staraz, M.; Péros, J.P.; This, P. Large-scale parentage analysis in an extended set of grapevine cultivars (Vitis vinifera L.). Theor. Appl. Genet. 2013, 126, 401–414. [Google Scholar] [CrossRef] [PubMed]
- Constantinescu, G. Ampelografia; Ministerul Agriculturii si Silviculturii, Editura Agro-Silvica da Stat: Bucurest, Romania, 1958. [Google Scholar]
- Bellin, D.; Peressotti, E.; Merdinoglu, D.; Wiedemann-Merdinoglu, S.; Adam-Blondon, A.F.; Cipriani, G.; Morgante, M.; Testolin, R.; Di Gaspero, G. Resistance to Plasmopara viticola in grapevine “Bianca” is controlled by a major dominant gene causing localised necrosis at the infection site. Theor. Appl. Genet. 2009, 120, 163–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pacifico, D.; Gaiotti, F.; Giust, M.; Tomasi, D. Performance of interspecific grapevine varieties in north-east Italy. Agric. Sci. 2013, 4, 91–101. [Google Scholar] [CrossRef] [Green Version]
- Alba, V.; Bergamini, C.; Genghi, R.; Gasparro, M.; Perniola, R.; Antonacci, D. Ampelometric Leaf Trait and SSR Loci Selection for a Multivariate Statistical Approach in Vitis vinifera L. Biodiversity Management. Mol. Biotechnol. 2015, 57, 709–719. [Google Scholar] [CrossRef]
- Crespan, M.; Calò, A.; Giannetto, S.; Sparacio, A.; Storchi, P.; Costacurta, A. “Sangiovese” and “Garganega” are two key varieties of the Italian grapevine assortment evolution. Vitis J. Grapevine Res. 2008, 47, 97–104. [Google Scholar] [CrossRef]
- Armanni, A.; Baldi, M.; Epifani, A.M.; Giannetti, F. Contributo alla valorizzazione del germoplasma viticolo: Valutazione di vitigni minori della provincia di Arezzo. Infowine Internet J. Vitic. Enol. 2012, 2/3, 1–7. [Google Scholar]
- Cosmo, I.; Dell’Olio, G.; Macaluso, R.; Ricci, P. Lambrusca di Alessandria. In Principali Vitigni da Vino Coltivati in Italia; Ministero dell’Agricoltura e delle Foreste: Rome, Italy, 1965. [Google Scholar]
- Raimondi, S.; Tumino, G.; Ruffa, P.; Boccacci, P.; Gambino, G.; Schneider, A. DNA-based genealogy reconstruction of Nebbiolo, Barbera and other ancient grapevine cultivars from northwestern Italy. Sci. Rep. 2020, 10, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Novello, V. Lo Stato Delle Conoscenze Viticole Sul ‘Barbera’. Infowine Internet J. Vitic. Enol. 2005, 2/2, 1–15. [Google Scholar]
- De Bosis, F. Lavori eseguiti dalla Commissione Ampelografica della provincia di Ravenna. In Bullettino Ampelografico; Tipografia Eredi Botta: Roma, Italy, 1879. [Google Scholar]
- Tartarotti, P.; Benciolini, L. Il vino più alto d’Europa: L’influenza della geologia e del clima nella coltura della vite in Valle d’Aosta. Boll. Della Soc. Geol. Ital. Suppl. 2006, 6, 107–114. [Google Scholar]
- Schneider, A.; Marinoni, D.T.; De Andrés, M.T.; Raimondi, S.; Cabello, F.; Ruffa, P.; Garcia-Muñoz, S.; Munoz-Organero, G. Prié Blanc and Legiruela: A unique grape cultivar grown in distant european regions. J. Int. Sci. Vigne Vin 2010, 44, 1–7. [Google Scholar] [CrossRef]
- Storchi, P.; Armanni, A.; Randellini, L.; Giannetto, S.; Meneghetti, S.; Crespan, M. Investigations on the identity of “Canaiolo bianco” and other white grape varieties of central Italy. Vitis J. Grapevine Res. 2011, 50, 59–64. [Google Scholar] [CrossRef]
- Di Vecchi Staraz, M.; Bandinelli, R.; Boselli, M.; This, P.; Boursiquot, J.M.; Laucou, V.; Lacombe, T.; Varès, D. Genetic structuring and parentage analysis for evolutionary studies in grapevine: Kin group and origin of the cultivar sangiovese revealed. J. Am. Soc. Hortic. Sci. 2007, 132, 514–524. [Google Scholar] [CrossRef]
- Maraš, V.; Bozovic, V.; Giannetto, S.; Crespan, M. SSR molecular marker analysis of the grapevine germplasm of montenegro. J. Int. Sci. Vigne Vin 2014, 48, 87–97. [Google Scholar] [CrossRef]
- Maraš, V.; Tello, J.; Gazivoda, A.; Mugoša, M.; Perišić, M.; Raičević, J.; Štajner, N.; Ocete, R.; Božović, V.; Popović, T.; et al. Population genetic analysis in old Montenegrin vineyards reveals ancient ways currently active to generate diversity in Vitis vinifera. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Kalinowski, S.T.; Taper, M.L.; Marshall, T.C. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007, 16, 1099–1106. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Huff, D.R.; Peakall, R.; Smouse, P.E. RAPD variation within and among natural populations of outcrossing buffalograss [Buchloë dactyloides (Nutt.) Engelm.]. Theor. Appl. Genet. 1993, 86, 927–934. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).