X-ray Fluorescence Techniques in Determining the Habitat Preferences of Species—Ulva pilifera (Ulvales, Chlorophyta) from Montenegro Case Study
Abstract
:1. Introduction
2. Results and Discussion
2.1. Habitat and Morphological Observations
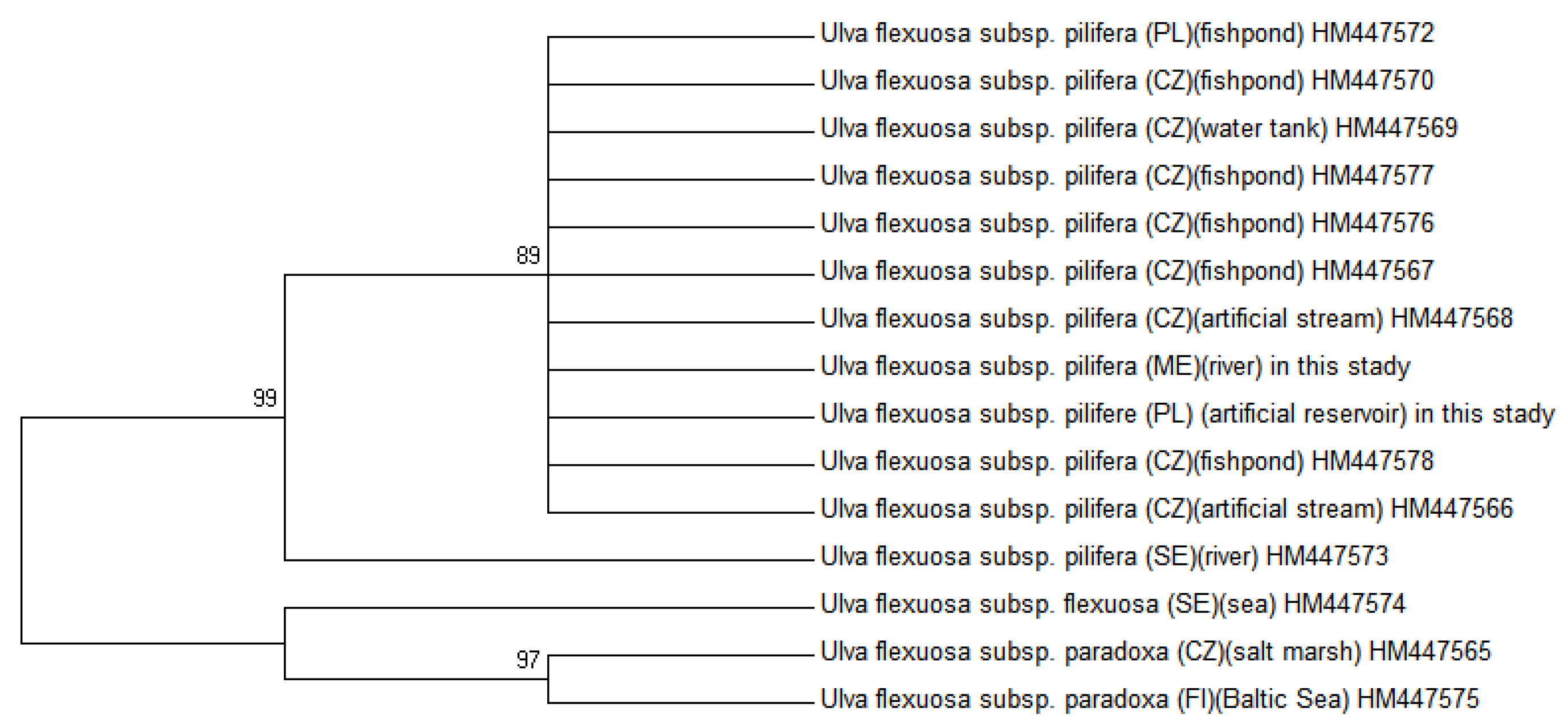
2.2. Molecular Phylogenetic Analysis
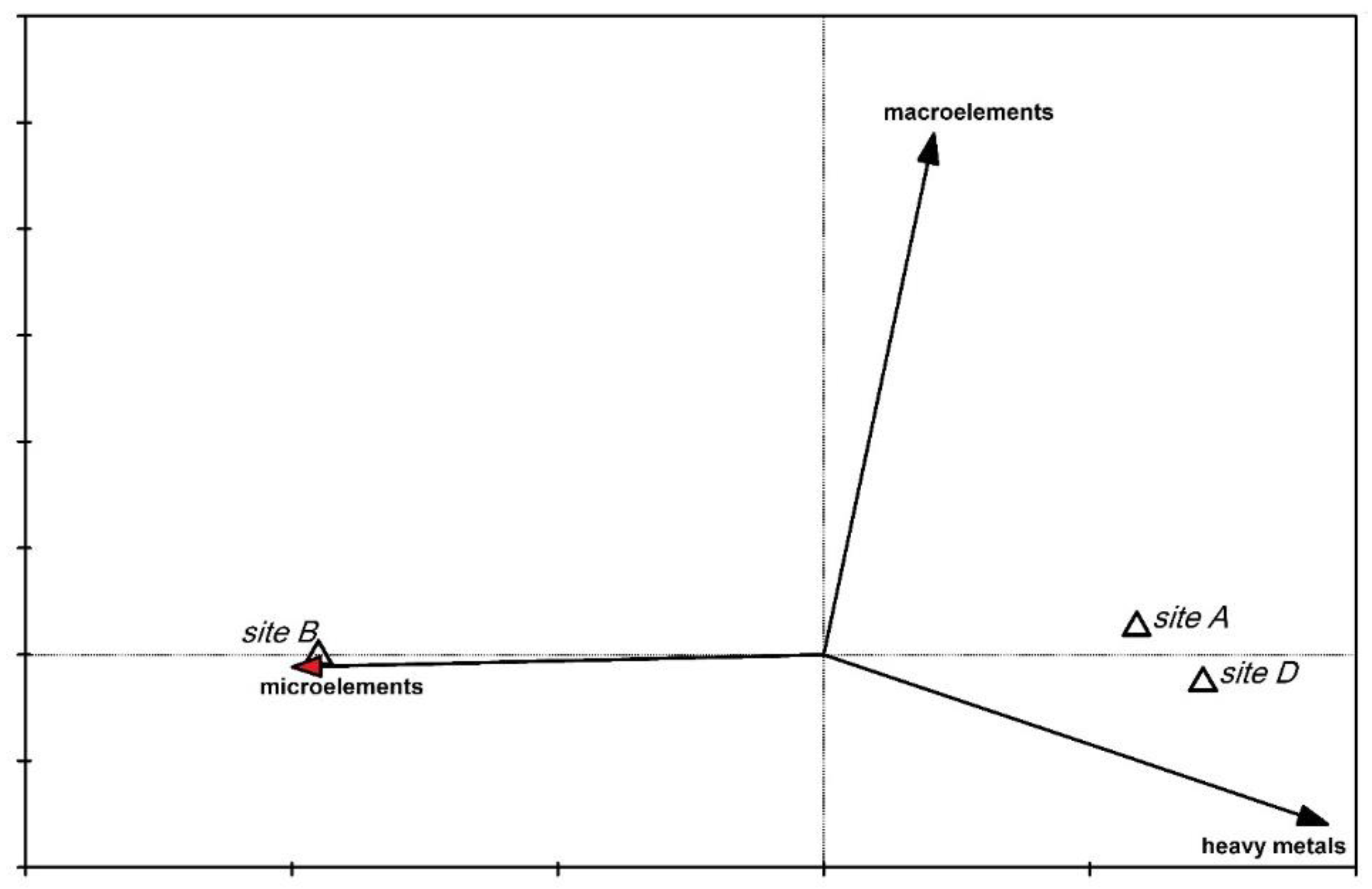
2.3. Elemental Analysis
2.4. Comparison of Data Analysis
2.5. Distribution and Habitat Preferences of the Ulva Species
3. Materials and Methods
3.1. Study Site
3.2. Samples Preparation
3.3. Physicochemical Water Quality
3.4. Molecular and Phylogenetic Analysis
3.5. Wavelength Dispersive X-ray Fluorescence (WDXRF) Technique
3.6. Total Reflection X-ray Fluorescence (TXRF) Technique
3.7. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- John, D.M.; Whitton, B.A.; Brook, A.J. The Freshwater Algal Flora of the British Isles: An Identification Guide to Freshwater and Terrestrial Algae, 2nd ed.; Cambridge University Press: Cambridge, UK, 2011; pp. 5–15. [Google Scholar]
- Wolf, M.A.; Sciuto, K.; Andreoli, C.; Moro, I. Ulva (Chlorophyta, Ulvales) biodiversity in the North Adriatic Sea (Mediterranean, Italy): Cryptic species and new introductions. J. Phycol. 2012, 48, 1510–1521. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, T.; Ohki, K.; Kamiya, M. Difference of spatial distribution and seasonal succession among Ulva species (Ulvophyceae) across salinity gradients. J. Phycol. 2013, 52, 637–651. [Google Scholar] [CrossRef]
- Saber, A.A.; Mareš, J.; Guella, G.; Anesi, A.; Štenclová, L.; Cantonati, M. Polyphasic approach to a characteristic Ulva population from a limno-rheocrenic, mineral (chloride, sodium, sulphate) in the Siwa Oasis (Western Desert of Egypt). Fottea 2018, 18, 227–242. [Google Scholar] [CrossRef] [Green Version]
- Merceron, M.; Antoine, V.; Auby, I.; Morand, P. In situ growth potential of the subtidal part of green tide forming Ulva spp. stocks. Sci. Total Environ. 2007, 384, 293–305. [Google Scholar] [CrossRef] [Green Version]
- Ye, N.; Zhang, X.; Mao, Y.; Liang, C.; Xu, D.; Zou, J.; Zhuang, Z.; Qing, W. Green tides are overwhelming the coastline of our blue planet: Taking the world’s largest example. Ecol. Res. 2011, 26, 477–485. [Google Scholar] [CrossRef]
- Hayden, H.S.; Blomster, J.; Maggs, C.A.; Silva, P.C.; Stanhope, M.J.; Waaland, J.R. Linnaeus was right all along: Ulva and Enteromorpha are not distinct genera. Eur. J. Phycol. 2003, 38, 277–294. [Google Scholar] [CrossRef]
- Shimada, S.; Hiraoka, M.; Nabata, S.; Iima, M.; Masuda, M. Molecular phylogenetic analyses of the Japanese Ulva and Enteromorpha (Ulvales, Ulvophyceae), with special reference to the free-floating Ulva. Phycol. Res. 2003, 51, 99–108. [Google Scholar] [CrossRef]
- Leskinen, E.; Alström-Rapaport, C.; Pamilo, P. Phytogeographical structure, distribution and genetic variation of the green algae Ulva intestinalis and U. compressa (Chlorophyta) in the Baltic Sea area. Mol. Ecol. 2014, 13, 2257–2265. [Google Scholar] [CrossRef]
- Rybak, A.S. Species of Ulva (Ulvophyceae, Chlorophyta) as indicators of salinity. Ecol. Ind. 2018, 85, 253–261. [Google Scholar] [CrossRef]
- Beach, K.S.; Smith, C.M.; Michael, T.; Shin, H.W. Photosynthesis in reproductive unicells of Ulva fasciata and Enteromorpha flexuosa: Implications for ecological success. Mar. Ecol. Prog. Ser. 1995, 125, 229–237. [Google Scholar] [CrossRef]
- Mareš, J.; Leskinen, E.; Sitkowska, M.; Skácelová, O.; Blomster, J. True identity of the European freshwater Ulva (Chlorophyta, Ulvophyceae) revealed by a combined molecular and morphological approach. J. Phycol. 2011, 47, 1177–1192. [Google Scholar] [CrossRef]
- Lougheed, V.L.; Stevenson, R.J. Exotic marine macroalga (Enteromorpha flexuosa) reaches bloom proportions in a coastal lake of Lake Michigan. J. G. Lak. Res. 2004, 30, 538–544. [Google Scholar] [CrossRef]
- Škaloud, P.; Rindi, F.; Boedeker, C.; Leliaert, F. Freshwater Flora of Central Europe, Chlorophyta: Ulvophyceae (Süßwasserflora von Mitteleuropa, Bd. 13: Chlorophyta: Ulvophyceae); Springer Spektrum: Berlin, Germany, 2018; Volume 13, 289p. [Google Scholar]
- Bliding, C.A. A critical survey of European taxa in Ulvales, Part Ulva, Ulvaria, Monostroma, Kornmannia. Bot. Not. 1968, 121, 535–629. [Google Scholar]
- Enteromorpha pilifera Kütz., 1856 in Gargominy 0. TAXREF; Version 4; UMS PatriNAT (OFB-CNRS-MNHN): Paris, France, 2020.
- Wynne, M.J. A checklist of benthic marine algae of the tropical and subtropical western Atlantic: Third revision. Nova Hedwig. Beih. 2011, 140, 7–166. [Google Scholar]
- Messyasz, B.; Czerwik-Marcinkowska, J.; Massalski, A.; Uher, B.; Rybak, A.; Szendzina, L.; Pikosz, M. Morphological and ultrastructural studies on Ulva flexuosa subsp. pilifera (Chlorophyta) from Poland. Acta Soc. Bot. Pol. 2013, 82, 157–163. [Google Scholar] [CrossRef] [Green Version]
- Richter, D.; Pietryka, M. New localities of Ulva flexuosa subsp. pilifera (Chlorophyta, Ulvophyceae) in Lower Silesia—Morphometric and habitat characteristics. Frag. Flor. Geobot. Pol. 2016, 23, 305–319. [Google Scholar]
- Kopp, R. Phytoplankton of the Zámecký Pond. Fottea 2006, 6, 111–125. [Google Scholar]
- Skácelová, O. Flóra Sinic a Řas Tůní v Inundačních Pásmech Řek; Faculty of Biological Science: Českeé Budějovice, Czech Republic, 2004; pp. 1–143. [Google Scholar]
- Kapetanović, R.; Sladić, D.; Popov, S.; Zlatović, M.; Kljajić, Z.; Gašić, M.J. Sterol composition of the Adriatic sea algae Ulva lactuca, Codium dichotonium, Cystoseira adriatica and Fucus virsoides. J. Serb. Chem. Soc. 2005, 70, 1395–1400. [Google Scholar] [CrossRef]
- Kosanić, M.; Ranković, B.; Stanjković, T. Biological activities of two macroalgae from Adriatic coast of Montenegro. Saudi J. Biol. Sci. 2016, 22, 390–397. [Google Scholar] [CrossRef] [Green Version]
- Apaydm, G.; Ayhkcr, V.; Cengiz, E.; Saydam, M.; Küp, N.; Tirasoglu, E. Analysis of metal contents of seaweed (Ulva lactuca) from Istanbul, Turkey by EDXRF. Turk. J. Fish. Aquat. Sci. 2010, 10, 215–220. [Google Scholar]
- Shaaban, A.E.S.; Mansour, H.A.; Saber, A.A. Soil algae of El-Farfara Oasis (The Western Desert, Egypt), and N2–fixation efficiency of five heterocytous cyanopytes. Egypt J. Bot. 2013, 57, 517–524. [Google Scholar]
- Wang, Y.; Qu, T.; Zhao, X.; Tang, X.; Xiao, H.; Tang, X. A comparative study of the photosynthetic capacity in two green tide macroalgae using chlorophyll fluorescence. Spring 2016, 5, 775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phang, S.M.; Yeong, H.Y.; Ganzon-Fortes, E.T.; Lewmanomont, K.; Prathep, A.; Hau, L.N.; Gerung, G.S.; Tan, K.S. Marine algae of the South China Sea bordered by Indonesia, Malaysia, Philippines, Singapore, Thailand and Vietnam. Raffles Bull. Zool. 2016, 40, 13–59. [Google Scholar]
- Shimada, S.; Yokoyama, N.; Arai, S.; Hiraoka, M. Phylogeography of the genus Ulva (Ulvophyceae, CHlorophyta), with special reference to the Japanese freshwater and brackish taxa. J. Appl. Phycol. 2008, 20, 979–989. [Google Scholar] [CrossRef]
- Lee, S.H.; Kang, P.J.; Nam, K.W. New record of two marine ulvalean species (Chlorophyta) in Korea. J. Ecol. Environ. 2014, 37, 375–389. [Google Scholar] [CrossRef] [Green Version]
- Taskin, E.; Öztürk, M.; Kurt, O.; Öztürk, M. Checklist of marine flora of Turkey. J. Phycol. 2018, 40, 71–72. [Google Scholar]
- Nguyen, T.V.; Le, N.H.; Lin, S.M.; Steen, F.; De Clerck, O. Checklist of the marine macroalgae of Vietnam. Bot. Mar. 2013, 56, 207–320. [Google Scholar] [CrossRef]
- Kraft, L.; Kraft, G.T.; Waller, R. Investigations into southern Australian Ulva (Ulvophyceae, Chlorophyta) taxonomy and molecular phylogeny indicate both cosmopolitanism and endemic cryptic species. J. Phycol. 2010, 46, 1257–1277. [Google Scholar] [CrossRef]
- Kirkendale-Saunders, L.; Winberg, P. A molecular survey of Ulva (Chlorophyta) in Temperate Australia reveals enhanced levels of cosmopolitanism. J. Phycol. 2012, 49, 324–328. [Google Scholar] [CrossRef]
- Messyasz, B.; Rybak, A. Abiotic factors affecting the development of Ulva sp. (Ulvophyceae; Chlorophyta) in freshwater ecosystems. Aquat Ecol. 2011, 45, 75–87. [Google Scholar] [CrossRef] [Green Version]
- Brodie, J.; Maggs, C.A.; John, D.M. Green Seaweeds of Britain and Ireland; British Phycological Society: London, UK, 2007; 250p. [Google Scholar]
- Tsiamis, K.; Panayotidis, P.; Economou-Amilli, A.; Katsaros, C. Seaweeds of the Greek coasts. II. Ulvophyceae. Med. Mar. Sci. 2014, 15, 449–461. [Google Scholar] [CrossRef] [Green Version]
- Anon, A. Inventaire National du Patrimoine Naturel; Muséum National d’Histoire Naturelle: Paris, France, 2017; pp. 45–51. [Google Scholar]
- Sfriso, A. Chlorophyta Multicellulari e Fanerogame Acquatiche. Ambiente di Transizione Italiani e Litorali Adiacenti. I Quaderni di ARPA; ARPA Emilia-Romagna: Bologna, Italy, 2011; 320p. [Google Scholar]
- Favot, G.; Engelen, A.H.; Cunha, M.E.; Serräo, M.E. Identification of Ulva sp. grown in multitropic aquaculture systems. J. Aquac Fish. 2019, 3, 24–31. [Google Scholar]
- De Zaixso, A.L.B. Elementos para el Estudio de las Macroalgas de Argentina; De Zaixso, C.C.J.M., Ed.; Editorial Universitaria de la Patagonia: Comodoro Rivadavia, Argentina, 2013; 204p. [Google Scholar]
- Rybak, S.A.; Czerwoniec, A.; Gąbka, M.; Messyasz, B. Ulva flexuosa (Ulvaceae, Chlorophyta) inhabiting inland aquatic ecosystems: Molecular, morphological and ecological discrimination of subspecies. Eur. J. Phycol. 2014, 49, 471–485. [Google Scholar] [CrossRef] [Green Version]
- Ichihara, K.; Arai, S.; Uchimura, M.; Fay, J.E.; Ebata, H.; Hiraoka, M.; Shimada, S. New species of freshwater Ulva, Ulva limnetica (Ulvales, Ulvophyceae) from the Ryukyu Islands, Japan. Phycol. Res. 2009, 57, 94–103. [Google Scholar] [CrossRef]
- Rybak, S.A. Revision of herbarium specimens of freshwater Enteromorpha-like Ulva (Ulvaceae, Chlorophyta) collected from Central Europe during the years 1849. Phytotaxa 2015, 1, 1–29. [Google Scholar] [CrossRef]
- Malta, E.J.; Draisma, S.G.A.; Kamermans, P. Free-floating Ulva in the southwest Netherlands; species or morphotypes? A morphological, molecular and ecological comparison. Eur. J. Phycol. 1999, 34, 443–454. [Google Scholar] [CrossRef]
- Loughnane, C.J.; McIvor, L.M.; Rindi, F.; Stengel, B.; Guiry, M. Morphology, rbcL phylogeny and distribution of distromatic Ulva (Ulvophyceae, Chlorophyta) in Ireland and southern Britan. J. Phycol. 2008, 47, 416–429. [Google Scholar] [CrossRef]
- Poole, L.J.J.; Raven, A. The biology of Enteromorpha. Prog. Phycol. Res. 1997, 12, 1–148. [Google Scholar]
- Imchen, T. Recruitment potential of a green alga Ulva flexuosa Wulfen dark preserved zoospore and its development. PLoS ONE 2012, 7, e32651. [Google Scholar] [CrossRef] [Green Version]
- Holzinger, A.; Herburger, K.; Kaplan, F.; Lewis, L.A. Desiccation tolerance the chlorophyte green alga Ulva compressa: Does cell wall architecture contribute to ecological success? Planta 2015, 242, 477–492. [Google Scholar] [CrossRef] [Green Version]
- Kirst, G.O. Salinity tolerance of eukaryotic marine algae. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 41, 21–53. [Google Scholar] [CrossRef]
- West, K.R.; Pitman, M.G. Ionic relations and ultrastructure in Ulva lactuca. Aust. J. Biol. Sci. 1967, 20, 901–914. [Google Scholar] [CrossRef] [Green Version]
- Ustunada, M.; Erdugan, H.; Yilmaz, S.; Akgul, R.; Aysel, V. Seasonal concentrations of some heavy metals (Cd, Pb, Zn, and Cu) in Ulva rigida J. Agardh (Chlorophyta) from Dardanelles (Canakkale, Turkey). Environ. Monit. Assess. 2011, 177, 337–342. [Google Scholar] [CrossRef]
- Villares, R.; Puente, X.; Carballeira, A. Ulva and Enteromorpha as indicators of heavy metal pollution. Hydrobiology 2001, 462, 221–232. [Google Scholar] [CrossRef]
- Clesceri, L.S.; Eaton, A.D.; Baird, R.B. Standard Methods for the Examination of Water and Waste Water; American Public Health Association: Washington, DC, USA, 2020; pp. 1–19. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. Molecular evolutionary genetics analyses version 6. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Kubala-Kukuś, A.; Arabski, M.; Stabrawa, I.; Banaś, D.; Różański, W.; Lipiński, M.; Majewska, U.; Wudarczyk-Moćko, J.; Braziewicz, J.; Pajek, M.; et al. Application of TXRF and XRPD techniques for analysis of elemental and chemical composition of human kidney stones. X-Ray Spec. 2017, 46, 412–420. [Google Scholar] [CrossRef] [Green Version]
- Arabski, M.; Stabrawa, I.; Kubala-Kukuś, A.; Gałczyńska, K.; Banaś, D.; Piskorz, Ł.; Forma, E.; Bryś, M.; Różański, W.; Lipiński, M. The correlation of crystalline and elemental composition of urinary stones with a history of bacterial infections: TXRF, XRPD and PCR-DGGE studies. Eur. Biophys. J. 2018, 48, 111–118. [Google Scholar] [CrossRef] [Green Version]
- Braak, C.J.F.T.; Šmilauer, P. Canoco Reference Manual and User’s Guide: Software for Ordination (Version 5.0); Microcomputer Power: Ithaca, NY, USA, 2012; pp. 1–496. [Google Scholar]
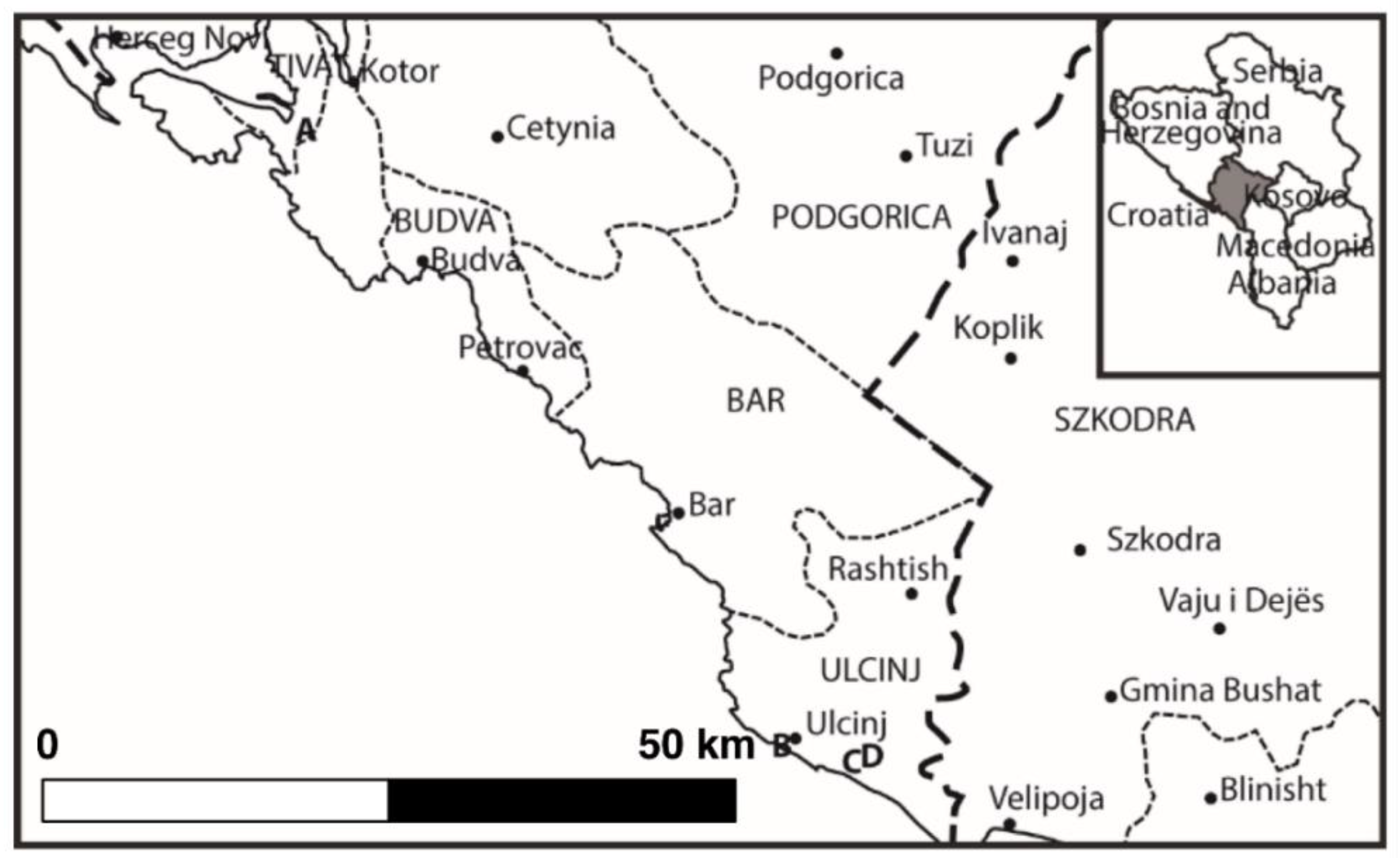


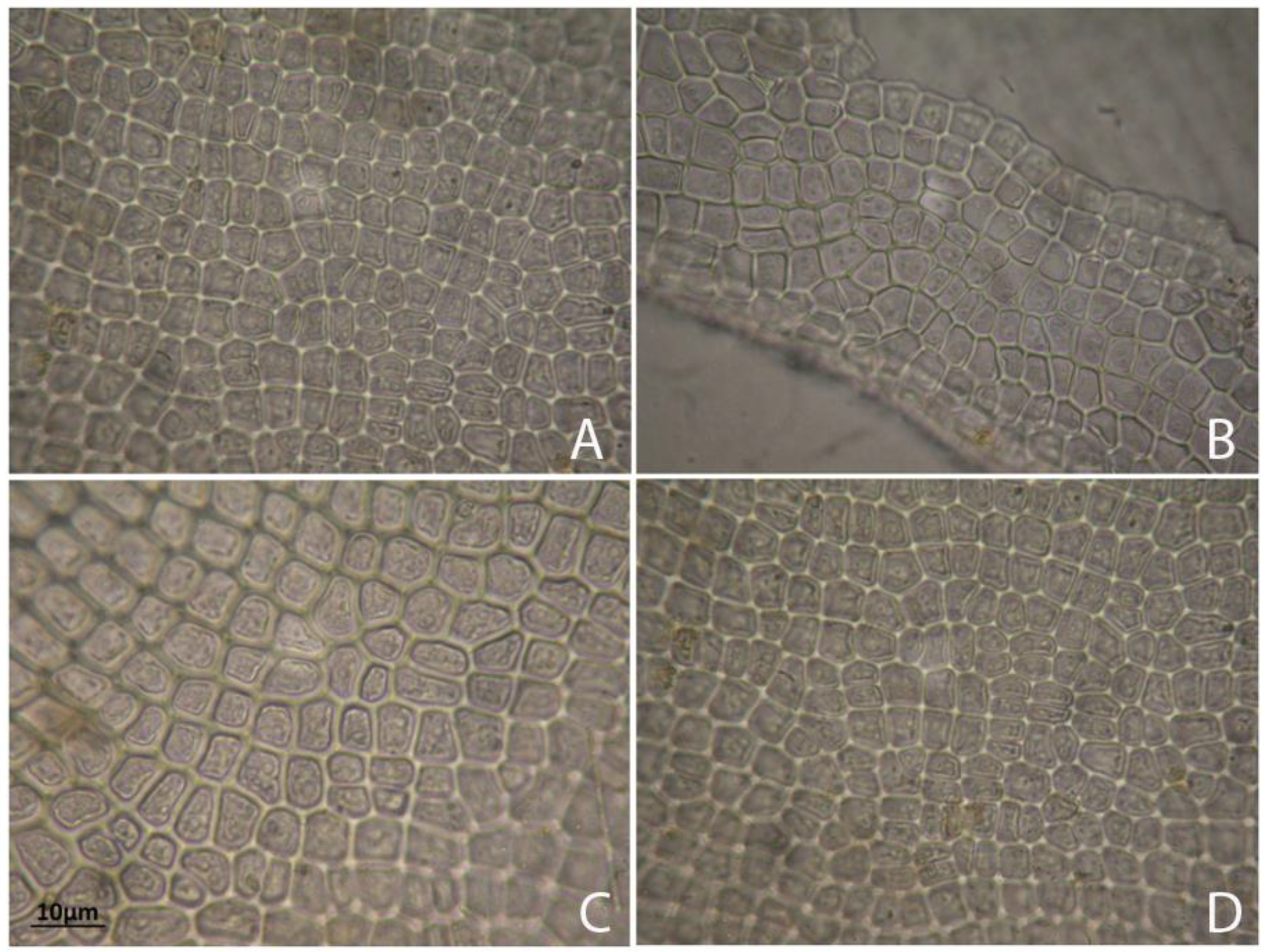


| Characters | Montenegro | Poland |
|---|---|---|
| morphology | tube-like | tubular, curled, and bubbled |
| thalli colour | bright to dark green (mature yellow-green) | bright green |
| branching | frequent | abundant to almost absent |
| uniseriate branches | frequent to rare, obtusely rounded ends | present |
| cell shape | rounded polygonal to quadrangular | rectangular, polygonal |
| structure of branch tips | globular cell on the apex | rectangular or square |
| number of pyrenoids | 2–3–4 | 2–4 |
| chloroplast shape | parietal | parietal, girdle-shaped |
| cell size: length of cells (μm) width of cells (μm) | 42.25–66.56 7.58–19.80 | 32.05–55.20 10.50–35.50 |
| thallus size: length of thalli (cm) width of thalli (mm) | 6.50–56.20 0.20–1.50 | 10.1–65.1 0.20–2.50 |
| cell arrangement | disordered, in small groups or short rows | disordered, in small groups or short rows |
| habitat | brackish and marine waters (sea coast) | enriched freshwaters (ponds, pools, small streams) |
| mode of life | attached or free floating | attached or free floating, occasionally in masses |
| Element | Concentration (%) ± Expanded Uncertainty | ||
|---|---|---|---|
| Site A | Site B | Site D | |
| Al | 0.186 ± 0.026 | 0.243 ± 0.030 | 0.310 ± 0.034 |
| Ba | 0.020 ± 0.008 | -------- | -------- |
| Br | -------- | 0.039 ± 0.012 | 0.013 ± 0.006 |
| Ca | 16.300 ± 0.200 | 0.854 ± 0.056 | 10.300 ± 0.200 |
| Cl | 0.560 ± 0.044 | 7.630 ± 0.170 | 0.498 ± 0.042 |
| Fe | 0.120 ± 0.020 | 0.129 ± 0.022 | 0.366 ± 0.036 |
| K | 0.698 ± 0.050 | 3.330 ± 0.110 | 1.450 ± 0.070 |
| Mg | 0.999 ± 0.060 | 1.390 ± 0.070 | 1.550 ± 0.060 |
| Mn | 0.299 ± 0.032 | -------- | 1.020 ± 0.060 |
| Na | 0.454 ± 0.040 | 4.990 ± 0.130 | 0.149 ± 0.024 |
| P | 0.070 ± 0.016 | 0.242 ± 0.030 | 0.372 ± 0.036 |
| S | 1.860 ± 0.080 | 1.860 ± 0.080 | 1.640 ± 0.080 |
| Si | 1.670 ± 0.080 | 1.250 ± 0.070 | 1.640 ± 0.080 |
| I | ----------- | 0.009 ± 0.006 | ------- |
| Element | Concentration (μg/g) ± Expanded Uncertainty | ||
|---|---|---|---|
| Site A | Site B | Site D | |
| Ti | 148.000 ± 2.000 | 82.600 ± 1.000 | 135.000 ± 2.000 |
| V | 13.800 ± 0.800 | 7.140 ± 0.540 | 8.090 ± 1.080 |
| Cr | 1.800 ± 0.640 | 9.190 ± 0.420 | 4.530 ± 0.900 |
| Ni | 22.200 ± 0.200 | 15.700 ± 0.200 | 39.200 ± 0.400 |
| Cu | 10.700 ± 0.200 | 9.030 ± 0.180 | 7.390 ± 0.240 |
| Zn | 11.300 ± 0.200 | 19.400 ± 0.200 | 20.100 ± 0.200 |
| As | 0.531 ± 0.138 | 11.100 ± 0.200 | 1.900 ± 0.140 |
| Rb | 5.680 ± 0.140 | 22.200 ± 0.200 | 6.920 ± 0.140 |
| Sr | 292.000 ± 2.000 | 541.000 ± 2.000 | 238.000 ± 2.000 |
| Hf | 58.400 ± 0.400 | 15.700 ± 0.200 | 27.200 ± 0.400 |
| Pb | 9.510 ± 0.180 | 6.030 ± 0.140 | 13.300 ± 0.200 |
| Region | Habitats | References | Notes |
|---|---|---|---|
| Africa | |||
| Egypt | limno-rheocrenic, thermal, mineral (chloride, sodium, sulphate) spring known as “Ain Abu Sherouf” in the Siwa Oasis; Red Sea coasts; brackish estuaries of the Bile River | Shaaban et al. [25] Saber et al. [4] | the Western Desert of Egypt |
| Asia | |||
| China | Yellow Sea, South China Sea | Wang et al. [26] Phang et al. [27] | the Subei Shoal coastal waters; bordered by Philippines |
| Japan | freshwater and brackish | Shimada et al. [28] | |
| Korea | marine | Lee et al. [29] | |
| Turkey | Aegean Sea | Taskin et al. [30] | |
| Vietnam | marine | Nguyen et al. [31] | mats |
| Australia | |||
| marine | Kraft et al. [32] Kirkendale-Saunders and Winberg [33] | algal blooms, southern Australia | |
| Europe | |||
| Czech Republic | lakes, ponds, streams, rivers in Central Europe | Mareš et al. [12] Messyasz and Rybak [34] | |
| Germany | pond | Kopp [20] Mareš et al. [12] | |
| Great Britain | near the top of the shore, on rocks or other algae, on open coasts or in estuaries and harbours | Brodie et al. [35] John et al. [1] | |
| Greece | Ionian Sea | Tsiamis et al. [36] | the Greek coasts |
| Hungary | freshwater | Mareš et al. [12] | |
| France | freshwater | Anon [37] | |
| Italy | marine | Sfriso [38] | Veneto, Mediterranean Sea |
| Montenegro | freshwater (a river, a ditch, the Milet Canal) and marine (on the rocky shore of the Adriatic Sea) | Czerwik-Marcinkowska et al. | |
| Poland | from freshwater to hyperhaline, and brine habitats | Mareš et al. [12] Messyasz et al. [18] Richter and Pietryka [19] | mats |
| Portugal | ponds | Favot et al. [39] | the Ria Formosa Lagoon |
| Slovakia | freshwater | Mareš et al. [12] | |
| Sweden | Mareš et al. [12] | ||
| North America | |||
| USA | freshwater | Lougheed and Stevenson [13] | Lake Michigan, Muskegon Lake |
| South America | |||
| Argentina | freshwater | Boraso and Zaixso [40] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Czerwik-Marcinkowska, J.; Piwowarczyk, R.; Uher, B.; Tomal, E.; Wojciechowska, A. X-ray Fluorescence Techniques in Determining the Habitat Preferences of Species—Ulva pilifera (Ulvales, Chlorophyta) from Montenegro Case Study. Molecules 2020, 25, 5022. https://doi.org/10.3390/molecules25215022
Czerwik-Marcinkowska J, Piwowarczyk R, Uher B, Tomal E, Wojciechowska A. X-ray Fluorescence Techniques in Determining the Habitat Preferences of Species—Ulva pilifera (Ulvales, Chlorophyta) from Montenegro Case Study. Molecules. 2020; 25(21):5022. https://doi.org/10.3390/molecules25215022
Chicago/Turabian StyleCzerwik-Marcinkowska, Joanna, Renata Piwowarczyk, Bohuslav Uher, Ewa Tomal, and Anna Wojciechowska. 2020. "X-ray Fluorescence Techniques in Determining the Habitat Preferences of Species—Ulva pilifera (Ulvales, Chlorophyta) from Montenegro Case Study" Molecules 25, no. 21: 5022. https://doi.org/10.3390/molecules25215022





