Modeling the Effects of the Environment and the Host Plant on the Ripe Rot of Grapes, Caused by the Colletotrichum Species
Abstract
1. Introduction
2. Results
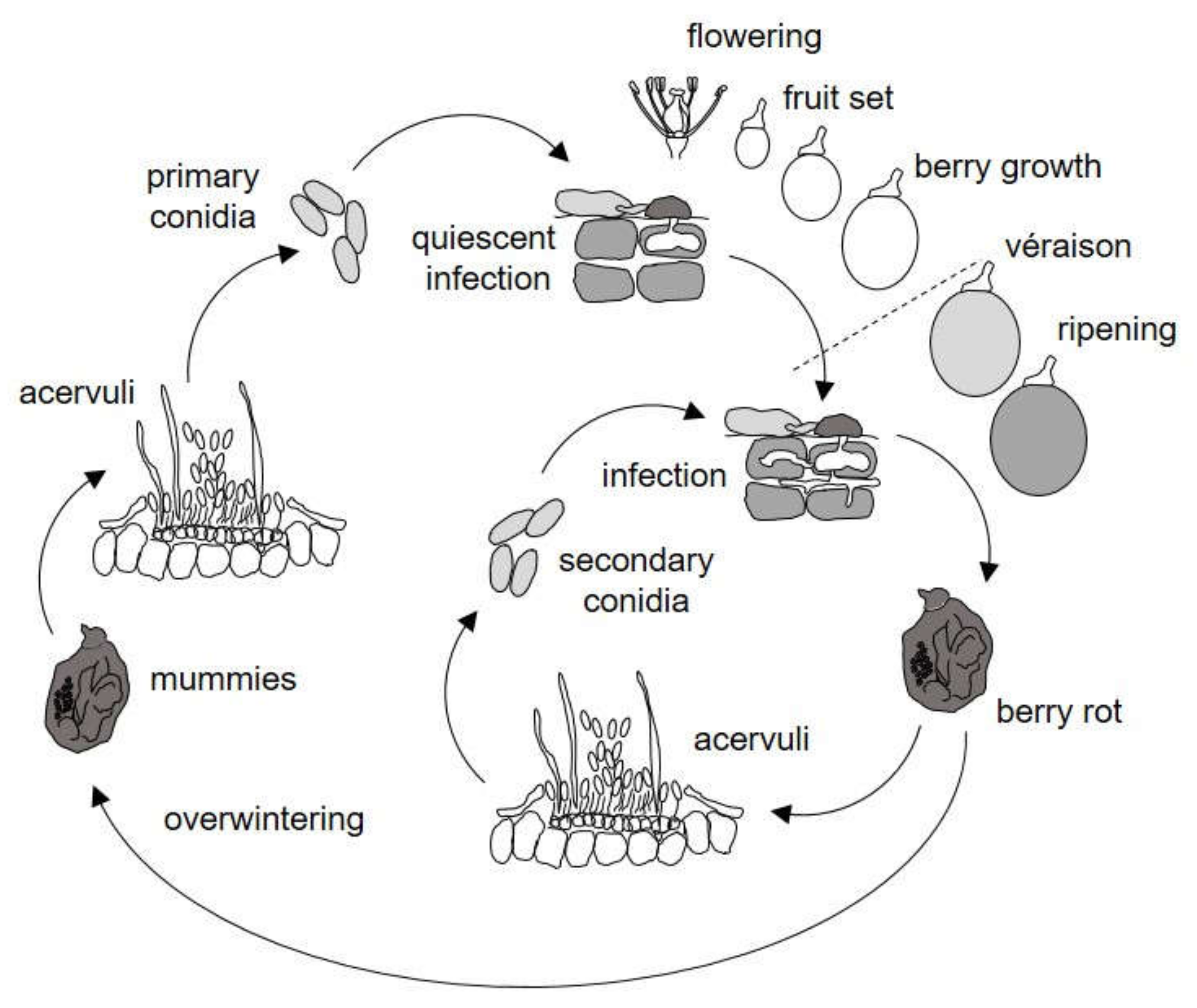
2.1. Model Description
2.1.1. Production and Maturation of Primary Inoculum
2.1.2. Spore Dispersal
2.1.3. Primary Infection Events
2.1.4. Production of Secondary Inoculum
2.1.5. Secondary Infection Events
2.2. Model Evaluation
2.2.1. Predictions and Disease Incidence
2.2.2. Prediction of Infection Events and Disease Progress
3. Discussion
4. Materials and Method
4.1. Literature Search
4.2. Systems Analysis and Model Development
4.3. Data for Model Evaluation
4.4. Data Analysis
4.4.1. Predictions of Disease Incidence
4.4.2. Prediction of Infection Events
4.4.3. Prediction of Disease Progress
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Meunier, M.; Steel, C.C. Effect of Colletotrichum acutatum ripe rot on the composition and sensory attributes of cabernet sauvignon grapes and wine. Aust. J. Grape Wine Res. 2009, 15, 223–227. [Google Scholar] [CrossRef]
- Hong, S.K.; Kim, W.G.; Yun, H.K.; Choi, K.J. Morphological variations, genetic diversity and pathogenicity of Colletotrichum species causing grape ripe rot in Korea. Plant Pathol. J. 2008, 24, 269–278. [Google Scholar] [CrossRef]
- Southworth, E.A. Ripe rot of grapes and apples. J. Mycol. 1981, 6, 164–173. [Google Scholar] [CrossRef]
- Milholland, R.D. Ripe rot. In Compendium of Grape Diseases; Pearson, R.C., Goheen, A.C., Eds.; American Phytopathological Society: Saint Paul, MN, USA, 1988; pp. 23–24. [Google Scholar]
- Greer, L.A.; Harper, J.D.I.; Savocchia, S.; Samuelian, S.K.; Steel, C.C. Ripe rot of south-eastern Australian wine grapes is caused by two species of Colletotrichum: C. acutatum and C. gloeosporioides with differences in infection and fungicide sensitivity. Aust. J. Grape Wine Res. 2011, 17, 123–128. [Google Scholar] [CrossRef]
- Huang, X.Q.; Kong, F.F.; Wang, Z.Y. Grape ripe rot. In Crop Diseases and Insect Pests in China; Guo, Y.Y., Wu, K.M., Chen, W.Q., Eds.; China Agricultural Press: Beijing, China, 2015; pp. 809–813. [Google Scholar]
- Damm, U.; Cannon, P.F.; Woudenberg, J.H.C.; Crous, P.W. The Colletotrichum acutatum species complex. Stud. Mycol. 2012, 73, 37–113. [Google Scholar] [CrossRef]
- Weir, B.S.; Johnston, P.R.; Damm, U. The Colletotrichum gloeosporioides species complex. Stud. Mycol. 2012, 73, 115–180. [Google Scholar] [CrossRef]
- Yan, J.Y.; Jayawardena, M.M.R.S.; Goonasekara, I.D.; Wang, Y.; Zhang, W.; Liu, M.; Huang, J.B.; Wang, Z.Y.; Shang, J.J.; Peng, Y.L.; et al. Diverse species of Colletotrichum associated with grapevine anthracnose in China. Fungal Divers. 2015, 71, 233–246. [Google Scholar] [CrossRef]
- Kim, J.S.; Hassan, O.; Go, M.; Chang, T. First report of Colletotrichum aenigma causing anthracnose of grape in Korea. Plant Dis. 2021. In Press. [Google Scholar] [CrossRef]
- Sawant, I.S.; Narkar, S.P.; Shetty, D.S.; Upadhyay, A.; Sawant, S.D. First report of Colletotrichum capsici causing anthracnose on grapes in Maharashtra, India. New Dis. Rep. 2012, 25, 2. [Google Scholar] [CrossRef]
- Pan, F.Y.; Huang, Y.; Lin, L.; Zhou, Y.M.; Wei, R.F.; Guo, W.F.; Lu, J. First report of Colletotrichum capsici causing grape ripe rot in Guangxi, China. Plant Dis. 2016, 100, 2531. [Google Scholar] [CrossRef]
- Peng, L.J.; Sun, T.; Yang, Y.L.; Cai, L.; Hyde, K.D.; Bahkali, A.H.; Liu, Z.Y. Colletotrichum species on grape in Guizhou and Yunnan provinces, China. Mycoscience 2013, 54, 29–41. [Google Scholar] [CrossRef]
- Lim, Y.S.; Hassan, O.; Chang, T. First report of anthracnose of shine muscat caused by Colletotrichum fructicola in Korea. Mycobiology 2020, 48, 75–79. [Google Scholar] [CrossRef] [PubMed]
- Zapparata, A.; da Lio, D.; Sarrocco, S.; Vannacci, G.; Baroncelli, R. First report of Colletotrichum godetiae causing grape (Vitis vinifera) berry rot in Italy. Plant Dis. 2017, 101, 1051. [Google Scholar] [CrossRef]
- Echeverrigaray, S.; Scariot, F.J.; Fontanella, G.; Favaron, F.; Sella, L.; Santos, M.C.; Schwambach, J.; Pedrotti, C.; Delamare, A.P.L. Colletotrichum species causing grape ripe rot disease in Vitis labrusca and V. vinifera varieties in the highlands of southern Brazil. Plant Pathol. 2020, 69, 1504–1512. [Google Scholar] [CrossRef]
- Steel, C.C.; Greer, L.A.; Savocchia, S. Grapevine inflorescences are susceptible to the bunch rot pathogens, Greeneria uvicola (bitter rot) and Colletotrichum acutatum (ripe rot). Eur. J. Plant Pathol. 2012, 133, 773–778. [Google Scholar] [CrossRef]
- Daykin, M.E.; Milholland, R.D. Ripe rot of muscadine grape caused by Colletotrichum gloeosporioides and its control. Phytopathology 1984, 74, 710–714. [Google Scholar] [CrossRef]
- Rossi, V.; Caffi, T.; Salinari, F. Helping farmers face the increasing complexity of decision-making for crop protection. Phytopathol. Mediterr. 2012, 51, 457–479. [Google Scholar]
- Caffi, T.; Rossi, V.; Bugiani, R. Evaluation of a warning system for controlling primary infections of grapevine downy mildew. Plant Dis. 2010, 94, 709–716. [Google Scholar] [CrossRef]
- Caffi, T.; Legler, S.E.; Rossi, V.; Bugiani, R. Evaluation of a warning system for early-season control of grapevine powdery mildew. Plant Dis. 2012, 96, 104–110. [Google Scholar] [CrossRef]
- Rossi, V.; Caffi, T.; Giosuè, S.; Bugiani, R. A mechanistic model simulating primary infections of downy mildew in grapevine. Ecol. Modell. 2008, 212, 480–491. [Google Scholar] [CrossRef]
- Rossi, V.; Giosuè, S.; Caffi, T. Modelling plant diseases for decision making in crop protection. In Precision Crop Protection—The Challenge and Use of Heterogeneity; Springer: Dordrecht, The Netherlands, 2010; pp. 241–258. [Google Scholar]
- Rossi, V.; Onesti, G.; Legler, S.E.; Caffi, T. Use of systems analysis to develop plant disease models based on literature data: Grape black-rot as a case-study. Eur. J. Plant Pathol. 2015, 141, 427–444. [Google Scholar] [CrossRef]
- Caffi, T.; Rossi, V.; Legler, S.E.; Bugiani, R. A mechanistic model simulating ascosporic infections by Erysiphe necator, the powdery mildew fungus of grapevine. Plant Pathol. 2011, 60, 522–531. [Google Scholar] [CrossRef]
- González-Domínguez, E.; Caffi, T.; Ciliberti, N.; Rossi, V. A mechanistic model of Botrytis cinerea on grapevines that includes weather, vine growth stage, and the main infection pathways. PLoS ONE 2015, 10, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Ji, T.; Caffi, T.; Carisse, O.; Li, M.; Rossi, V. Development and evaluation of a model that predicts grapevine anthracnose caused by Elsinoë ampelina. Phytopathology 2021. In Press. [Google Scholar] [CrossRef]
- Yun, S.C.; Park, E.W. Effects of temperature and wetness period on infection of grape by Colletotrichum gloeosporioides. Korean J. Plant Pathol. 1990, 6, 219–228. [Google Scholar]
- Park, E.W.; Hur, J.S.; Yun, S.C. A forecasting system for scheduling fungicide sprays to control grape ripe rot caused by Colletotrichum gloeosporioides. Korean J. Plant Pathol. 1992, 8, 177–184. [Google Scholar]
- Lorenz, D.H.; Eichhorn, K.W.; Bleiholder, H.; Klose, R.; Meier, U.; Weber, E. Growth stages of the grapevine: Phenological growth stages of the grapevine (Vitis vinifera L. ssp. vinifera)—Codes and descriptions according to the extended BBCH scale. Aust. J. Grape Wine Res. 1995, 1, 100–103. [Google Scholar]
- Fukaya, M. Studies on etiology and control of grapevine ripe rot. I. Primary infection of grapevine ripe rot. Bull. Akita Fruit-Tree Exp. Stn. 2001, 27, 24–35. [Google Scholar]
- Greer, L.A.; Harper, J.D.I.; Steel, C.C. Infection of Vitis vinifera (cv Chardonnay) inflorescences by Colletotrichum acutatum and Greeneria uvicola. J. Phytopathol. 2014, 162, 407–410. [Google Scholar] [CrossRef]
- Daykin, M.E.; Milholland, R.D. Histopathology of ripe rot caused by Colletotrichum gloeosporioides on muscadine grape. Phytopathology 1984, 74, 1339–1341. [Google Scholar] [CrossRef]
- Steel, C.C.; Greer, L.A.; Samuelian, S.; Savocchia, S. Two species of fungus Colletotrichum responsible for ripe rot of grapes. Wine Vitic. J. 2011, 26, 48–58. [Google Scholar]
- Steel, C.C.; Greer, L.A.; Savocchia, S. Studies on Colletotrichum acutatum and Greeneria uvicola: Two fungi associated with bunch rot of grapes in sub-tropical Australia. Aust. J. Grape Wine Res. 2007, 13, 23–29. [Google Scholar] [CrossRef]
- Es-Soufi, R.; El Kbiach, M.; Errabii, T.; Saidi, R.; Badoc, A.; Chaveriat, L.; Martin, P.; Lamarti, A. Biology and physiology of Colletotrichum acutatum strains causing strawberry’s anthracnose. Agric. Sci. 2018, 9, 974–990. [Google Scholar]
- Everett, K.R.; Pushparajah, I.P.S.; Timudo, O.E.; Ah Chee, A.; Scheper, R.W.A.; Shaw, P.W.; Spiers, T.M.; Taylor, J.T.; Wallis, D.R.; Wood, P.N. Infection criteria, inoculum sources and splash dispersal pattern of Colletotrichum acutatum causing bitter rot of apple in New Zealand. Eur. J. Plant Pathol. 2018, 152, 367–383. [Google Scholar] [CrossRef]
- Fernando, T.H.P.S.; Jayasinghe, C.K.; Wijesundera, R.L.C. Factors affecting spore production, germination and viability of Colletotrichum acutatum isolates from Hevea brasiliensis. Mycol. Res. 2000, 104, 681–685. [Google Scholar] [CrossRef]
- King, W.T.; Madden, L.V.; Ellis, M.A.; Wilson, L.L. Effects of temperature on sporulation and latent period of Colletotrichum spp. infecting strawberry fruit. Plant Dis. 1997, 81, 77–84. [Google Scholar] [CrossRef]
- Liu, A.Y.; Chen, W.X.; Gu, H.; Shi, J.Y.; Li, J. Biological characteristic of pathogenic fungus causing anthracnose of loquat fruit. Acta Hortic. 2007, 750, 447–465. [Google Scholar] [CrossRef]
- Fitzell, R.D.; Peak, C.M. The epidemiology of anthracnose disease of mango: Inoculum sources, spore production and dispersal. Ann. Appl. Biol. 1984, 104, 53–59. [Google Scholar] [CrossRef]
- Mello, A.F.S.; Machado, A.C.Z.; Bedendo, I.P. Development of Colletotrichum gloeosporioides isolated from green pepper in different culture media, temperatures, and light regimes. Sci. Agric. 2004, 61, 542–544. [Google Scholar] [CrossRef]
- Pandey, R.R.; Arora, D.K.; Dubey, R.C. Effect of environmental conditions and inoculum density on infection of guava fruits by Colletotrichum glososporioides. Mycopathologia 1997, 137, 165–172. [Google Scholar] [CrossRef]
- Wastie, R.L. Secondary leaf fall of Hevea brasiliensis: Factors affecting the production, germination and viability of spores of Colletotrichum gloeosporioides. Ann. Appl. Biol. 1972, 72, 273–282. [Google Scholar] [CrossRef]
- Wang, P.S.; Liu, X.Q.; Wang, Y.Z.; Luan, B.H.; Zhang, W. Study on the biological characteristics of grape ripe rot. Jiangsu Agric. Sci. 2009, 01, 128–129. [Google Scholar]
- Veloso, J.S.; Lima, W.G.; Reis, A.; Doyle, V.P.; Michereff, S.J.; Câmara, M.P.S. Factors influencing biological traits and aggressiveness of Colletotrichum species associated with cashew anthracnose in Brazil. Plant Pathol. 2021, 70, 167–180. [Google Scholar] [CrossRef]
- Lv, X.; Wang, S.; Li, J.; Li, Y.; Li, X.; Wang, Q. Investigation on the occurrence of main diseases and pests at organic wine vineyards in Beijing. China Fruits 2013, 82–84. [Google Scholar] [CrossRef]
- Li, D.; Qi, H.; Li, S.; Cao, L.; Hu, Z.; Li, X. The survey for wine grape disease kinds and the analysis for epidemic factors. North. Hortic. 2010, 9, 174–177. [Google Scholar]
- Pang, J. The Research about the Character of Disease and Insect Pest Occurrence of Cabernet Sauvignon in Changli. Master’s Thesis, Northwest A&F University, Yangling, China, 2016. Available online: https://kns.cnki.net/KCMS/detail/detail.aspx?dbname=CMFD201602&filename=1016161423.nh (accessed on 6 August 2021).
- Zadoks, J.C. Systems analysis and the dynamics of epidemics. Phytopathology 1971, 61, 600–610. [Google Scholar]
- Levin, S.A.; Grenfell, B.; Hastings, A.; Perelson, A.S. Mathematical and computational challenges in population biology and ecosystems science. Science 1997, 275, 334–343. [Google Scholar] [CrossRef]
- Zadoks, J.C.; Schein, R.D. Epidemiology and Plant Disease Management; Oxford University Press Inc.: New York, NY, USA, 1979. [Google Scholar]
- Bove, F.; Savary, S.; Willocquet, L.; Rossi, V. Designing a modelling structure for the grapevine downy mildew pathosystem. Eur. J. Plant Pathol. 2020, 157, 251–268. [Google Scholar] [CrossRef]
- Quimio, T.H.; Quimio, A.J. Notes on Philippine grape and guava anthracnose. Plant Dis. Report. 1975, 59, 221–224. [Google Scholar]
- Oliver, C. Phylogeny, Histological Observation, and In Vitro Fungicide Screening and Field Trials of Multiple Colletotrichum Species, the Causal Agents of Grape Ripe Rot. Doctoral Dissertation, Virginia Polytechnic Institute and State University, Blacksburg, VA, USA, 2019. [Google Scholar]
- Nita, M.; Pathologist, G.P.; Tech, V.; Khang, C. Final Report to Virginia Wine Board 2014 FY (14-1690-02) Project Title: Investigating the Lifecycle of Ripe Rot of Grape Caused by Colletotrichum Species. 2014. Available online: https://cdn.ymaws.com/vawine.site-ym.com/resource/resmgr/docs/2015_Wine_Board_Research_Reports/REPORT_14_-_Nita_Final_Ripe_.pdf (accessed on 6 August 2021).
- Cannon, P.F.; Damm, U.; Johnston, P.R.; Weir, B.S. Colletotrichum—Current status and future directions. Stud. Mycol. 2012, 73, 181–213. [Google Scholar] [CrossRef] [PubMed]
- Ntahirnpera, N.; Wilson, L.L.; Ellis, M.A.; Madden, L.V. Comparison of rain effects on splash dispersal of three Colletotrichum species infecting strawberry. Phytopathology 1999, 89, 555–563. [Google Scholar] [CrossRef]
- Peres, N.A.; Timmer, L.W.; Adaskaveg, J.E.; Correll, J.C. Lifestyles of Colletotrichum acutatum. Plant Dis. 2005, 89, 784–796. [Google Scholar] [CrossRef] [PubMed]
- Gregory, P.H.; Guthrie, E.J.; Bunce, M.E. Experiments on splash dispersal of fungus spores. J. Gen. Microbiol. 1959, 20, 328–354. [Google Scholar] [CrossRef]
- Lax, A.R.; Templeton, G.E. Isolation, purification, and biological activity of a self-inhibitor from conidia of Colletotrichum gloeosporioides. Phytopathology 1985, 75, 386–390. [Google Scholar] [CrossRef]
- Rajasab, A.H.; Chawda, H.T. Dispersal of the conidia of Colletotrichum gloeosporioides by rain and the development of anthracnose on onion. Grana 1994, 33, 162–165. [Google Scholar] [CrossRef]
- Madden, L.V.; Yang, X.; Wilson, L.L. Effects of rain intensity on splash dispersal of Colletotrichum acutatum. Phytopathology 1996, 86, 864–874. [Google Scholar] [CrossRef]
- Hamada, N.A.; Moreira, R.R.; Nesi, C.N.; May de Mio, L.L. Pathogen dispersal and Glomerella leaf spot progress within apple canopy in Brazil. Plant Dis. 2019, 103, 3209–3217. [Google Scholar] [CrossRef]
- Guyot, J.; Ntawanga Omanda, E.; Pinard, F. Some epidemiological investigations on Colletotrichum leaf disease on rubber tree. Crop Prot. 2005, 24, 65–77. [Google Scholar] [CrossRef]
- Oliver, C. Investigation of Wine Grape Cultivar and Cluster Developmental Stage Susceptibility to Grape Ripe Rot Caused by Two Fungal Species Complexes, Colletotrichum gloeosporioides, and C. acutatum, and the Evaluation of Potential Controls. Master’s Thesis, Virginia Polytechnic Institute and State University, Blacksburg, VA, USA, 2016. [Google Scholar]
- Nasrollahiazar, E.; Miles, T.; Sabbatini, P. Critical Practices to Control Late Season Grape Diseases and the Potential Effects of Fungicides on Fermentation. Available online: https://www.canr.msu.edu/news/late-season-grape-diseases-and-fungicides-on-fermentation (accessed on 6 August 2021).
- Khilare, V.C.; Chavan, S.S. Effect of passage on the development of carbendazim resistance in Gloeosporium ampelophagum causing anthracnose of grapes. Biosci. Biotechnol. Res. Asia. 2010, 8, 501–504. [Google Scholar] [CrossRef]
- Sawant, I.S.; Narkar, S.P.; Shetty, D.S.; Upadhyay, A.; Sawant, S.D. Emergence of Colletotrichum gloeosporioides sensu lato as the dominant pathogen of anthracnose disease of grapes in India as evidenced by cultural, morphological and molecular data. Australas. Plant Pathol. 2012, 41, 493–504. [Google Scholar] [CrossRef]
- Xu, X.F.; Lin, T.; Yuan, S.K.; Dai, D.J.; Shi, H.J.; Zhang, C.Q.; Wang, H.D. Characterization of baseline sensitivity and resistance risk of Colletotrichum gloeosporioides complex isolates from strawberry and grape to two demethylation-inhibitor fungicides, prochloraz and tebuconazole. Australas. Plant Pathol. 2014, 43, 605–613. [Google Scholar] [CrossRef]
- López-Zapata, S.P.; Castaño-Zapata, J. In vitro effect of four fungicides on Colletotrichum gloeosporioides causing anthracnosis on the Red Globe grape variety. Rev. Acad. Colomb. Cienc. Exactas Fis. Nat. 2020, 44, 747–758. [Google Scholar] [CrossRef]
- Bozdogan, H. Model selection and Akaike’s information criterion (AIC): The general theory and its analytical extensions. Psychometrika 1987, 52, 345–370. [Google Scholar] [CrossRef]
- Wang, H.Y.; Guo, S.Y. Study on the annual occurrence of grape ripe rot and downy mildew in Henan Province. J. Henan Agric. Sci. 1999, 01, 30–32. [Google Scholar]
- Du, F.; Deng, W.; Yang, M.; Wang, H.; Mao, R.; Shao, J.; Fan, J.; Chen, Y.; Fu, Y.; Li, C.; et al. Protecting grapevines from rainfall in rainy conditions reduces disease severity and enhances profitability. Crop Prot. 2015, 67, 261–268. [Google Scholar] [CrossRef]
- González-Domínguez, E.; Rossi, V.; Farina, V.; Gianguzzi, G.; Berbegal, M.; Armengol, J. Validation of a mechanistic model for predicting fruit scab infection on different loquat cultivars. Phytopathol. Mediterr. 2017, 56, 400–408. [Google Scholar]
- Onesti, G.; González-Domínguez, E.; Rossi, V. Accurate prediction of black rot epidemics in vineyards using a weather-driven disease model. Pest Manag. Sci. 2016, 72, 2321–2329. [Google Scholar] [CrossRef]
- Lin, L. A concordance correlation coefficient to evaluate reproducibility. Biometrics 1989, 45, 255. [Google Scholar] [CrossRef]
- Nash, J.E.; Sutcliffe, J.V. River flow forecasting through conceptual models part I—A discussion of principles. J. Hydrol. 1970, 10, 282–290. [Google Scholar] [CrossRef]









| Abbreviation | Description | Unit |
|---|---|---|
| State variables | ||
| S1 | Conidia from overwintered primary sources | N per plant |
| S2 | Conidia deposited on berries | N per plant |
| S3 | Conidia causing infection in berries | N per plant |
| S4 | Conidia leading to berry rot | N per plant |
| S5 | Colonies in berries producing acervuli (i.e., fertile colonies) | N per plant |
| S6 | Conidia produced on rotten berries | N per plant |
| Rate variables | ||
| DELR | Delay rate at which quiescent infections become visible | 0 to 1 |
| DEPR | Deposition rate of conidia on flowers and berries | 0 to 1 |
| INCR | Rate at which infected berries become visible | 0 to 1 |
| INFR | Infection rate on flowers and berries | 0 to 1 |
| LATR | Rate at which infected berries become fertile | 0 to 1 |
| MATR | Rate at which primary conidia mature | 0 to 1 |
| SPOR | Sporulation rate on affected and fertile berries | 0 to 1 |
| Parameters, counters, and auxiliary variables | ||
| k | Seasonal number of conidia from overwintered primary sources | N per plant |
| i | Counter of the day | N |
| j | Counter of the cohorts of primary conidia | N |
| t | Time required by quiescent infections to cause berry rot after véraison | days |
| GS | Growth stage of vines based on Lorenz et al. (1995) | 0 to 99 |
| IP | Incubation period | Days |
| LP | Latent period | Days |
| LA | Leaf area per plant | m2 leaf |
| BA | Berry area per plant | m2 berries |
| BLR | Berry to leaf area: BA/(BA+LA) | 0 to 1 |
| DD | Degree-days accumulated on moist days | °C day |
| Driving variables | ||
| R | Hourly rainfall | mm |
| T | Hourly air temperature | °C |
| Tmax | Maximum temperature for sporulation | °C |
| Tmin | Minimum temperature for sporulation | °C |
| TWD | Mean temperature of the wet period | °C |
| WD | Duration of wet period | N hours |
| Location (Literature Citation) | Year | Epidemic Abbreviation | Cultivar | Disease at Harvest | |
|---|---|---|---|---|---|
| Incidence 1 | Severity 2 | ||||
| Akita, Japan (Fukaya [31]) | 1986 | AK-86 | Campbell early | 34.0 | -3 |
| 1987 | AK-87 | 97.6 | - | ||
| 1988 | AK-88 | 66.7 | - | ||
| 1989 | AK-89 | 47.6 | - | ||
| 1990 | AK-90 | 75.4 | - | ||
| 1991 | AK-91 | 36.4 | - | ||
| 1992 | AK-92 | 56.0 | - | ||
| 1993 | AK-93 | 35.0 | - | ||
| 1994 | AK-94 | 55.2 | - | ||
| 1995 | AK-95 | 94.7 | - | ||
| 1996 | AK-96 | 52.0 | - | ||
| 1997 | AK-97 | 34.0 | - | ||
| Castle Hayne, USA (Daykin and Miholland [18]) | 1980 | CH-80 | Carlos | 53.8 | - |
| 1981 | CH-81 | 23.7 | - | ||
| Beijing, China (Lv et al. [47]) | 2011 | BJ-11 | Merlot | 29.0 | - |
| Changli, China (Li et al. [48]) | 2009 | CL-09 | Cabernet Sauvignon | 29.5 | 9.7 |
| Changli, China (Pang [49]) | 2012 | CL-12 | Cabernet Sauvignon | 25.8 | 17.3 |
| 2013 | CL-13 | 31.4 | 33.5 | ||
| 2014 | CL-14 | 26.1 | 29.4 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, T.; Salotti, I.; Dong, C.; Li, M.; Rossi, V. Modeling the Effects of the Environment and the Host Plant on the Ripe Rot of Grapes, Caused by the Colletotrichum Species. Plants 2021, 10, 2288. https://doi.org/10.3390/plants10112288
Ji T, Salotti I, Dong C, Li M, Rossi V. Modeling the Effects of the Environment and the Host Plant on the Ripe Rot of Grapes, Caused by the Colletotrichum Species. Plants. 2021; 10(11):2288. https://doi.org/10.3390/plants10112288
Chicago/Turabian StyleJi, Tao, Irene Salotti, Chaoyang Dong, Ming Li, and Vittorio Rossi. 2021. "Modeling the Effects of the Environment and the Host Plant on the Ripe Rot of Grapes, Caused by the Colletotrichum Species" Plants 10, no. 11: 2288. https://doi.org/10.3390/plants10112288
APA StyleJi, T., Salotti, I., Dong, C., Li, M., & Rossi, V. (2021). Modeling the Effects of the Environment and the Host Plant on the Ripe Rot of Grapes, Caused by the Colletotrichum Species. Plants, 10(11), 2288. https://doi.org/10.3390/plants10112288








