Structural Characterization of Black Widow Spider Dragline Silk Proteins CRP1 and CRP4
Abstract
:1. Introduction
2. Results
2.1. Purification of Recombinantly Expressed CRPs from Bacteria
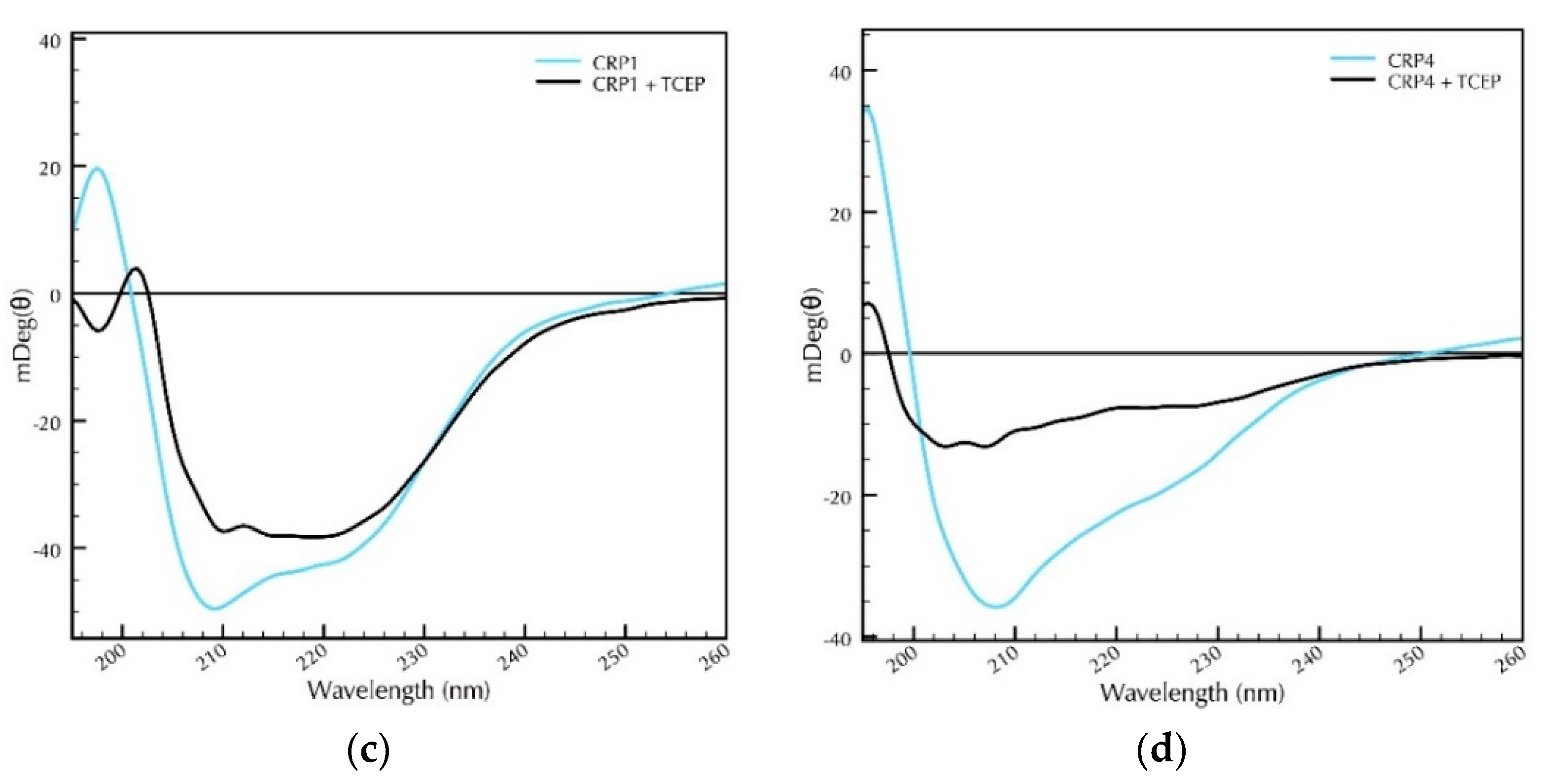
2.2. Secondary Structural Studies of Recombinant CRP1 and CRP4: The Impact of pH and Reducing Agents on Protein Folding
2.3. Secondary Structural Analysis of CRPs Under Different Temperature and pH
2.4. XL-MS Studies Support Molecular Modeling of CRP1 as a Cystine Slipknot Member
3. Discussion
4. Materials and Methods
4.1. Plasmid Construction and Expression of Recombinant CRP Proteins in Bacteria
4.2. Purification of CRP1 and CRP4 using Ni-NTA Agarose Affinity Purification and Removal of the 10xHis-SUMO-tag
4.3. MS/MS Analysis Using a NanoLC Orbitrap Fusion™ Tribrid™ Mass Spectrometer
4.4. Circular Dichroism Spectroscopy and DichroWeb
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gosline, J.M.; DeMont, M.E.; Denny, M.W. The structure and properties of spider silk. Endeavour 1986, 10, 31–43. [Google Scholar] [CrossRef]
- Moore, A.M.; Tran, K. Material properties of cobweb silk from the black widow spider Latrodectus hesperus. Int. J. Biol. Macromol. 1999, 24, 277–282. [Google Scholar] [CrossRef]
- Foelix, R. Biology of Spiders; Oxford University Press: New York, NY, USA, 1996. [Google Scholar]
- Guerette, P.A.; Ginzinger, D.G.; Weber, B.H.; Gosline, J.M. Silk properties determined by gland-specific expression of a spider fibroin gene family. Science 1996, 272, 112–115. [Google Scholar] [CrossRef]
- Andersson, M.; Holm, L.; Ridderstrale, Y.; Johansson, J.; Rising, A. Morphology and composition of the spider major ampullate gland and dragline silk. Biomacromolecules 2013, 14, 2945–2952. [Google Scholar] [CrossRef] [PubMed]
- Vollrath, F.; Knight, D.P. Liquid crystalline spinning of spider silk. Nature 2001, 410, 541–548. [Google Scholar] [CrossRef]
- Jin, H.J.; Kaplan, D.L. Mechanism of silk processing in insects and spiders. Nature 2003, 424, 1057–1061. [Google Scholar] [CrossRef]
- Xu, M.; Lewis, R.V. Structure of a protein superfiber: Spider dragline silk. Proc. Natl. Acad. Sci. USA 1990, 87, 7120–7124. [Google Scholar] [CrossRef] [Green Version]
- Hinman, M.B.; Lewis, R.V. Isolation of a clone encoding a second dragline silk fibroin. Nephila clavipes dragline silk is a two-protein fiber. J. Biol. Chem. 1992, 267, 19320–19324. [Google Scholar]
- Ayoub, N.A.; Garb, J.E.; Tinghitella, R.M.; Collin, M.A.; Hayashi, C.Y. Blueprint for a high-performance biomaterial: Full-length spider dragline silk genes. PLoS ONE 2007, 2, e514. [Google Scholar] [CrossRef]
- Hagn, F.; Eisoldt, L.; Hardy, J.G.; Vendrely, C.; Coles, M.; Scheibel, T.; Kessler, H. A conserved spider silk domain acts as a molecular switch that controls fibre assembly. Nature 2010, 465, 239–242. [Google Scholar] [CrossRef] [Green Version]
- Landreh, M.; Askarieh, G.; Nordling, K.; Hedhammar, M.; Rising, A.; Casals, C.; Astorga-Wells, J.; Alvelius, G.; Knight, S.D.; Johansson, J.; et al. A pH-dependent dimer lock in spider silk protein. J. Mol. Biol. 2010, 404, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Kronqvist, N.; Otikovs, M.; Chmyrov, V.; Chen, G.; Andersson, M.; Nordling, K.; Landreh, M.; Sarr, M.; Jornvall, H.; Wennmalm, S.; et al. Sequential pH-driven dimerization and stabilization of the N-terminal domain enables rapid spider silk formation. Nat. Commun. 2014, 5, 3254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trancik, J.E.; Czernuszka, J.T.; Bell, F.I.; Viney, C. Nanostructural features of a spider dragline silk as revealed by electron and X-ray diffraction studies. Polymer 2006, 47, 5633–5642. [Google Scholar] [CrossRef]
- Holland, G.P.; Jenkins, J.E.; Creager, M.S.; Lewis, R.V.; Yarger, J.L. Solid-state NMR investigation of major and minor ampullate spider silk in the native and hydrated states. Biomacromolecules 2008, 9, 651–657. [Google Scholar] [CrossRef]
- Jenkins, J.E.; Sampath, S.; Butler, E.; Kim, J.; Henning, R.W.; Holland, G.P.; Yarger, J.L. Characterizing the secondary protein structure of black widow dragline silk using solid-state NMR and X-ray diffraction. Biomacromolecules 2013, 14, 3472–3483. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Dong, Q.; Yu, Y.; Niu, B.; Ji, D.; Li, M.; Huang, Y.; Chen, X.; Tan, A. Mass spider silk production through targeted gene replacement in Bombyx mori. Proc. Natl. Acad. Sci. USA 2018, 115, 8757–8762. [Google Scholar] [CrossRef] [Green Version]
- Bowen, C.H.; Dai, B.; Sargent, C.J.; Bai, W.; Ladiwala, P.; Feng, H.; Huang, W.; Kaplan, D.L.; Galazka, J.M.; Zhang, F. Recombinant spidroins fully replicate primary mechanical properties of natural spider silk. Biomacromolecules 2018, 19, 3853–3860. [Google Scholar] [CrossRef] [Green Version]
- Rammensee, S.; Slotta, U.; Scheibel, T.; Bausch, A.R. Assembly mechanism of recombinant spider silk proteins. Proc. Natl. Acad. Sci. USA 2008, 105, 6590–6595. [Google Scholar] [CrossRef] [Green Version]
- Lane, A.K.; Hayashi, C.Y.; Whitworth, G.B.; Ayoub, N.A. Complex gene expression in the dragline silk producing glands of the western black widow (Latrodectus hesperus). BMC Genomics 2013, 14, 846. [Google Scholar] [CrossRef] [Green Version]
- Clarke, T.H.; Garb, J.E.; Hayashi, C.Y.; Haney, R.A.; Lancaster, A.K.; Corbett, S.; Ayoub, N.A. Multi-tissue transcriptomics of the black widow spider reveals expansions, co-options, and functional processes of the silk gland gene toolkit. BMC Genomics 2014, 15, 365. [Google Scholar] [CrossRef] [Green Version]
- Lloyd, J.U. A treatise on apis (the bee), tella araneae (cobweb), spongia and cantharis; Lloyd Brothers: Cincinnati, OH, USA, 1911. [Google Scholar]
- Pham, T.; Chuang, T.; Lin, A.; Joo, H.; Tsai, J.; Crawford, T.; Zhao, L.; Hsia, Y.; Williams, C.; Vierra, C.A. Dragline silk: A fiber assembled with low-molecular-weight cysteine-rich proteins. Biomacromolecules 2014, 11, 4073. [Google Scholar] [CrossRef] [PubMed]
- Larracas, C.; Hekman, R.; Dyrness, S.; Arata, A.; Williams, C.; Vierra, C. Comprehensive proteomic analysis of spider dragline silk from black widows: A recipe to build synthetic silk fibers. Int. J. Mol. Sci. 2016, 17, 1537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sikora, M.; Sulkowska, J.I.; Cieplak, M. Mechanical strength of 17,134 model proteins and cysteine slipknots. PLoS Comput Biol 2009, 5, e1000547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vollrath, F.; Wen Hu, X.; Knight, D.P. Silk production in a spider involves acid bath treatment. Proc. R. Soc. B 1998, 263, 817–820. [Google Scholar] [CrossRef]
- Nentwig, W. Ecophysiology of Spiders; Springer-Verlag: Berlin, Germany, 1987; p. 448. [Google Scholar]
- Touw, W.; Baakman, C.; Black, J.; Beek, T.; Kreiger, E.; Joosten, R.P.; Vriend, G. A series of PDB related databases for everyday needs. Nucleic Acids Res. 2015, 43, 364–368. [Google Scholar] [CrossRef]
- Kabsch, W.; Sander, C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef]
- Dicko, C.; Vollrath, F.; Kenney, J.M. Spider silk protein refolding is controlled by changing ph. Biomacromolecules 2004, 5, 704–710. [Google Scholar] [CrossRef]
- Dicko, C.; Kenney, J.M.; Knight, D.; Vollrath, F. Transition to a beta-sheet-rich structure in spidroin in vitro: The effects of pH and cations. Biochemistry 2004, 43, 14080–14087. [Google Scholar] [CrossRef]
- Askarieh, G.; Hedhammar, M.; Nordling, K.; Saenz, A.; Casals, C.; Rising, A.; Johansson, J.; Knight, S.D. Self-assembly of spider silk proteins is controlled by a pH-sensitive relay. Nature 2010, 465, 236–238. [Google Scholar] [CrossRef]
- Bauer, J.; Schaal, D.; Eisoldt, L.; Schweimer, K.; Schwarzinger, S.; Scheibel, T. Acidic residues control the dimerization of the N-terminal domain of black widow spiders’ major ampullate spidroin 1. Sci. Rep. 2016, 6, 34442. [Google Scholar] [CrossRef] [Green Version]
- Hagn, F.; Thamm, C.; Scheibel, T.; Kessler, H. pH-dependent dimerization and salt-dependent stabilization of the N-terminal domain of spider dragline silk--implications for fiber formation. Angew. Chem. 2011, 50, 310–313. [Google Scholar] [CrossRef]
- Schwarze, S.; Zwettler, F.U.; Johnson, C.M.; Neuweiler, H. The N-terminal domains of spider silk proteins assemble ultrafast and protected from charge screening. Nat. Commun. 2013, 4, 2815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clarke, T.H.; Garb, J.E.; Haney, R.A.; Crystal Chaw, R.; Hayashi, C.Y.; Ayoub, N. Evolutionary shifts in gene expression decoupled from gene duplication across functionally distinct spider silk glands. Sci. Rep. 2017, 7, 8393. [Google Scholar] [CrossRef] [PubMed]
- Scheufler, C.; Sebald, W.; Hulsmeyer, M. Crystal structure of human bone morphogenetic protein-2 at 2.7 Å resolution. J. Mol. Biol. 1999, 287, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Vitt, U.A.; Hsu, S.Y.; Hsueh, A.J. Evolution and classification of cystine knot-containing hormones and related extracellular signaling molecules. Mol. Endocrinol. 2001, 15, 681–694. [Google Scholar] [CrossRef] [PubMed]
- Pugno, N. The “Egg of Columbus” for making the world’s toughest fibres. PloS ONE 2014, 9, e93079. [Google Scholar] [CrossRef]
- Muller, Y.A.; Heiring, C.; Misselwitz, R.; Welfle, K.; Welfle, H. The cystine knot promotes folding and not thermodynamic stability in vascular endothelial growth factor. J. Biol. Chem. 2002, 277, 43410–43416. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, A.; Funkner, A.; Neumann, P.; Juhnke, S.; Walther, M.; Schierhorn, A.; Weininger, U.; Balbach, J.; Reuter, G.; Stubbs, M.T. Biophysical characterization of refolded Drosophila spatzle, a cystine knot protein, reveals distinct properties of three isoforms. J. Biol. Chem. 2008, 283, 32598–32609. [Google Scholar] [CrossRef]
- Klykov, O.; Steigenberger, B.; Pektas, S.; Fasci, D.; Heck, A.J.R.; Scheltema, R.A. Efficient and robust proteome-wide approaches for cross-linking mass spectrometry. Nat. Protoc. 2018, 13, 2964–2990. [Google Scholar] [CrossRef]
- Liu, F.; Lossl, P.; Scheltema, R.; Viner, R.; Heck, A.J.R. Optimized fragmentation schemes and data analysis strategies for proteome-wide cross-link identification. Nat. Commun. 2017, 8, 15473. [Google Scholar] [CrossRef]
Sample Availability: Samples of CRP1 and CRP4 are available from the authors. |








| Percentage of Each Characteristic at 25 °C for CRP1 | ||||
|---|---|---|---|---|
| Random Coil | Beta Sheet | Turns | Helix | |
| pH 5.8 | 45.6 | 31.35 | 11.95 | 10.6 |
| pH 6.8 | 45.1 | 31.1 | 11.95 | 11.4 |
| pH 7.0 | 41.85 | 35.25 | 11.75 | 10.15 |
| pH 7.0 + TCEP | 37.9 | 33.8 | 15.2 | 12.6 |
| Predicted * | 43.5 | 30.7 | 18.2 | 7.9 |
| DSSP algorithm | 44.4 | 23.9 | 21.6 | 10.2 |
| Percentage of Each Characteristic at 25 °C for CRP4 | ||||
|---|---|---|---|---|
| Random Coil | Beta Sheet | Turns | Helix | |
| pH 5.8 | 44.95 | 33 | 11.25 | 10.3 |
| pH 6.8 | 42.85 | 34.3 | 11.65 | 10.65 |
| pH 7.0 | 45 | 32.35 | 11.15 | 11.05 |
| pH 7.0 + TCEP | 38.8 | 43.2 | 13.3 | 3.7 |
| Predicted * | 45.5 | 33.3 | 17.8 | 3.3 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shanafelt, M.; Rabara, T.; MacArt, D.; Williams, C.; Hekman, R.; Joo, H.; Tsai, J.; Vierra, C. Structural Characterization of Black Widow Spider Dragline Silk Proteins CRP1 and CRP4. Molecules 2020, 25, 3212. https://doi.org/10.3390/molecules25143212
Shanafelt M, Rabara T, MacArt D, Williams C, Hekman R, Joo H, Tsai J, Vierra C. Structural Characterization of Black Widow Spider Dragline Silk Proteins CRP1 and CRP4. Molecules. 2020; 25(14):3212. https://doi.org/10.3390/molecules25143212
Chicago/Turabian StyleShanafelt, Mikayla, Taylor Rabara, Danielle MacArt, Caroline Williams, Ryan Hekman, Hyun Joo, Jerry Tsai, and Craig Vierra. 2020. "Structural Characterization of Black Widow Spider Dragline Silk Proteins CRP1 and CRP4" Molecules 25, no. 14: 3212. https://doi.org/10.3390/molecules25143212
APA StyleShanafelt, M., Rabara, T., MacArt, D., Williams, C., Hekman, R., Joo, H., Tsai, J., & Vierra, C. (2020). Structural Characterization of Black Widow Spider Dragline Silk Proteins CRP1 and CRP4. Molecules, 25(14), 3212. https://doi.org/10.3390/molecules25143212






