Abstract
An outbreak of bacterial soft rot and blackleg of potato has occurred since 2014 with the epicenter being in the northeastern region of the United States. Multiple species of Pectobacterium and Dickeya are causal agents, resulting in losses to commercial and seed potato production over the past decade in the Northeastern and North Central United States. To clarify the pathogen present at the outset of the epidemic in 2015 and 2016, a phylogenetic study was made of 121 pectolytic soft rot bacteria isolated from symptomatic potato; also included were 27 type strains of Dickeya and Pectobacterium species, and 47 historic reference strains. Phylogenetic trees constructed based on multilocus sequence alignments of concatenated dnaJ, dnaX and gyrB fragments revealed the epidemic isolates to cluster with type strains of D. chrysanthemi, D. dianthicola, D. dadantii, P. atrosepticum, P. brasiliense, P. carotovorum, P. parmentieri, P. polaris, P. punjabense, and P. versatile. Genetic diversity within D. dianthicola strains was low, with one sequence type (ST1) identified in 17 of 19 strains. Pectobacterium parmentieri was more diverse, with ten sequence types detected among 37 of the 2015–2016 strains. This study can aid in monitoring future shifts in potato soft rot pathogens within the U.S. and inform strategies for disease management.
1. Introduction
In 2020, potatoes were, by weight, the fifth most produced food crop globally, behind sugar cane, maize, wheat, and rice [1]. The United States was the fifth top country, producing 41.5 billion pounds of potatoes valued at $3.9 billion in 2020 [1,2]. Pectolytic soft rot diseases cause annual field and storage losses in potato production [3,4,5]. Severe outbreaks of soft rot in 2014 in particular led to yield and crop losses across the Northeastern and North Central U.S. [6].
Pectolytic soft rot bacteria can infect potato at any stage in production, from planting to post-harvest storage. Blackleg disease results from infections in mother tubers that spread through vascular tissue and eventually cause dark greasy lesions in the lower stem. Aerial soft rots can be caused by infections of fleshy above-ground tissues including stems and leaves. In potato production, soft rot usually refers to tuber decay in storage, but is also used in general to describe all forms of disease. Infested potatoes, water supplies, equipment, and storage facilities serve as inoculum sources [7]. Symptom development depends on environmental conditions favorable to host susceptibility and to pathogen growth and virulence [6,7,8,9].
Soft rot diseases of potato are caused by several bacterial taxa. Paine described the first known report of the disease in Great Britain [10]. The causal agent was identified as Bacillus atrosepticus and the descriptions of morphological characteristics and disease symptoms are almost exactly as described today. Several soft rot bacteria were later classified in the Enterobacteriaceae and named as Erwinia spp. Currently, most pectolytic soft rot bacteria affecting potato are classified in the genera Pectobacterium and Dickeya within the family Pectobacteriaceae [3,11]. Numerous species of Pectobacterium and Dickeya are of concern in potato production. The importance of each species varies. Some, like Dickeya solani van der Wolf et al. 2014 sp. nov., which emerged in Europe and caused severe losses for several years, have not yet been reported in the U.S. [12]. Zero tolerance laws and international quarantines were established to limit the spread of such highly virulent pathogens.
The severe 2014 outbreak of blackleg in Maine brought attention to the significance of soft rot diseases in the U.S. potato industry [13]. In 2016, increased losses due to blackleg were reported relative to 2015. In Maine, economic losses in the seed industry resulted from reduced seed emergence and seed disqualifications [14]. In New York, blackleg and aerial soft rot were found at multiple locations, with Long Island incurring significant yield losses [15].
To address continued concerns about the impacts of soft rot diseases on potato production and to increase awareness of the soft rot pathogens present in the U.S., a multi-state survey was conducted to identify the taxa of Pectobacterium and Dickeya present in the Northeastern and North Central U.S. during 2015–2016. Dickeya solani was not detected in any samples collected during the survey; however, the survey led to multiple state-level new, first reports of bacterial species in the region. Dickeya dianthicola [5] is now attributed to the devastating losses seen in Maine, New Jersey, and New York [5,13,15,16]. It has since been isolated in Texas and Hawaii [17,18]. Dickeya solani has not been isolated from any of the samples collected in 2015–2016. Pectobacterium parmentieri [19] emerged from the surveys as another pectolytic soft rot pathogen associated with recent aerial soft rot outbreaks in Maine, New York, Minnesota, North Dakota, and Michigan [15,19,20,21,22]. Although the species P. parmentieri was only recently described, it has been in Wisconsin since at least 2001 and is also present in Hawaii [19,23,24,25]. Other Pectobacterium species identified from the survey included P. atrosepticum and P. carotovorum in Maine, and P. brasiliense in Minnesota [15,26].
Since 2018, several additional valid species and amended names have been validly published or proposed for Dickeya and Pectobacterium [27,28,29,30,31,32,33,34,35,36,37]. To increase awareness of the currently recognized soft rot species present in the U.S., we conducted phylogenetic analyses of type strains of several newly named species of Dickeya and Pectobacterium and soft rot strains from the 2015–2016 survey. To gain insights on the possibility that some of the newly named species were present in older collections, strains isolated prior to 2014 were evaluated as references. The relatedness among strains of Dickeya dianthicola and of P. parmentieri isolated from different states was also assessed. Our findings confirm that soft rot diseases in the U.S potato industry are caused by a wide range of Dickeya and Pectobacterium species.
2. Materials and Methods
2.1. Bacterial Strains
Soft rot bacteria analyzed in this study included 113 strains isolated in 2015 and 2016 from symptomatic potato tissues collected in production and testing fields in Northeastern and North Central USA. Six of the 113 strains, CIR1009, CIR1011, CIR1058, CIR1182, CIR1183, and CIR1185, were obtained from decaying tubers in Minnesota. Eight strains isolated from pond water were also included, giving a total of 121 strains in the 2015–2016 collection (Table 1). The number of strains varied by state: Florida (1), Hawaii (5), Maine (11), Massachusetts (1), Michigan (2), Minnesota (50), New Jersey (1), New York (20), North Dakota (25), and Pennsylvania (5) (Table 1). Isolation and initial classification and identification of bacteria were conducted as previously described [13,15,22,38]. A total of 26 type strains were included in the study (Table 2). Cultures of type and pathotype strains of nine Dickeya spp. and six Pectobacterium spp. were obtained from the Belgian Co-Ordinated Collections of Micro-Organisms (BCCM/LMG) (Table 2). DNA sequence data of additional type strains of Dickeya spp. and Pectobacterium spp. were obtained from GenBank (Table 2). DNA or sequences of an additional 40 reference strains of Dickeya spp. and eight strains of Pectobacterium spp. were obtained from GenBank and ASAP [39], or provided by A. Charkowski from the A. Kelman collection or R.S. Dickey collection (Table 3).

Table 1.
Description and MLSA clade of Dickeya and Pectobacterium strains collected in 2015 and 2016 from production and testing areas associated with potato production in Northeastern and North Central U.S.

Table 2.
Description of type and pathotype strains used in this study.

Table 3.
Description of reference strains of Dickeya and Pectobacterium and MLSA clades predicted by dnaJ, dnaX, and gyrB multilocus sequence alignments.
2.2. DNA Extraction and Amplification
DNA extractions were performed using DNeasy Blood and Tissue kit (Qiagen). Three loci were targeted for analysis: dnaJ, dnaX, and gyrB [56]. PCR mixtures contained 5 μL GoTaq master mix (Promega, Madison, WI, USA), 0.5 μL dNTPs, 0.125 μL GoTaq polymerase (Promega), 1 μL each forward and reverse primer (10 μM), 16.875 μL sterile H2O, and 0.5 μL template DNA. Thermal cycler programs varied for each locus. For dnaX, amplification cycles included an initial denaturation for 3 min at 94 °C, 35 cycles consisting of 1 min at 94 °C, 1 min of annealing at 59 °C, a 2 min extension at 72 °C, and a final extension of 5 min at 72 °C. For dnaJ, cycle settings consisted of an initial denaturation of 3 min at 94 °C, 35 cycles of 30 s at 94 °C, 30 s annealing at 55 °C, 1 min extension at 72 °C, and a final extension of 10 min at 72 °C. For gyrB, the thermal cycler program included a 4 min initial denaturation at 94 °C, 35 cycles of 1 min at 94 °C, 1 min of annealing at 56 °C, 2 min extension at 72 °C, and a final extension of 10 min at 72 °C. PCR products were visualized on 1.0% TBE agarose gel stained with ethidium bromide and shipped to McLabs (San Francisco, CA, USA) for PCR purification and Sanger sequencing.
2.3. Multilocus Sequence Analysis (MLSA)
Nucleotide sequences obtained by direct amplification as described above and from GenBank, ASAP, and collaborators, were aligned, trimmed to a consistent length, and concatenated using CLC Main Workbench (Qiagen, Germantown, MD, USA). Fragment lengths for each gene locus were selected as previously described [56]. Sequence fragments and sequences were concatenated in the following order: dnaJ (672 bp), dnaX (450 bp), gyrB (822 bp for Dickeya spp., 711 bp for Pectobacterium spp.), with a total concatenated length of 1944 bp for Dickeya spp. and 1833 bp for Pectobacterium spp. Evolutionary model testing was run in CLC Main Workbench which determined the general time reversible model (GTR+G+T) to be the best fit model for this data set. Phylogenies were inferred via Bayesian analysis using Bayesian Evolutionary Analysis Sampling Trees (BEAST 1.8.2) assuming a strict molecular clock and 10 million generations [80]. Output from BEAST was analyzed in Tracer v1.6.0 [81] and phylogenetic trees were constructed in FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree, accessed on 11 August 2021).
2.4. Multilocus Sequence Typing (MLST)
Sequence types were assigned for 25 strains of D. dianthicola and 39 strains of P. parmentieri to further characterize diversity within these groups. Unique haplotypes for each locus and for concatenated sequences were identified using DnaSP v5 [82]. Minimum spanning trees were constructed in PHYLOViZ 2.0 to infer distance-based relativity of STs and predict founder STs within clonal complexes [83].
2.5. Nucleotide Accession Numbers
Sequences were deposited in GenBank and assigned to following accession numbers: MW978791 to MW979234.
3. Results
3.1. Phylogeny of 2015–2016 and Reference Strains of Dickeya spp.
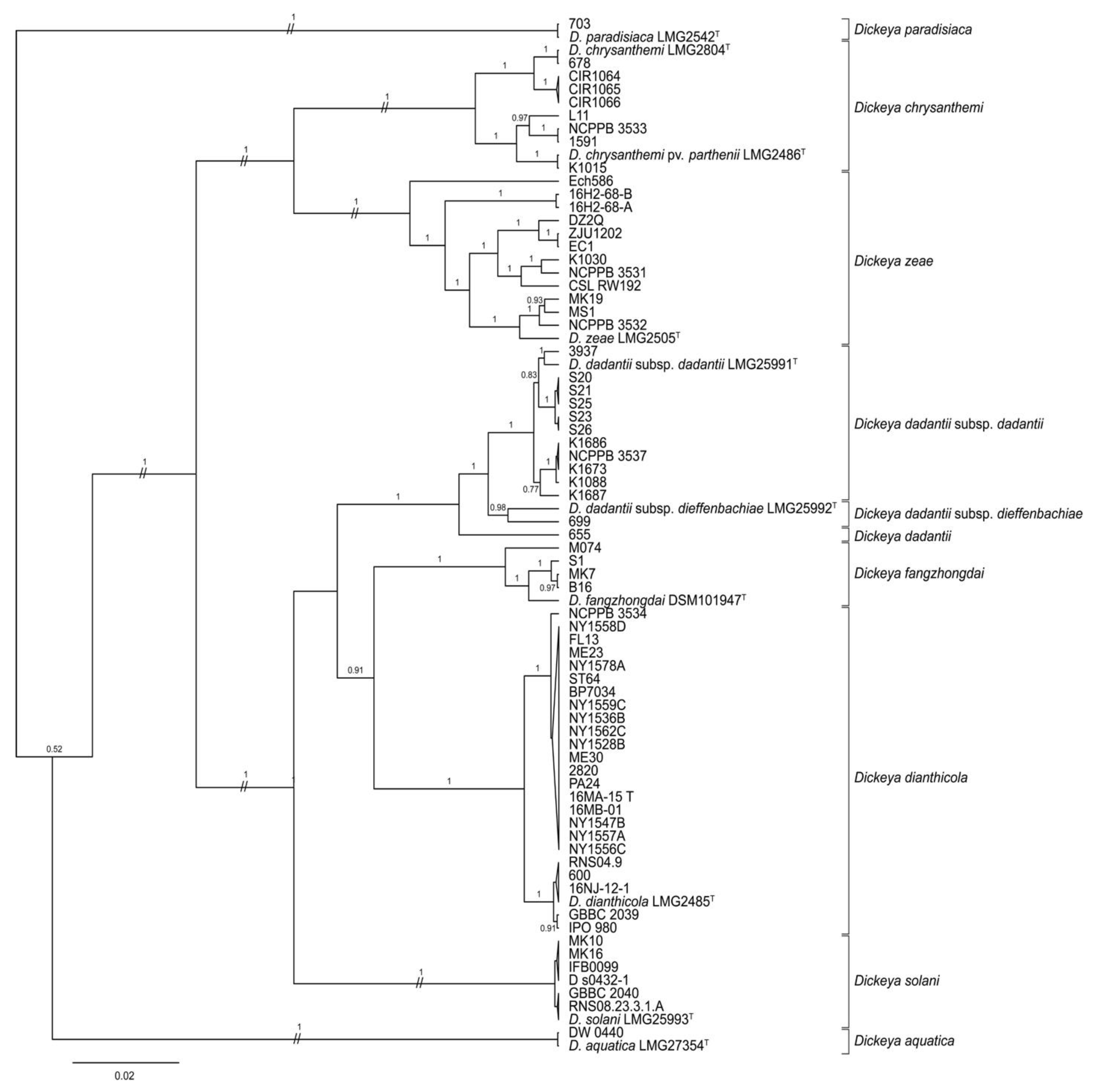
The phylogeny predicted by concatenated dnaJ, dnaX, and gyrB sequences placed several reference and 2015–2016 strains, some previously identified only to genus, within clades containing type strains of D. chrysanthemi, D. dianthicola, D. dadantii, and D. zeae (Figure 1; Table 1, Table 2 and Table 3). All strains identified previously as D. dianthicola were grouped in the same MLSA clade as the D. dianthicola type strain, LMG 2485T [13,15,22]. Seven strains in the 2015–2016 collection identified by their providers as Dickeya sp. Were also grouped with LMG 2485T, as was reference strain Dickeya sp. 600, which was isolated from a sweet potato in Georgia. Five potato strains from Hawaii were most closely related to the type strain of D. dadantii subsp. dadantii, as were four strains from maize. Two strains isolated from pond water adjacent to potato fields, PA24 and ST64, in the 2015–2016 collection were related to D. dianthicola, while two others, 16H2-68-A and 16H2-68-B were most closely related to D. zeae.
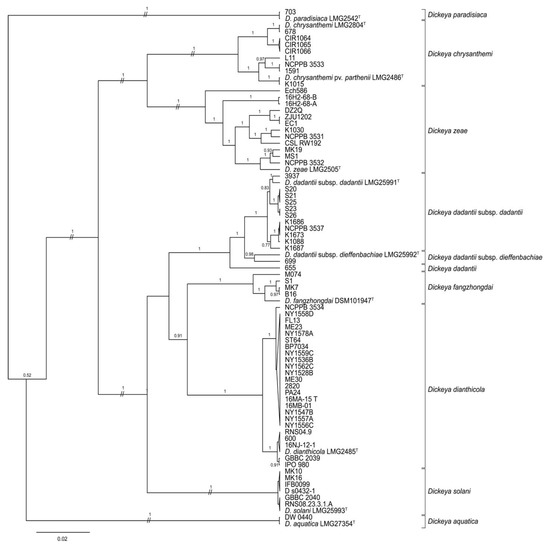
Figure 1.
Bayesian tree of Dickeya spp. constructed from concatenated sequences of fragments of the genes dnaJ, dnaX, and gyrB of 29 strains collected in 2015–2016, 10 type strains, and 40 historic reference strains. The superscript letter T indicates the species type strain. Posterior probabilities > 0.6 are shown at the corresponding node. Branch lengths are drawn to scale and represent sequence changes since the common ancestor.
Some reference strains, but none of the 2015–2016 strains, were grouped with type strains of D. aquatica, D. chrysanthemi pv. parthenii, D. solani, D. dadantii subsp. dieffenbachiae, D. fangzhongdai, or D. paradisiaca (Figure 1 and Table 1 and Table 3). Only D. aquatica DW 0440, a reference strain isolated from river water in Finland, was identified as D. aquatica [54]. In our analyses, one reference strain, Dickeya sp. 699, was identified as D. dadantii subsp. dieffenbachiae, while the subspecies status of D. dadantii strain 655 was ambiguous. Reference strain Dickeya sp. K1015 was most closely related to D. chrysanthemi pv. parthenii. The D. solani clade contained all six reference strains initially identified as D. solani: MK10, MK16, IFB 0099, GBBC 2040, D s0432-1, and RNS08.23.3.1.A. The MLSA phylogeny placed four reference strains recently classified as D. fangzhongdai within the same clade as the type strain of this species DSM 101947T (Figure 1, Table 3).
3.2. Phylogeny of 2015–2016 and Reference Strains of Pectobacterium spp.
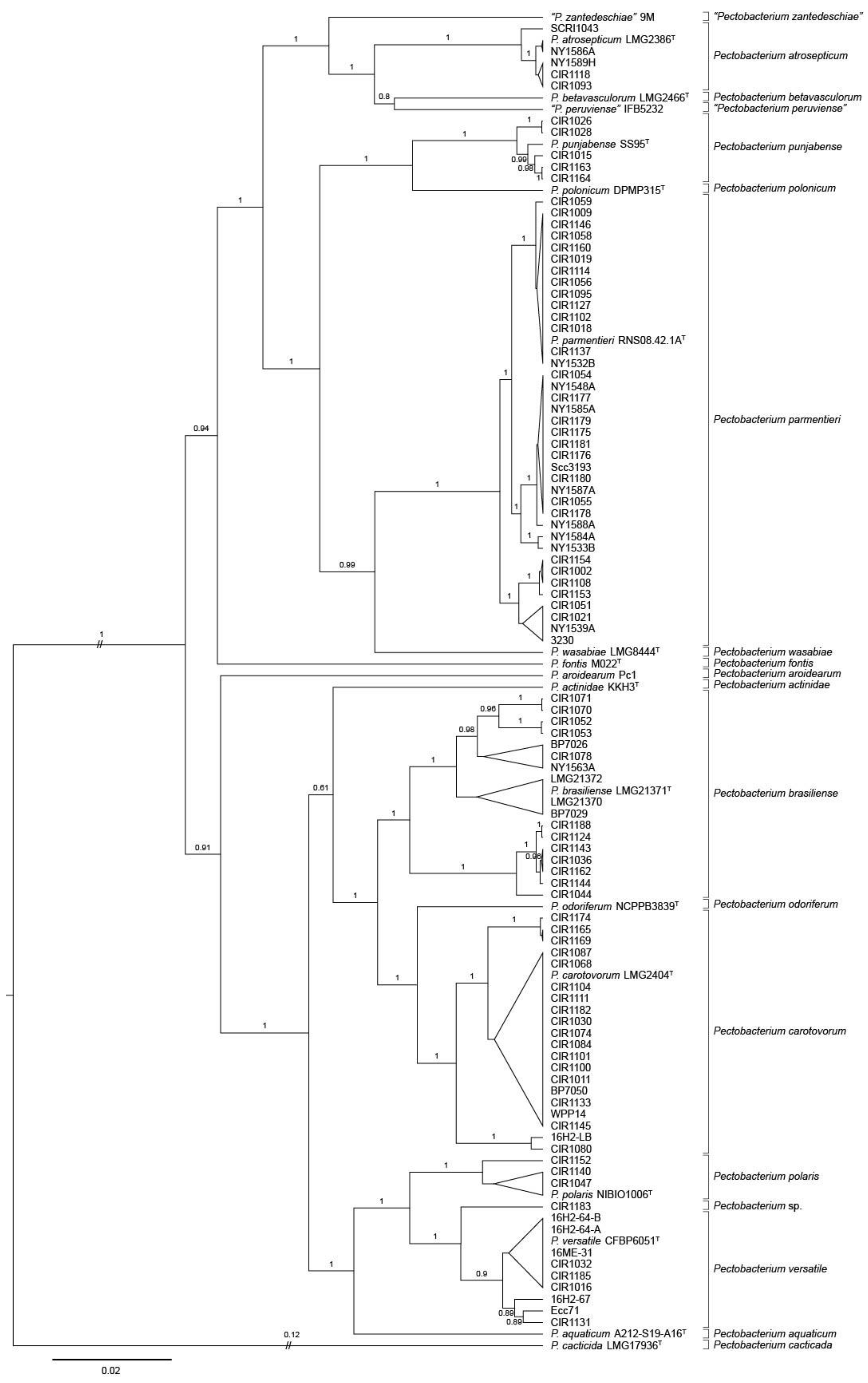
The phylogenetic tree of Pectobacterium placed several strains from the 2015–2016 and reference collections into well-supported clades containing the type strains of P. atrosepticum, P. brasiliense, P. carotovorum, and P. parmentieri (Figure 2). In addition, strains of the 2015–2016 collection were most closely related to type strains of P. polaris, P. punjabense, and P. versatile. Minnesota strains CIR1140, CIR1152 and CIR1152 from potato were grouped with P. polaris NIBIO 1006T. Five strains (CIR1015, CIR1026, CIR1028, CIR1163, CIR1164), all from potato originating in Minnesota or North Dakota, were most closely related to type strain P. punjabense SS95T. P. versatile was the closest relative of three water strains from Maine (16H2-64-A, 16H2-64-B, 16ME-31) and four potato strains isolated in Maine, Minnesota, and North Dakota. Five strains isolated from potato tubers in Minnesota clustered with type strains of either P. versatile, P. parmentieri, or P. carotovorum. The other tuber isolate, CIR1183 was most closely related to P. versatile but was on a separate branch nearly as divergent from P. versatile as that of P. polaris and P. versatile. None of the 2015–2016 strains were found in MLSA clades corresponding to P. actinidae, P. aquatica, P. aroidearum, P. betavasculorum, P. cacticida, P. fontis, P. odoriferum, P. peruviense, P. polonicum, P. wasabiae, or P. zantedeschiae.
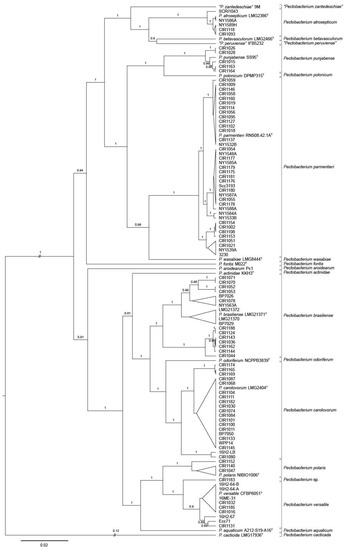
Figure 2.
Bayesian tree of Pectobacterium spp. constructed from concatenated sequences of fragments of the genes dnaJ, dnaX, and gyrB of 92 strains collected in 2015–2016, 17 type strains, and seven historic reference strains. The superscript letter T indicates the species type strain. Posterior probabilities > 0.6 are shown at the corresponding node. Branch lengths are drawn to scale and represent sequence changes since the common ancestor.
Some of the major phylogenetic clades predicted by concatenated dnaJ, dnaX, and gyrB sequence alignments were further subdivided into well-supported sub-lineages. This was especially evident in P. brasiliense and P. carotovorum, which contained sub-branching of greater divergence than detected within other Pectobacterium species. Two major branches of P. brasiliense were evident. One, which was further subdivided into four lineages, contained the type strain, eight strains from the 2015–2016 collection, and reference strains LMG 21370 and LMG 21372. The other was comprised of seven strains collected in 2015 and 2016 from Minnesota or North Dakota. Three groups of P. carotovorum were detected. Two strains, 16H2-LB and CIR 1080 were located on a branch that was distinct from the major clade containing P. carotovorum LMG 2402T (Figure 2, Table 1).
3.3. Diversity within Dickeya dianthicola
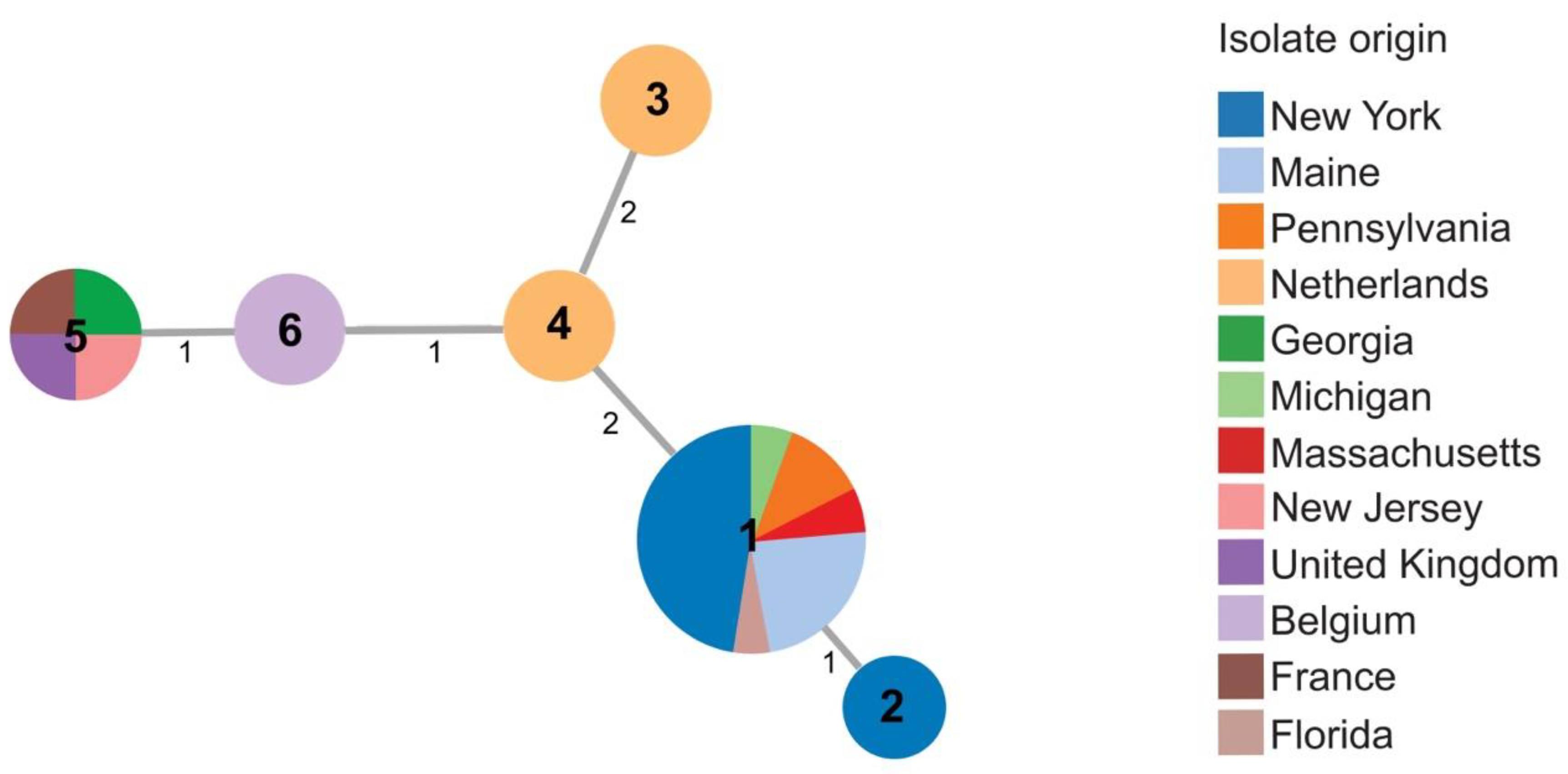
Twenty-five Dickeya dianthicola isolates were included in this study, with six STs identified (Table S1). Most (17/19) D. dianthicola strains from the 2015–2016 collection were assigned sequence type 1 (ST1). ST1 strains came from Maine, New York, Pennsylvania, Massachusetts, and Florida. None of the strains from Netherlands, France, Belgium, or the United Kingdom were ST1. ST2 was found in only one strain isolated in 2016 from New York. The sweet potato strain, 600, from R. S. Dickey’s collection, previously identified as Dickeya sp. and identified here as D. dianthicola, shared the same sequence type (ST5) as New Jersey strain 16NJ-12 1, and strains RNS0.4.9 and LMG 2485T from France and from the United Kingdom, respectively (Figure 3 and Table S1).
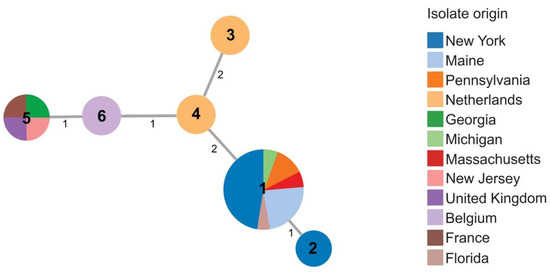
Figure 3.
Minimal spanning tree showing relatedness of 25 strains of D. dianthicola including 19 strains obtained in 2015–2016. Each circle represents a unique sequence type (ST) derived from the concatenated sequences of three housekeeping genes (dnaJ, dnaX, and gyrB). Sequence type numbers are given in black in each circle. Sizes of circles are relative to the number of individuals sharing the same ST. The relatedness between strains is indicated by relative distance as indicated by branch lengths. Geographic origins of strains are depicted by color: New York (blue); Maine (light blue); Pennsylvania (orange); Netherlands (light orange); Georgia (green); Michigan (light green); Massachusetts (red); New Jersey (pink); United Kingdom (purple); Belgium (light purple); France (brown); Florida (light brown).
3.4. Diversity within Pectobacterium parmentieri
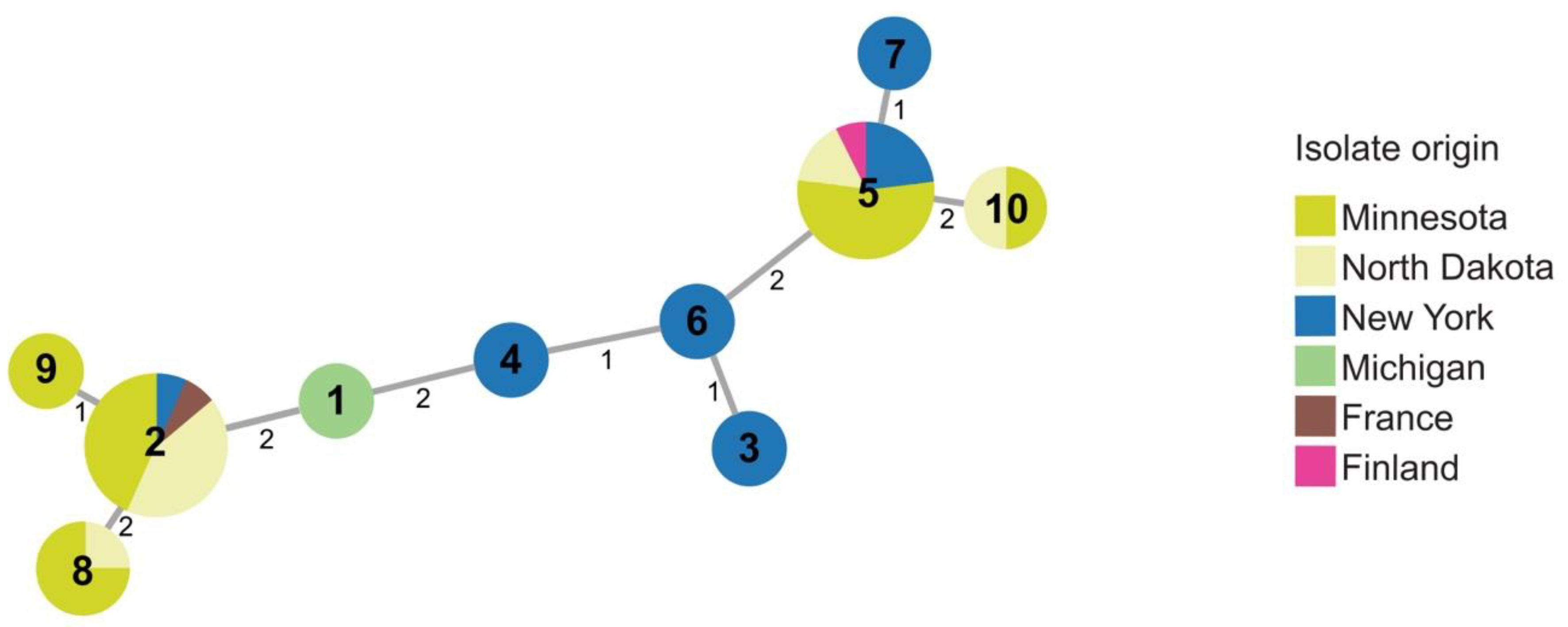
Several (37) strains in the 2015–2016 collection clustered with type strain, RNS08.42.1A, of P. parmentieri, including strains from Michigan, Minnesota, New York, and North Dakota. Ten sequence types were identified within P. parmentieri (Figure 4 and Table S2). P. parmentieri RNS08.42.1AT from France was assigned ST2. ST2 was also detected in Minnesota, New York, and North Dakota. Strains with ST5 originated in Finland, Minnesota, New York, and North Dakota. Multiple sequence types were found in some states. For example, strains from Minnesota and New York separated into five and six sequence types, respectively. While some STs, such as ST2 and ST5, were found in multiple states, ST8 and ST10 appeared only in Minnesota and North Dakota, and ST1, ST3, ST4, ST6, ST7, and ST9 were unique to particular states.
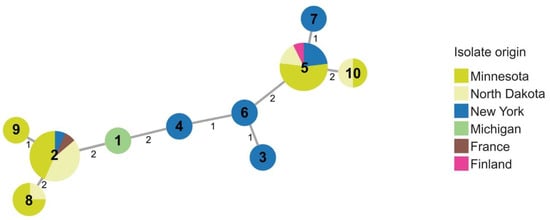
Figure 4.
Minimal spanning tree showing relatedness of 39 strains of P. parmentieri including 37 strains obtained in 2015–2016. Each circle represents a sequence type based on concatenated STs from three housekeeping genes (dnaJ, dnaX, and gyrB). Sequence type numbers are given in black in each circle. Sizes of circles are relative to the number of individuals sharing the same ST. The relatedness between strains is indicated by relative distance as indicated by branch lengths. Geographic origins of strains are depicted by color: Minnesota (lime green); North Dakota (yellow); New York (blue); Michigan (green); France (brown); Finland (pink).
4. Discussion
The taxonomy of Pectobacterium and Dickeya has been refined substantially in the past ten years by the widespread adaption of DNA sequencing strategies in soft rot research. Classification and identification of soft rot bacteria by multilocus sequence alignments of concatenated sequences and by whole genome sequence comparisons has added clarification to the complex polyphyletic nature of some earlier-described species of Pectobacterium and Dickeya. In this study, all strains of Pectobacterium and Dickeya previously identified to species level by whole genome sequencing grouped with the corresponding type strains in phylogeny predicted by MLSA of dnaJ, dnaX, and gyrB. The clades and phylogenies predicted in this study using concatenated sequences of dnaJ, dnaX, and gyrB are also similar to those generated by single and multiple sequence alignments [15,35,37,51,56,58,63,84,85,86]. The consistency among tree topologies generated by single, multiple, or entire genome sequences enables presumptive identification of unknown soft rot bacteria to species and provides insights on the diversity, ecology, and epidemiology of specific taxa [6,54,85,87,88].
The inclusion of several reference and type strains in our MLSA study provided new insights on species diversity of soft rot bacteria prior to the 2014 blackleg outbreak. MLSA was sufficient for identification of several reference strains previously identified only to the genus level. Reference strains identified as Dickeya sp. in Kelman’s and Dickey’s collections were assigned to MLSA clades D. chrysanthemi, D. dadantii subsp. dadantii, D. dadantii subsp. dieffenbachia, D. dianthicola, and D. zeae. The placement of the sweet potato strain Dickeya sp. 600 within D. dianthicola is evidence that D. dianthicola was present in the USA prior to the 2014 blackleg outbreak. The four strains identified as D. fangzhongdai in the reference collection all originated outside the U.S. D. fangzhongdai has very recently been identified in New York as the cause of soft rot of onion [89]. Historic isolates of Pectobacterium versatile have been reported from the U.S. In a retrospective study of soft rot bacteria, Portier et al. identified four historic strains from the USA as P. versatile: three from potato isolated in 2001 and one from Iris sp. isolated in 1973 [37]. Reference strain Pectobacterium sp. Ecc71 was recently reclassified as P. versatile [30]. In our study, Ecc71 grouped with other P. versatile strains. The taxon includes soft rot strains isolated in Russia from potato and cabbage unofficially named “Candidatus Pectobacterium maceratum” [30,90].
MLSA and MLST of the 2015–2016 collection provides a broad view of the species of pectolytic soft rot bacteria present in the Northeastern and North Central U.S. immediately following the 2014 blackleg outbreak. The identification of P. parmentieri, P. brasiliense, P. carotovorum, and P. atrosepticum is consistent with prior reports of the occurrences of these species in the U.S. [15,20,21,23,24,26]. We did not expect to find nine strains of P. versatile in the 2015–2016 collection. P. versatile was recently reported in potato from New York and appears to represent a shift in 2017 from the P. parmentieri that predominated in 2016 [91]. Less expected was finding P. polaris and P. punjabense, as these have not been reported in the U.S. P. polaris was validly reported in 2017 and is found in Norway [52] and Morocco [85]. P. punjabense was reported in Pakistan [36]. No strains of P. polaris or P. punjabense originating from the U.S. were noted in the original species description or in recent examinations of other species of Pectobacterium [30,31,35,37,52,84]. Further polyphasic analyses are warranted to validate the MLSA identification of P. polaris or P. punjabense within the 2015–2016 collection.
The 2015–2016 collection contained D. dianthicola strains originating from multiple states. D. dianthicola has been reported previously in New York, Maine, Michigan, and New Jersey [6,13,15,16,22]. Our findings confirmed that D. dianthicola is also present in Florida, Massachusetts, and Pennsylvania. The majority (17/19) of D. dianthicola isolates in the 2015–2016 collection from Maine, New York, Pennsylvania, Massachusetts, and Florida are sequence type 1 (ST1). This result might be expected if soft rot outbreaks in the Northeastern U.S. were caused by a single (or limited) introduction of one strain of D. dianthicola. However, isolates of ST2 and ST5, from New York and New Jersey, respectively, were also identified within the 2015–2016 collection, which suggests the outbreak was not only clonal. A genetic diversity analysis of 256 isolates of D. dianthicola collected over 5 years concluded that the blackleg outbreak in Northeastern U.S. was caused by multiple strains [41]. Ge et al., 2021 identified three genotypes within D. dianthicola, the frequencies of which varied by year and by state [41]. Some D. dianthicola isolates reported by Ge et al. were also included in our study. Based on overlapping results, ST1 in our study corresponds well to Type I of Ge et al., 2021, while ST 2–6 strains are represented in Type II and III [41].
The 2015–2016 collection is not comprehensive, in that the number of strains from the contributing states varied and no systematic sampling strategy was applied for obtaining isolates over states. The variation in coverage by state resulted in different findings. For example, the deep coverage of strains from Minnesota and North Dakota led to first reports of some species and the likely identification of P. polaris and P. punjabense in the U.S. The abundance of strains of D. dianthicola and P. parmentieri in the collection enabled sequence type analyses, from which we can conclude that the genetic diversity within P. parmentieri was greater than that of D. dianthicola during those years and across states.
Most strains in the 2015-2016 collection originated from plants. The eight strains from water were enough to show that water can serve as a source of D. dianthicola, D. zeae, P. actinidae, and P. versatile. In a large comprehensive study of Dickeya in temperate regions of Central Europe, the diversity of Dickeya species recovered from water was different than that obtained from potato [45]. In that study, D. dianthicola was not recovered from water. Future studies involving larger numbers of aquatic strains could improve our understanding of the relative diversity of soft rot species in water and plant sources in the U.S. [6,13,15,16,22,41].
Four newly described species of Dickeya were validly published as we completed our analyses [27,29,86,92]. Future studies with two of these, D. oryzae and D. poaceiphila, would aid in resolution of isolates within the MLSA clade D. zeae. Since D. oryzae ZYY5T was not included in our study, we did not delineate a clade of D. oryzae. NCPPB 3531, CSL RW192, DZ2Q, ZJU 1202, EC1, and the reference strain K1030 from maize clustered together in the MLSA phylogeny presented here. All these strains are currently identified as D. oryzae [86]. We also note that two water strains from Maine, 16H2-68-A and 16H2-68-B, and the reference strain, Ech586, are divergent of other strains within the D. zeae clade. This might reflect the natural diversity within D. zeae or an unresolved species assignment. In a phylogenetic study that included NCPPB 569, the type strain of D. poaceiphila [92] and the reference strain Ech586, Ech586 was placed in D. zeae [31,93]. It is likely that the water strains are also correctly placed within D. zeae; however, because D. zeae, D. poaceiphila, and D. chrysanthemi are closely related species within Dickeya, future studies of water strains 16H2-68-A and 16H2-68-B including the type strain of D. poaceiphila could verify their species assignment.
P. parvum is the only validly published and currently used species name of Pectobacterium not included in our study [84]. The inclusion of P. parvum might have provided insights on the identity of Pectobacterium sp. CIR1183, the 2016 isolate from Minnesota, which by dnaJ, dnaX, and gyrB phylogeny represented a separate branch within the P. polaris/P. versatile clade. The closest relatives of P. parvum are P. polaris and P. versatile. Isolates of P. parvum were described as a group of atypical, less virulent strains closely related to, but distinguished from P. polaris [84]. In the future, whole genomic sequence comparisons, evaluation of differentiating biochemical traits, toxicity tests in insects, and soft rot virulence assays that include the type strain of P. parvum might enable more accurate placement of Pectobacterium sp. CIR1183 within the P. versatile/P. polaris/P. parvum super clade.
While much progress has been made toward defining a stable, monophyletic taxonomy for species of Dickeya and Pectobacterium, some taxa remain notably complex and certain strains of soft rot bacteria do not align well with known species [37,63,84]. Two strains, classified here as P. carotovorum, CIR1080 and 16H2-LB, clustered together on a separate branch of P. carotovorum. These might belong to other species related to P. carotovorum. Similarly, the diverging branches of P. brasiliense strains within the 2015–2016 collection and reference strains is consistent with the conclusion that P. brasiliense is less homogenous than other species of Pectobacterium [87].
Phylogenetic relationships predicted by the MLSA schema of dnaJ, dnaX, and gyrB support and extend recent evidence that several newly described species of Dickeya and Pectobacterium cause soft rot diseases of potato in the U.S. The importance of specific species on disease occurrence, symptomology, and severity remains unclear. MLSA/MLST studies could be extended to study the contributions of within- and between-location genetic variability, multiple species and genotype combinations, and cultivar on disease severity. Comprehensive genomic comparisons of large numbers of representative strains within a given species will continue to advance stable, well-delineated species definitions with the goal of improving pathogen detection and disease management [6,41,87].
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microorganisms9081733/s1, Table S1: Haplotypes of dnaJ, dnaX, and gyrB and sequence types (ST) of strains within the MLSA clade of Dickeya dianthicola, Table S2: Allelic variation in dnaJ, dnaX, and gyrB and sequence types (ST) of strains within the MLSA clade of Pectobacterium parmentieri.
Author Contributions
Conceptualization and methodology, formal analysis, investigation, and writing—original draft preparation, C.A.I. and R.D.C.; resources and writing—review and editing, A.M., K.L.P., J.H., A.O.C., C.T.B., R.R.M., S.B.J., N.R., G.A.S., R.P.L., B.K.G.; supervision, project administration, and funding acquisition, C.A.I. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by United States Department of Agriculture/Agricultural Research Service State Partnership Potato Program 8030-62660-003-00D. Funding for the project came from Minnesota Department of Agriculture/USDA Farm Bill, Northern Plains Potato Growers Association, Cavendish Farms, MN Area II Potato Research and Promotion Council, and Minnesota Agricultural Experiment Station, and the United States Department of Agriculture National Institute of Food and Agriculture Federal Appropriations (USDA-NIFA), United States Department of Agriculture/Agricultural Research Service State Partnership Potato Program 8030-62660-003-00D, Pennsylvania Department of Agriculture, and USDA-NIFA-Hatch ME022010 through the Maine Agricultural and Forest Experiment Station, and Maine Agricultural and Forest Experiment Publication Number 3827.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank Blake Webster and Hilary Snyder for technical assistance and A. P. Robinson, K. Sather, and J. Miller for providing soft rot samples from commercial and seed potato fields in Minnesota and North Dakota. The helpful discussions and agency of Kromroy and J. Ciborowski, Minnesota Department of Agriculture, are especially acknowledged. Thank you to the Pennsylvania Co-Operative Potato Growers for connecting growers with symptomatic potatoes to researchers.
Conflicts of Interest
The authors declare no conflict of interest.
References
- FAO. FAOSTAT Statistical Database; Food and Agricultural Organization of the United Nations: Rome, Italy, 2020.
- NASS. Agricultural Statistics Board; United States Department of Agriculture (USDA), National Agricultural Statistics Service: Washington, DC, USA, 2021. [Google Scholar]
- Hauben, L.; Moore, E.R.; Vauterin, L.; Steenackers, M.; Mergaert, J.; Verdonck, L.; Swings, J. Phylogenetic Position of Phytopathogens within the Enterobacteriaceae. Syst. Appl. Microbiol. 1998, 21, 384–397. [Google Scholar] [CrossRef]
- Gardan, L.; Gouy, C.; Christen, R.; Samson, R. Elevation of Three Subspecies of Pectobacterium carotovorum to Species Level: Pectobacterium atrosepticum sp. nov., Pectobacterium betavasculorum sp. nov. and Pectobacterium wasabiae sp. nov. Int. J. Syst. Evol. Microbiol. 2003, 53, 381–391. [Google Scholar] [CrossRef]
- Samson, R.; Legendre, J.B.; Christen, R.; Saux, M.F.; Achouak, W.; Gardan, L. Transfer of Pectobacterium chrysanthemi (Burkholder Et Al. 1953) Brenner Et Al. 1973 and Brenneria paradisiaca to the genus Dickeya gen. nov. as Dickeya chrysanthemi comb. nov. and Dickeya paradisiaca comb. nov. and Delineation of Four Novel Species, Dickeya dadantii sp. nov., Dickeya dianthicola sp. nov., Dickeya dieffenbachiae sp. nov. and Dickeya zeae sp. nov. Int. J. Syst. Evol. Microbiol. 2005, 55, 1415–1427. [Google Scholar] [PubMed]
- Charkowski, A.O. The Changing Face of Bacterial Soft-Rot Diseases. Annu. Rev. Phytopathol. 2018, 56, 269–288. [Google Scholar] [CrossRef]
- Czajkowski, R.; Pérombelon, M.C.M.; van Veen, J.A.; van der Wolf, J.M. Control of Blackleg and Tuber Soft Rot of Potato Caused by Pectobacterium and Dickeya Species: A Review. Plant Pathol. 2011, 60, 999–1013. [Google Scholar] [CrossRef]
- Glasner, J.D.; Marquez-Villavicencio, M.; Kim, H.S.; Jahn, C.E.; Ma, B.; Biehl, B.S.; Rissman, A.I.; Mole, B.; Yi, X.; Yang, C.H.; et al. Niche-Specificity and the Variable Fraction of the Pectobacterium Pan-Genome. Mol. Plant-Microbe Interact. 2008, 21, 1549–1560. [Google Scholar] [CrossRef]
- Golanowska, M.; Potrykus, M.; Motyka-Pomagruk, A.; Kabza, M.; Bacci, G.; Galardini, M.; Bazzicalupo, M.; Makalowska, I.; Smalla, K.; Mengoni, A.; et al. Comparison of Highly and Weakly Virulent Dickeya solani Strains, with a View on the Pangenome and Panregulon of This Species. Front. Microbiol. 2018, 9, 1940. [Google Scholar] [CrossRef]
- Paine, S.G. Studies in Bacteriosis: I. “Blackleg” of the Potato. J. Agric. Sci. 1917, 8, 480. [Google Scholar] [CrossRef]
- Adeolu, M.; Alnajar, S.; Naushad, S.; Gupta, R.S. Genome-Based Phylogeny and Taxonomy of the ’Enterobacteriales’: Proposal for Enterobacterales ord. nov. Divided into the Families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 5575–5599. [Google Scholar]
- Van der Wolf, J.M.; Nijhuis, E.H.; Kowalewska, M.J.; Saddler, G.S.; Parkinson, N.; Elphinstone, J.G.; Pritchard, L.; Toth, I.K.; Lojkowska, E.; Potrykus, M.; et al. Dickeya solani sp. nov., a Pectinolytic Plant-Pathogenic Bacterium Isolated from Potato (Solanum tuberosum). Int. J. Syst. Evol. Microbiol. 2014, 64, 768–774. [Google Scholar] [CrossRef]
- Jiang, H.H.; Hao, J.J.; Johnson, S.B.; Brueggeman, R.S.; Secor, G. First Report of Dickeya dianthicola Causing Blackleg and Bacterial Soft Rot on Potato in Maine. Plant Dis. 2016, 100, 2320. [Google Scholar] [CrossRef]
- Johnson, S.B. Novel Approach for Identification of Potato Seed Lots Suspected to Be Effected by Dickeya. Plant Pathol. J. 2017, 16, 96–100. [Google Scholar] [CrossRef]
- Ma, X.; Schloop, A.; Swingle, B.; Perry, K.L. Pectobacterium and Dickeya Responsible for Potato Blackleg Disease in New York State in 2016. Plant Dis. 2018, 102, 1834–1840. [Google Scholar] [CrossRef] [PubMed]
- Patel, N.; Baldwin, A.C.; Patel, R.D.; Kobayashi, D.Y.; Wyenandt, C.A. First Report of Dickeya dianthicola Causing Blackleg and Soft Rot on Potato (Solanum tuberosum) in New Jersey, USA. Plant Dis. 2019, 103, 146. [Google Scholar] [CrossRef]
- Nasaruddin, A.S.; Charkowski, A.O.; Babler, B.N.; Perna, N.T.; Glasner, J.D. First Report of Dickeya dianthicola Causing Blackleg on Potato in Texas. Plant Dis. 2019, 103, 2121. [Google Scholar] [CrossRef]
- Boluk, G.; Arif, M. First Report of Dickeya dianthicola as a Causal Agent of Bacterial Soft Rot of Potato in Hawaii. Plant Dis. 2019, 103, 2943. [Google Scholar] [CrossRef]
- Khayi, S.; Cigna, J.; Chong, T.M.; Quêtu-Laurent, A.; Chan, K.G.; Hélias, V.; Faure, D. Transfer of the Potato Plant Isolates of Pectobacterium wasabiae to Pectobacterium parmentieri sp. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 5379–5383. [Google Scholar] [CrossRef]
- Ge, T.L.; Jiang, H.H.; Hao, J.J.; Johnson, S.B. First Report of Pectobacterium parmentieri Causing Bacterial Soft Rot and Blackleg on Potato in Maine. Plant Dis. 2018, 102, 437. [Google Scholar] [CrossRef]
- McNally, R.R.; Curland, R.D.; Webster, B.T.; Robinson, A.P.; Ishimaru, C.A. First Report of Blackleg and Tuber Soft Rot of Potato Caused by Pectobacterium parmentieri in Minnesota and North Dakota. Plant Dis. 2017, 101, 2144. [Google Scholar] [CrossRef]
- Rosenzweig, N.; Steere, L.; Kirk, W.W.; Mambetova, S.; Long, C.; Schafer, R.; Dangi, S.; Byrne, J. First Report of Dickeya dianthicola and Pectobacterium wasabiae Causing Aerial Stem Rot of Potato in Michigan, USA. New. Dis. Rep. 2016, 33, 10. [Google Scholar] [CrossRef]
- Arizala, D.; Dobhal, S.; Paudel, S.; Gunarathne, S.; Boluk, G.; Arif, M. First Report of Bacterial Soft Rot and Blackleg on Potato Caused by Pectobacterium parmentieri in Hawaii. Plant Dis. 2020, 104, 970. [Google Scholar] [CrossRef]
- Ma, B.; Hibbing, M.E.; Kim, H.S.; Reedy, R.M.; Yedidia, I.; Breuer, J.; Breuer, J.; Glasner, J.D.; Perna, N.T.; Kelman, A.; et al. Host Range and Molecular Phylogenies of the Soft Rot Enterobacterial Genera Pectobacterium and Dickeya. Phytopathology 2007, 97, 1150–1163. [Google Scholar] [CrossRef]
- Kim, H.S.; Ma, B.; Perna, N.T.; Charkowski, A.O. Phylogeny and Virulence of Naturally Occurring Type Iii Secretion System-Deficient Pectobacterium Strains. Appl. Environ. Microbiol. 2009, 75, 4539–4549. [Google Scholar] [CrossRef]
- McNally, R.R.; Curland, R.D.; Webster, B.T.; Robinson, A.P.; Ishimaru, C.A. First Report of Pectobacterium carotovorum subsp. Brasiliensis Causing Blackleg and Stem Rot in Commercial and Seed Potato Fields in Minnesota and North Dakota. Plant Dis. 2017, 101, 1672. [Google Scholar] [CrossRef]
- Oulghazi, S.; Pédron, J.; Cigna, J.; Lau, Y.Y.; Moumni, M.; Van Gijsegem, F.; Chan, K.-G.; Faure, D. Dickeya undicola sp. nov., a Novel Species for Pectinolytic Isolates from Surface Waters in Europe and Asia. Int. J. Syst. Evol. Microbiol. 2019, 69, 2440–2444. [Google Scholar] [CrossRef] [PubMed]
- Duprey, A.; Taib, N.; Leonard, S.; Garin, T.; Flandrois, J.-P.; Nasser, W.; Brochier-Armanet, C.; Reverchon, S. The Phytopathogenic Nature of Dickeya aquatica 174/2 and the Dynamic Early Evolution of Dickeya Pathogenicity. Environ. Microbiol. 2019, 21, 2809–2835. [Google Scholar] [CrossRef]
- Hugouvieux-Cotte-Pattat, N.; Jacot-des-Combes, C.; Briolay, J. Dickeya lacustris sp. nov., a Water-Living Pectinolytic Bacterium Isolated from Lakes in France. Int. J. Syst. Evol. Microbiol. 2019, 69, 721–726. [Google Scholar] [CrossRef]
- Portier, P.; Pédron, J.; Taghouti, G.; Fischer-Le Saux, M.; Caullireau, E.; Bertrand, C.; Laurent, A.; Chawki, K.; Oulgazi, S.; Moumni, M.; et al. Elevation of Pectobacterium carotovorum subsp. Odoriferum to Species Level as Pectobacterium odoriferum sp. nov., Proposal of Pectobacterium brasiliense sp. nov. and Pectobacterium actinidiae sp. nov., Emended Description of Pectobacterium carotovorum and Description of Pectobacterium versatile sp. nov., Isolated from Streams and Symptoms on Diverse Plants. Int. J. Syst. Evol. Microbiol. 2019, 69, 3207–3216. [Google Scholar] [PubMed]
- Pédron, J.; Bertrand, C.; Taghouti, G.; Portier, P.; Barny, M.-A. Pectobacterium aquaticum sp. nov., Isolated from Waterways. Int. J. Syst. Evol. Microbiol. 2019, 69, 745–751. [Google Scholar] [CrossRef]
- Oulghazi, S.; Cigna, J.; Lau, Y.Y.; Moumni, M.; Chan, K.G.; Faure, D. Transfer of the Waterfall Source Isolate Pectobacterium carotovorum M022 to Pectobacterium fontis sp. nov., a Deep-Branching Species within the Genus Pectobacterium. Int. J. Syst. Evol. Microbiol. 2019, 69, 470–475. [Google Scholar] [CrossRef]
- Waleron, M.; Misztak, A.; Waleron, M.; Franczuk, M.; Wielgomas, B.; Waleron, K. Transfer of Pectobacterium carotovorum Subsp. Carotovorum Strains Isolated from Potatoes Grown at High Altitudes to Pectobacterium peruviense sp. nov. Syst. Appl. Microbiol. 2018, 41, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Waleron, M.; Misztak, A.; Waleron, M.; Franczuk, M.; Jonca, J.; Wielgomas, B.; Mikiciński, A.; Popović, T.; Waleron, K. Pectobacterium zantedeschiae sp. nov. A New Species of a Soft Rot Pathogen Isolated from Calla Lily (Zantedeschia Spp.). Syst. Appl. Microbiol. 2019, 42, 275–283. [Google Scholar] [CrossRef]
- Waleron, M.; Misztak, A.; Waleron, M.; Jonca, J.; Furmaniak, M.; Waleron, K. Pectobacterium polonicum sp. nov. Isolated from Vegetable Fields. Int. J. Syst. Evol. Microbiol. 2019, 69, 1751–1759. [Google Scholar] [CrossRef]
- Sarfraz, S.; Riaz, K.; Oulghazi, S.; Cigna, J.; Sahi, S.T.; Khan, S.H.; Faure, D. Pectobacterium punjabense sp. nov., Isolated from Blackleg Symptoms of Potato Plants in Pakistan. Int. J. Syst. Evol. Microbiol. 2018, 68, 3551–3556. [Google Scholar] [CrossRef] [PubMed]
- Portier, P.; Pédron, J.; Taghouti, G.; Dutrieux, C.; Barny, M.A. Updated Taxonomy of Pectobacterium Genus in the Cirm-Cfbp Bacterial Collection: When Newly Described Species Reveal “Old” Endemic Population. Microorganisms 2020, 8, 1441. [Google Scholar] [CrossRef] [PubMed]
- McNally, R.R.; Curland, R.D.; Webster, B.T.; Robinson, A.P.; Ishimaru, C.A. First Report of Stem Rot on Potato Caused by Dickeya chrysanthemi in Minnesota. Plant Dis. 2018, 102, 238. [Google Scholar] [CrossRef]
- Glasner, J.D.; Liss, P.; Plunkett, I.G.; Darling, A.; Prasad, T.; Rusch, M.; Byrnes, A.; Gilson, M.; Biehl, B.; Blattner, F.R.; et al. Asap, a Systematic Annotation Package for Community Analysis of Genomes. Nucleic Acids Res. 2003, 31, 147–151. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Perna, N.T.; Glasner, J.D.; Hao, J.; Johnson, S.; Nasaruddin, A.S.; Charkowski, A.O.; Wu, S.; Fei, Z.; Perry, K.L.; et al. Complete Genome Sequence of Dickeya dianthicola Me23, a Pathogen Causing Blackleg and Soft Rot Diseases of Potato. Microbiol. Res. Announc. 2019, 8, e01526-18. [Google Scholar] [CrossRef] [PubMed]
- Ge, T.; Jiang, H.; Johnson, S.B.; Larkin, R.; Charkowski, A.O.; Secor, G.; Hao, J. Genotyping Dickeya dianthicola Causing Potato Blackleg and Soft Rot Outbreak Associated with Inoculum Geography in the United States. Plant Dis. 2021. [Google Scholar] [CrossRef]
- Parkinson, N.; DeVos, P.; Pirhonen, M.; Elphinstone, J. Dickeya aquatica sp. nov., Isolated from Waterways. Int. J. Syst. Evol. Microbiol. 2014, 64, 2264–2266. [Google Scholar] [CrossRef] [PubMed]
- Van Vaerenbergh, J.; Baeyen, S.; De Vos, P.; Maes, M. Sequence Diversity in the Dickeya flic Gene: Phylogeny of the Dickeya Genus and Taqman (R) Pcr for ’D. Solani’, New Biovar 3 Variant on Potato in Europe. PLoS ONE 2012, 7, e35738. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, L.; Humphris, S.; Saddler, G.S.; Elphinstone, J.G.; Pirhonen, M.; Toth, I.K. Draft Genome Sequences of 17 Isolates of the Plant Pathogenic Bacterium Dickeya. Genome Announc. 2013, 1, e00978-13. [Google Scholar] [CrossRef] [PubMed]
- Potrykus, M.; Golanowska, M.; Sledz, W.; Zoledowska, S.; Motyka, A.; Kolodziejska, A.; Butrymowicz, J.; Lojkowska, E. Biodiversity of Dickeya spp. Isolated from Potato Plants and Water Sources in Temperate Climate. Plant Dis. 2016, 100, 408–417. [Google Scholar] [CrossRef]
- Brady, C.L.; Cleenwerck, I.; Denman, S.; Venter, S.N.; Rodríguez-Palenzuela, P.; Coutinho, T.A.; De Vos, P. Proposal to Reclassify Brenneria quercina (Hildebrand and Schroth 1967) Hauben Et Al. 1999 into a New Genus, Lonsdalea gen. nov., as Lonsdalea quercina comb. nov., Descriptions of Lonsdalea quercina Subsp. Quercina comb. nov., Lonsdalea quercina subsp. Iberica subsp. nov. and Lonsdalea quercina subsp. Britannica subsp. nov., Emendation of the Description of the Genus Brenneria, Reclassification of Dickeya dieffenbachiae as Dickeya dadantii subsp. Dieffenbachiae comb. nov., and Emendation of the Description of Dickeya dadantii. Int. J. Syst. Evol. Microbiol. 2012, 62, 1592–1602. [Google Scholar] [PubMed]
- Tian, Y.; Zhao, Y.; Yuan, X.; Yi, J.; Fan, J.; Xu, Z.; Hu, B.; De Boer, S.H.; Li, X. Dickeya fangzhongdai sp. nov., a Plant-Pathogenic Bacterium Isolated from Pear Trees (Pyrus pyrifolia). Int. J. Syst. Evol. Microbiol. 2016, 66, 2831–2835. [Google Scholar] [CrossRef]
- Khayi, S.; Blin, P.; Chong, T.M.; Chan, K.-G.; Faure, D. Complete Genome Anatomy of the Emerging Potato Pathogen Dickeya solani Type Strain Ipo 2222(T). Stand. Genom. Sci. 2016, 11, 87–93. [Google Scholar] [CrossRef]
- Koh, Y.J.; Kim, G.H.; Lee, Y.S.; Sohn, S.H.; Koh, H.S.; Kwon, S.; Heu, S.; Jung, J.S. Pectobacterium carotovorum subsp. Actinidiae subsp. nov., a New Bacterial Pathogen Causing Canker-Like Symptoms in Yellow Kiwifruit, Actinidia chinensis. N. Z. J. Crop Hort. Sci. 2012, 40, 269–279. [Google Scholar] [CrossRef][Green Version]
- Thomson, S.V.; Hildebrand, D.C.; Schroth, M.N. Identificaiton and Nutritional Differentiation of the Erwinia Sugar Beet Pathogen from Members of Erwinia carotovora and Erwinia chrysanthemi. Phytopathology 1981, 71, 1037–1042. [Google Scholar] [CrossRef]
- Pritchard, L.; Glover, R.H.; Humphris, S.; Elphinstone, J.G.; Toth, I.K. Genomics and Taxonomy in Diagnostics for Food Security: Soft-Rotting Enterobacterial Plant Pathogens. Anal. Meth. 2016, 8, 12–24. [Google Scholar] [CrossRef]
- Dees, M.W.; Lysøe, E.; Rossmann, S.; Perminow, J.; Brurberg, M.B. Pectobacterium polaris sp. nov., Isolated from Potato (Solanum tuberosum). Int. J. Syst. Evol. Microbiol. 2017, 67, 5222–5229. [Google Scholar] [CrossRef] [PubMed]
- De Boer, S.H.; Copeman, R.J.; Vruggink, H. Serogroups of Erwinia carotovora Potato Strains Determined with Diffusible Somatic Antigens. Phytopathology 1979, 69, 316–319. [Google Scholar] [CrossRef]
- Laurila, J.; Ahola, V.; Lehtinen, A.; Joutsjoki, T.; Hannukkala, A.; Rahkonen, A.; Pirhonen, M. Characterization of Dickeya strains Isolated from Potato and River Water Samples in Finland. Eur. J. Plant Pathol. 2008, 122, 213–225. [Google Scholar] [CrossRef]
- Chan, K.-G.; Kher, H.-L.; Chang, C.-Y.; Yin, W.-F.; Tan, K.-H. Analysis of Pectate Lyase Genes in Dickeya chrysanthemi Strain L11, Isolated from a Recreational Lake in Malaysia: A Draft Genome Sequence Perspective. Genome Announc. 2015, 3, e00145-15. [Google Scholar] [CrossRef]
- Marrero, G.; Schneider, K.L.; Jenkins, D.M.; Alvarez, A.M. Phylogeny and Classification of Dickeya Based on Multilocus Sequence Analysis. Int. J. Syst. Evol. Microbiol. 2013, 63, 3524–3539. [Google Scholar] [CrossRef]
- Glasner, J.D.; Yang, C.H.; Reverchon, S.; Hugouvieux-Cotte-Pattat, N.; Condemine, G.; Bohin, J.P.; Van Gijsegem, F.; Yang, S.; Franza, T.; Expert, D.; et al. Genome Sequence of the Plant-Pathogenic Bacterium Dickeya dadantii 3937. J. Bacteriol. 2011, 193, 2076–2077. [Google Scholar] [CrossRef] [PubMed]
- Cigna, J.; Dewaegeneire, P.; Beury, A.; Gobert, V.; Faure, D. A Gapa Pcr-Sequencing Assay for Identifying the Dickeya and Pectobacterium Potato Pathogens. Plant Dis. 2017, 101, 1278–1282. [Google Scholar] [CrossRef]
- Pritchard, L.; Humphris, S.; Baeyen, S.; Maes, M.; Van Vaerenbergh, J.; Elphinstone, J.; Saddler, G.; Toth, I. Draft Genome Sequences of Four Dickeya dianthicola and Four Dickeya solani Strains. Genome Announc. 2013, 1, e00087-12. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, L.; Humphris, S.; Saddler, G.S.; Parkinson, N.M.; Bertrand, V.; Elphinstone, J.G.; Toth, I.K. Detection of Phytopathogens of the Genus Dickeya Using a Pcr Primer Prediction Pipeline for Draft Bacterial Genome Sequences. Plant Pathol. 2013, 62, 587–596. [Google Scholar] [CrossRef]
- Raoul des Essarts, Y.; Mondy, S.; Hélias, V.; Faure, D. Genome Sequence of the Potato Plant Pathogen Dickeya dianthicola Strain Rns04.9. Genome Announc. 2015, 3, e00581-15. [Google Scholar] [PubMed]
- Alič, Š.; Pédron, J.; Dreo, T.; Van Gijsegem, F. Genomic Characterisation of the New Dickeya fangzhongdai Species Regrouping Plant Pathogens and Environmental Isolates. BMC Genom. 2019, 20, 34–52. [Google Scholar] [CrossRef]
- Zhang, Y.; Fan, Q.; Loria, R. A Re-Evaluation of the Taxonomy of Phytopathogenic Genera Dickeya and Pectobacterium Using Whole-Genome Sequencing Data. Syst. Appl. Microbiol. 2016, 39, 252–259. [Google Scholar] [CrossRef]
- Alič, Š.; Naglič, T.; Llop, P.; Toplak, N.; Koren, S.; Ravnikar, M.; Dreo, T. Draft Genome Sequences of Dickeya sp. Isolates B16 (Nib Z 2098) and S1 (Nib Z 2099) Causing Soft Rot of Phalaenopsis Orchids. Genome Announc. 2015, 3, e00973-15. [Google Scholar] [CrossRef] [PubMed]
- Golanowska, M.; Galardini, M.; Bazzicalupo, M.; Hugouvieux-Cotte-Pattat, N.; Mengoni, A.; Potrykus, M.; Slawiak, M.; Lojkowska, E. Draft Genome Sequence of a Highly Virulent Strain of the Plant Pathogen Dickeya solani, Ifb0099. Genome Announc. 2015, 3, e00109–e00115. [Google Scholar] [CrossRef]
- Garlant, L.; Koskinen, P.; Rouhiainen, L.; Laine, P.; Paulin, L.; Auvinen, P.; Holm, L.; Pirhonen, M. Genome Sequence of Dickeya solani, a New Soft Rot Pathogen of Potato, Suggests Its Emergence May Be Related to a Novel Combination of Non-Ribosomal Peptide/Polyketide Synthetase Clusters. Diversity 2013, 5, 824–842. [Google Scholar] [CrossRef]
- Khayi, S.; Mondy, S.; Beury-Cirou, A.; Moumni, M.; Hélias, V.; Faure, D. Genome Sequence of the Emerging Plant Pathogen Dickeya solani Strain Rns 08.23.3.1a. Genome Announc. 2014, 2, e01270-13. [Google Scholar]
- Bertani, I.; Passos da Silva, D.; Abbruscato, P.; Piffanelli, P.; Venturi, V. Draft Genome Sequence of the Plant Pathogen Dickeya zeae Dz2q, Isolated from Rice in Italy. Genome Announc. 2013, 1, e00905–e00913. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Cheng, Y.; Lv, M.; Liao, L.; Chen, Y.; Gu, Y.; Liu, S.; Jiang, Z.; Xiong, Y.; Zhang, L. The Complete Genome Sequence of Dickeya zeae Ec1 Reveals Substantial Divergence from Other Dickeya Strains and Species. BMC Genom. 2015, 16, 571–586. [Google Scholar] [CrossRef] [PubMed]
- Hussain, M.B.B.M.; Zhang, H.-B.; Xu, J.-L.; Liu, Q.; Jiang, Z.; Zhang, L.-H. The Acyl-Homoserine Lactone-Type Quorum-Sensing System Modulates Cell Motility and Virulence of Erwinia chrysanthemi pv. Zeae. J. Bacteriol. 2008, 190, 1045–1053. [Google Scholar] [CrossRef]
- Lin, B.R.; Shen, H.F.; Pu, X.M.; Tian, X.S.; Zhao, W.J.; Zhu, S.F.; Dong, M.M. First Report of a Soft Rot of Banana in Mainland China Caused by a Dickeya sp. (Pectobacterium chrysanthemi). Plant Dis. 2010, 94, 640. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-X.; Lin, B.-R.; Shen, H.-F.; Pu, X.-M. Genome Sequence of the Banana Pathogen Dickeya zeae Strain Ms1, which Causes Bacterial Soft Rot. Genome Announc. 2013, 1, e00317-13. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Shi, Y.; Ibrahim, M.; Liu, H.; Shan, C.; Wang, Y.; Kube, M.; Xie, G.-L.; Sun, G. Genome Sequence of the Rice Pathogen Dickeya zeae Strain Zju1202. J. Bacteriol. 2012, 194, 4452–4453. [Google Scholar] [CrossRef]
- Nabhan, S.; De Boer, S.H.; Maiss, E.; Wydra, K. Pectobacterium aroidearum sp. nov., a Soft Rot Pathogen with Preference for Monocotyledonous Plants. Int. J. Syst. Evol. Microbiol. 2013, 63, 2520–2525. [Google Scholar] [CrossRef] [PubMed]
- Bell, K.S.; Sebaihia, M.; Pritchard, L.; Holden, M.T.G.; Hyman, L.J.; Holeva, M.C.; Thomson, N.R.; Bentley, S.D.; Churcher, L.J.C.; Mungall, K.; et al. Genome Sequence of the Enterobacterial Phytopathogen Erwinia carotovora subsp. Atroseptica and Characterization of Virulence Factors. Proc. Natl. Acad. Sci. USA 2004, 101, 11105–11110. [Google Scholar] [CrossRef] [PubMed]
- Duarte, V.; De Boer, S.H.; Ward, L.J.; de Oliveira, A.M.R. Characterization of Atypical Erwinia carotovora Strains Causing Blackleg of Potato in Brazil. J. Appl. Microbiol. 2004, 96, 535–545. [Google Scholar] [CrossRef]
- Yap, M.-N.; Barak, J.D.; Charkowski, A.O. Genomic Diversity of Erwinia carotovora subsp. Carotovora and Its Correlation with Virulence. Appl. Environ. Microbiol. 2004, 70, 3013–3023. [Google Scholar] [CrossRef]
- Koskinen, J.P.; Laine, P.; Niemi, O.; Nykyri, J.; Harjunpää, H.; Auvinen, P.; Paulin, L.; Pirhonen, M.; Palva, T.; Holm, L. Genome Sequence of Pectobacterium sp. Strain Scc3193. J. Bacteriol. 2012, 194, 6004. [Google Scholar] [CrossRef] [PubMed]
- Pirhonen, M.; Heino, P.; Helander, I.; Harju, P.; Palva, E.T. Bacteriophage T4 Resistant Mutants of the Plant Pathogen Erwinia carotovora. Microb. Pathogen. 1988, 4, 359–367. [Google Scholar] [CrossRef]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian Phylogenetics with Beauti and the Beast 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef]
- Rambaut, A.; Suchard, M.A.; Xie, D.; Drummond, A.J. Tracer V1.6. Available online: https://bioweb.pasteur.fr/packages/pack@Tracer@v1.6 (accessed on 11 August 2021).
- Rozas, J.; Sánchez-DelBarrio, J.C.; Messeguer, X.; Rozas, R. Dnasp, DNA Polymorphism Analyses by the Coalescent and Other Methods. Bioinformatics 2003, 19, 2496–2497. [Google Scholar] [CrossRef] [PubMed]
- Francisco, A.P.; Vaz, C.; Monteiro, P.T.; Melo-Cristino, J.; Ramirez, M.; Carriço, J.A. Phyloviz: Phylogenetic Inference and Data Visualization for Sequence Based Typing Methods. BMC Bioinform. 2012, 13, 87. [Google Scholar] [CrossRef]
- Pasanen, M.; Waleron, M.; Schott, T.; Cleenwerck, I.; Misztak, A.; Waleron, K.; Pritchard, L.; Bakr, R.; Degefu, Y.; van der Wolf, J.; et al. Pectobacterium parvum sp. nov., Having a Salmonella Spi-1-Like Type Iii Secretion System and Low Virulence. Int. J. Syst. Evol. Microbiol. 2020, 70, 2440–2448. [Google Scholar] [CrossRef]
- Oulghazi, S.; Moumni, M.; Khayi, S.; Robic, K.; Sarfraz, S.; Lopez-Roques, C.; Vandecasteele, C.; Faure, D. Diversity of Pectobacteriaceae Species in Potato Growing Regions in Northern Morocco. Microorganisms 2020, 8, 895–911. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; He, S.W.; Guo, H.B.; Han, J.G.; Thin, K.K.; Gao, J.S.; Wang, Y.; Zhang, X.X. Dickeya oryzae sp. nov., Isolated from the Roots of Rice. Int. J. Syst. Evol. Microbiol. 2020, 70, 4171–4178. [Google Scholar] [CrossRef] [PubMed]
- Oulghazi, S.; Sarfraz, S.; Zaczek-Moczydłowska, M.A.; Khayi, S.; Ed-Dra, A.; Lekbach, Y.; Campbell, K.; Novungayo Moleleki, L.; O’Hanlon, R.; Faure, D. Pectobacterium brasiliense: Genomics, Host Range and Disease Management. Microorganisms 2021, 9, 106–132. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ma, Y.; Liang, S.; Tian, Y.; Yin, S.; Xie, S.; Xie, H. Comparative Genomics of 84 Pectobacterium Genomes Reveals the Variations Related to a Pathogenic Lifestyle. BMC Genom. 2018, 19, 889–911. [Google Scholar] [CrossRef]
- Ma, X.; Bonasera, J.M.; Asselin, J.A.E.; Beer, S.V.; Swingle, B. First Report of Dickeya fangzhongdai Causing Soft Rot of Onion in New York State. Plant Dis. 2020, 104, 1251. [Google Scholar] [CrossRef]
- Shirshikov, F.V.; Korzhenkov, A.A.; Miroshnikov, K.K.; Kabanova, A.P.; Barannik, A.P.; Ignatov, A.N.; Miroshnikov, K.A. Draft Genome Sequences of New Genomospecies “Candidatus Pectobacterium Maceratum” Strains, which Cause Soft Rot in Plants. Genome Announc. 2018, 6, e00260-18. [Google Scholar] [CrossRef]
- Ma, X.; Stodghill, P.; Gao, M.; Perry, K.L.; Swingle, B. Identification of Pectobacterium versatile Causing Blackleg of Potato in New York State. Plant Dis. 2021. [Google Scholar] [CrossRef] [PubMed]
- Hugouvieux-Cotte-Pattat, N.; Brochier-Armanet, C.; Flandrois, J.-P.; Reverchon, S. Dickeya poaceiphila sp. nov., a Plant-Pathogenic Bacterium Isolated from Sugar Cane (Saccharum officinarum). Int. J. Syst. Evol. Microbiol. 2020, 70, 4508–4514. [Google Scholar] [CrossRef]
- Pédron, J.; Van Gijsegem, F. Diversity in the Bacterial Genus Dickeya Grouping Plant Pathogens and Waterways Isolates. OBM Genet. 2019, 3, 22. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).