Effects of a Reserve Protein on Spodoptera frugiperda Development: A Biochemical and Molecular Approach to the Entomotoxic Mechanism
Abstract
1. Introduction
2. Results
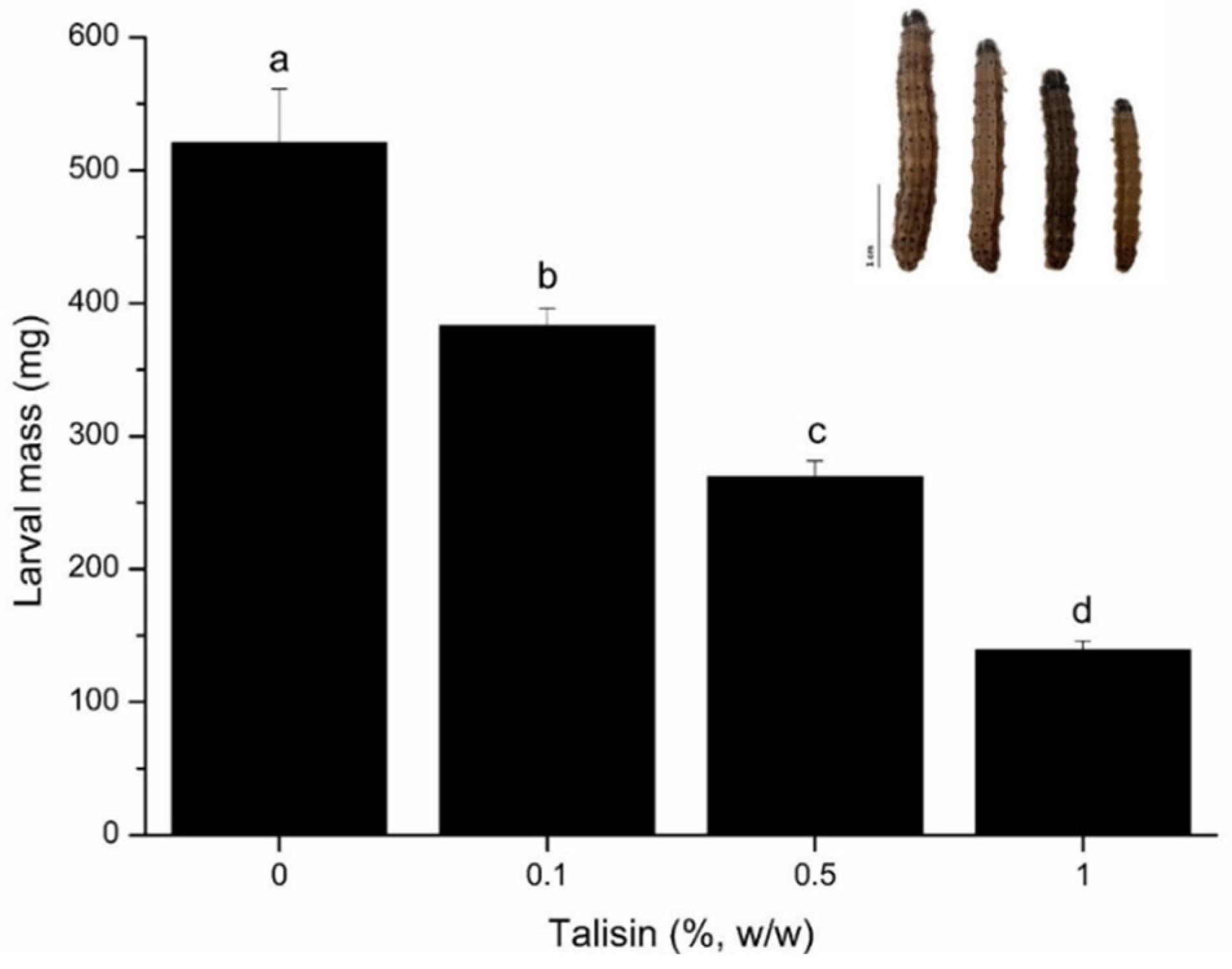
2.1. Effects of talisin on Insect Development
2.2. Nutritional Data
2.3. Digestive Enzyme Activity in Larvae Fed on Talisin-Amended Diet
2.4. In-Gel Visualization of Peptidase Activity
2.5. Talisin is Resistant to Hydrolysis by Midgut Peptidase
2.6. Microscopy Analysis
2.7. Real-Time PCR
2.8. Molecular Modeling and Docking
3. Discussion
4. Material and Methods
4.1. Talisin Extraction and Purification
4.2. Insects
4.3. In Vivo Insect Assays
4.4. Midgut and Frass Preparation
4.5. Nutritional Parameters
4.6. Protein Quantification
4.7. Enzymatic Assays
4.8. Peptidase Activity of Midgut Juice Extract in Native Polyacrylamide Gel Containing 1% Casein
4.9. Talisin Digestion
4.10. Microscopy Analysis
4.11. Quantitative Real-Time PCR
4.12. Molecular Modeling
4.13. Molecular Docking
4.14. Statistical Analysis
Author Contributions
Funding
Conflicts of Interest
References
- United Nations Population Division. The 2015 Revision of the UN’s World Population Projections. Popul. Dev. Rev. 2015, 41, 557–561. [Google Scholar] [CrossRef]
- Carzoli, A.K.; Aboobucker, S.I.; Sandall, L.L.; Lübberstedt, T.T.; Suza, W.P. Risks and opportunities of GM crops: Bt maize example. Glob. Food Secur. 2018, 19, 84–91. [Google Scholar] [CrossRef]
- De Santis, B.; Stockhofe, N.; Wal, J.-M.; Weesendorp, E.; Lallès, J.-P.; van Dijk, J.; Kok, E.; De Giacomo, M.; Einspanier, R.; Onori, R.; et al. Case studies on genetically modified organisms (GMOs): Potential risk scenarios and associated health indicators. Food. Chem. Toxicol. 2018, 117, 36–65. [Google Scholar] [CrossRef]
- Dunse, K.M.; Stevens, J.A.; Lay, F.T.; Gaspar, Y.M.; Heath, R.L.; Anderson, M.A. Coexpression of potato type I and II proteinase inhibitors gives cotton plants protection against insect damage in the field. Proc. Natl. Acad. Sci. USA 2010, 107, 15011–15015. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Qureshi, J.A.; Meagher, R.L., Jr.; Reisig, D.D.; Head, G.P.; Andow, D.A.; Ni, X.; Kerns, D.; Buntin, G.D.; Niu, Y.; et al. Cry1F resistance in fall armyworm Spodoptera frugiperda: Single gene versus pyramided Bt maize. PLoS ONE 2014, 9, e112958. [Google Scholar] [CrossRef] [PubMed]
- Jakka, S.R.K.; Gong, L.; Hasler, J.; Banerjee, R.; Sheets, J.J.; Narva, K.; Blanco, C.A.; Jurat-Fuentes, J.L. Field-evolved mode 1 resistance of the fall armyworm to transgenic Cry1fa-expressing corn associated with reduced Cry1fa toxin binding and midgut alkaline phosphatase expression. Appl. Environ. Microbiol. 2016, 82, 1023–1034. [Google Scholar] [CrossRef]
- Freire, M.d.G.M.; Vasconcelos, I.M.; Oliveira, M.V.; Filho, G.A.d.S.; Macedo, M.L.R. Characterization of a saccharide-binding protein from Talisia esculenta seeds with trypsin inhibitory activity. Protein Peptide Lett. 2009, 16, 1557–1564. [Google Scholar] [CrossRef]
- Dang, L.Y.; van Damme, E.J.M. Toxic proteins in plants. Phytochemistry 2015, 117, 51–64. [Google Scholar] [CrossRef]
- Freire, M.; Franco, O.L.; Kubo, C.E.; Migliolo, L.; Vargas, R.H.; de Oliveira, C.F.; Parra, J.R.; Macedo, M.L. Structural insights regarding an insecticidal Talisia esculenta protein and its biotechnological potential for Diatraea saccharalis larval control. Comp. Biochem. Phys. B 2012, 161, 86–92. [Google Scholar] [CrossRef]
- Macedo, M.; Freire, M.; Kubo, C.; Parra, J. Bioinsecticidal activity of Talisia esculenta reserve protein on growth and serine digestive enzymes during larval development of Anticarsia gemmatalis. Comp. Biochem. Phys. C 2010, 153, 24–33. [Google Scholar] [CrossRef]
- Macedo, M.; das Graças Machado Freire, M.; da Silva, M.; Coelho, L. Insecticidal action of Bauhinia monandra leaf lectin (BmoLL) against Anagasta kuehniella (Lepidoptera: Pyralidae), Zabrotes subfasciatus and Callosobruchus maculatus (Coleoptera: Bruchidae). Comp. Biochem. Phys. A 2007, 146, 486–498. [Google Scholar] [CrossRef] [PubMed]
- Dias, R.O.; Via, A.; Brandão, M.M.; Tramontano, A.; Silva-Filho, M.C. Digestive peptidase evolution in holometabolous insects led to a divergent group of enzymes in Lepidoptera. Insect Biochem. Molec. 2015, 58, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Montezano, D.G.; Specht, A.; Sosa-Gómez, D.R.; Roque-Specht, V.F.; Sousa-Silva, J.C.; Paula-Moraes, S.V.; Peterson, J.A.; Hunt, T.E. Host plants of Spodoptera frugiperda (Lepidoptera: Noctuidae) in the Americas. Afr. Entomol. 2018, 26, 286–300. [Google Scholar] [CrossRef]
- Javaid, S.; Naz, S.; Amin, I.; Jander, G.; Ul-Haq, Z.; Mansoor, S. Computational and biological characterization of fusion proteins of two insecticidal proteins for control of insect pests. Sci. Rep. 2018, 8, 4837. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef]
- Chen, V.B.; Arendall, W.B., 3rd; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 12–21. [Google Scholar] [CrossRef]
- War, A.R.; Paulraj, M.G.; Ahmad, T.; Buhroo, A.A.; Hussain, B.; Ignacimuthu, S.; Sharma, H.C. Mechanisms of plant defense against insect herbivores. Plant Signal. Behav. 2012, 7, 1306–1320. [Google Scholar] [CrossRef]
- Macedo, M.L.R.; Oliveira, C.F.R.; Oliveira, C.T. Insecticidal activity of plant lectins and potential application in crop protection. Molecules 2015, 20, 2014–2033. [Google Scholar] [CrossRef]
- Stevens, J.A.; Dunse, K.M.; Guarino, R.F.; Barbeta, B.L.; Evans, S.C.; West, J.A.; Anderson, M.A. The impact of ingested potato type II inhibitors on the production of the major serine proteases in the gut of Helicoverpa armigera. Insect Biochem. Molec. 2013, 43, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Howe, G.A.; Herde, M. Interaction of plant defense compounds with the insect gut: New insights from genomic and molecular analyses. Curr. Opin. Insect Sci. 2015, 9, 62–68. [Google Scholar] [CrossRef]
- De Oliveira, C.F.R.; de Oliveira Flores, T.M.; Henrique Cardoso, M.; Garcia Nogueira Oshiro, K.; Russi, R.; de França, A.F.J.; dos Santos, E.A.; Luiz Franco, O.; de Oliveira, A.S.; Migliolo, L. Dual insecticidal effects of Adenanthera pavonina kunitz-type inhibitor on Plodia interpunctella is mediated by digestive enzymes inhibition and chitin-binding properties. Molecules 2019, 24, 4344. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, C.T.; Kunz, D.; Silva, C.P.; Macedo, M.L.R. Entomotoxic properties of Dioclea violacea lectin and its effects on digestive enzymes of Anagasta kuehniella (Lepidoptera). J. Insect. Physiol. 2015, 81, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Rath, S.S.; Prasad, B.C.; Sinha, B.R.R.P. Food utilization efficiency in fifth instar larvae of Antheraea mylitta (Lepidoptera: Saturniidae) infected with Nosema sp. and its effect on reproductive potential and silk production. J. Invertebr. Pathol. 2003, 83, 1–9. [Google Scholar] [CrossRef]
- Li, Y.; Romeis, J. Impact of snowdrop lectin (Galanthus nivalis agglutinin; GNA) on adults of the green lacewing, Chrysoperla carnea. J. Insect. Physiol. 2009, 55, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Gahloth, D.; Shukla, U.; Birah, A.; Gupta, G.P.; Kumar, P.A.; Dhaliwal, H.S.; Sharma, A.K. Bioinsecticidal activity of Murraya koenigii miraculin-like protein against Helicoverpa armigera and Spodoptera litura. Arch. Insect Biochem. Physiol. 2011, 78, 132–144. [Google Scholar] [CrossRef]
- Chen, H.; Wilkerson, C.G.; Kuchar, J.A.; Phinney, B.S.; Howe, G.A. Jasmonate-inducible plant enzymes degrade essential amino acids in the herbivore midgut. Proc. Natl. Acad. Sci. USA 2005, 102, 19237. [Google Scholar] [CrossRef]
- Chen, M.-S. Inducible direct plant defense against insect herbivores: A review. Insect Sci. 2008, 15, 101–114. [Google Scholar] [CrossRef]
- Girard, C.; Le Métayer, M.; Bonadé-Bottino, M.; Pham-Delègue, M.-H.; Jouanin, L. High level of resistance to proteinase inhibitors may be conferred by proteolytic cleavage in beetle larvae. Insect Biochem. Mol. 1998, 28, 229–237. [Google Scholar] [CrossRef]
- Macedo, M.; de Castro, M.; Freire, M. Mechanisms of the insecticidal action of TEL (Talisia esculenta lectin) against Callosobruchus maculatus (Coleoptera: Bruchidae). Arch. Insect Biochem. Physiol. 2004, 56, 84–96. [Google Scholar] [CrossRef]
- Oliveira, C.F.R.; Paula Souza, T.; Parra, J.R.P.; Marangoni, S.; Castro Silva-Filho, M.; Macedo, M.L.R. Insensitive trypsins are differentially transcribed during Spodoptera frugiperda adaptation against plant protease inhibitors. Comp. Biochem. Phys. B 2013, 165, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Kuwar, S.S.; Pauchet, Y.; Vogel, H.; Heckel, D.G. Adaptive regulation of digestive serine proteases in the larval midgut of Helicoverpa armigera in response to a plant protease inhibitor. Insect Biochem. Mol. 2015, 59, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.E.; Salzman, R.A.; Braunagel, S.C.; Koiwa, H.; Zhu-Salzman, K. Functional roles of specific bruchid protease isoforms in adaptation to a soybean protease inhibitor. Insect Mol. Biol. 2004, 13, 649–657. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.E.; Lovingshimer, M.R.; Salzman, R.A.; Presnail, J.K.; Lu, A.L.; Koiwa, H.; Zhu-Salzman, K. Cowpea bruchid Callosobruchus maculatus counteracts dietary protease inhibitors by modulating propeptides of major digestive enzymes. Insect Mol. Biol. 2007, 16, 295–304. [Google Scholar] [CrossRef]
- Zhu-Salzman, K.; Koiwa, H.; Salzman, R.A.; Shade, R.E.; Ahn, J.E. Cowpea bruchid Callosobruchus maculatus uses a three-component strategy to overcome a plant defensive cysteine protease inhibitor. Insect Mol. Biol. 2003, 12, 135–145. [Google Scholar] [CrossRef]
- Brioschi, D.; Nadalini, L.D.; Bengtson, M.H.; Sogayar, M.C.; Moura, D.S.; Silva-Filho, M.C. General up regulation of Spodoptera frugiperda trypsins and chymotrypsins allows its adaptation to soybean proteinase inhibitor. Insect Biochem. Mol. 2007, 37, 1283–1290. [Google Scholar] [CrossRef]
- Hivrale, V.K.; Lomate, P.R.; Basaiyye, S.S.; Kalve, N.D. Compensatory proteolytic responses to dietary proteinase inhibitors from Albizia lebbeck seeds in the Helicoverpa armigera larvae. Arthropod-Plant Interact. 2013, 7, 259–266. [Google Scholar] [CrossRef]
- Terra, W.R.; Ferreira, C. Biochemistry and Molecular Biology of Digestion. In Insect Molecular Biology and Biochemistry; Gilbert, L.I., Ed.; Academic Press: San Diego, CA, USA, 2012; pp. 365–418. [Google Scholar] [CrossRef]
- Bown, D.P.; Wilkinson, H.S.; Gatehouse, J.A. Regulation of expression of genes encoding digestive proteases in the gut of a polyphagous lepidopteran larva in response to dietary protease inhibitors. Physiol. Entomol. 2004, 29, 278–290. [Google Scholar] [CrossRef]
- Souza, T.P.; Dias, R.O.; Castelhano, E.C.; Brandão, M.M.; Moura, D.S.; Silva-Filho, M.C. Comparative analysis of expression profiling of the trypsin and chymotrypsin genes from Lepidoptera species with different levels of sensitivity to soybean peptidase inhibitors. Comp. Biochem. Phys. B 2016, 196–197, 67–73. [Google Scholar] [CrossRef]
- Migliolo, L.; Oliveira, A.S.; Santos, E.A.; Franco, O.L.; Sales, M.P. Structural and mechanistic insights into a novel non-competitive Kunitz trypsin inhibitor from Adenanthera pavonina L. seeds with double activity toward serine- and cysteine-proteinases. J. Mol. Graph. Model. 2010, 29, 148–156. [Google Scholar] [CrossRef]
- Bendre, A.D.; Ramasamy, S.; Suresh, C.G. Analysis of Kunitz inhibitors from plants for comprehensive structural and functional insights. Int. J. Biol. Macromol. 2018, 113, 933–943. [Google Scholar] [CrossRef]
- Van Damme, E.J.M.; Hao, Q.; Barre, A.; Rougé, P.; Van Leuven, F.; Peumans, W.J. Major protein of resting rhizomes of Calystegia sepium (hedge bindweed) closely resembles plant RNAses but has no enzymatic activity. Plant Physiol. 2000, 122, 433–446. [Google Scholar] [CrossRef] [PubMed]
- Troncoso, M.F.; Zolezzi, P.C.; Hellman, U.; Wolfenstein-Todel, C. A novel trypsin inhibitor from Peltophorum dubium seeds, with lectin-like properties, triggers rat lymphoma cell apoptosis. Arch. Biochem. Biophys. 2003, 411, 93–104. [Google Scholar] [CrossRef]
- Macedo, M.L.R.; Freire, M.G.M.; Martins, L.T.; Martinez, D.S.; Gomes, V.M.; Smolka, M.B.; Toyama, M.H.; Marangoni, S.; Coelho, L.C.B.B. Novel protein from Labramia bojeri A. DC. seeds homologue to Kunitz-type trypsin inhibitor with lectin-like properties. J. Agric. Food Chem. 2004, 52, 7548–7554. [Google Scholar] [CrossRef] [PubMed]
- Shah, K.R.; Patel, D.K.; Pappachan, A.; Prabha, C.R.; Singh, D.D. Characterization of a Kunitz-type serine protease inhibitor from Solanum tuberosum having lectin activity. Int. J. Biol. Macromol. 2016, 83, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Macedo, M.L.R.; Freire, M.d.G.M.; Novello, J.C.; Marangoni, S. Talisia esculenta lectin and larval development of Callosobruchus maculatus and Zabrotes subfasciatus (Coleoptera: Bruchidae). Biochim. Biophys. Acta 2002, 1571, 83–88. [Google Scholar] [CrossRef]
- Silva, W.; Cardoso, C.; Ribeiro, A.F.; Terra, W.R.; Ferreira, C. Midgut proteins released by microapocrine secretion in Spodoptera frugiperda. J. Insect Physiol. 2013, 59, 70–80. [Google Scholar] [CrossRef][Green Version]
- Hegedus, D.; Erlandson, M.; Gillott, C.; Toprak, U. New Insights into peritrophic matrix synthesis, architecture, and function. Annu. Rev. Entomol. 2009, 54, 285–302. [Google Scholar] [CrossRef]
- Bolognesi, R.; Terra, W.R.; Ferreira, C. Peritrophic membrane role in enhancing digestive efficiency: Theoretical and experimental models. J. Insect Physiol. 2008, 54, 1413–1422. [Google Scholar] [CrossRef]
- Bolognesi, R.; Ribeiro, A.F.; Terra, W.R.; Ferreira, C. The peritrophic membrane of Spodoptera frugiperda: Secretion of peritrophins and role in immobilization and recycling digestive enzymes. Arch. Insect Biochem. 2001, 47, 62–75. [Google Scholar] [CrossRef]
- Caccia, S.; Van Damme, E.J.M.; De Vos, W.H.; Smagghe, G. Mechanism of entomotoxicity of the plant lectin from Hippeastrum hybrid (Amaryllis) in Spodoptera littoralis larvae. J. Insect Physiol. 2012, 58, 1177–1183. [Google Scholar] [CrossRef] [PubMed]
- Minoo Sajjadian, S.; Hosseininaveh, V. Destruction of peritrophic membrane and its effect on biological characteristics and activity of digestive enzymes in larvae of the Indian meal moth, Plodia interpunctella (Lepidoptera: Pyralidae). EJE 2015, 112, 245–250. [Google Scholar] [CrossRef]
- Kariyat, R.R.; Smith, J.D.; Stephenson, A.G.; De Moraes, C.M.; Mescher, M.C. Non-glandular trichomes of Solanum carolinense deter feeding by Manduca sexta caterpillars and cause damage to the gut peritrophic matrix. Proc. R. Soc. B-Biol. Sci. 2017, 284, 20162323. [Google Scholar] [CrossRef] [PubMed]
- Pechan, T.; Cohen, A.; Williams, W.P.; Luthe, D.S. Insect feeding mobilizes a unique plant defense protease that disrupts the peritrophic matrix of caterpillars. Proc. Natl. Acad. Sci. USA 2002, 99, 13319. [Google Scholar] [CrossRef]
- Senthilkumar, R.; Cheng, C.P.; Yeh, K.W. Genetically pyramiding protease-inhibitor genes for dual broad-spectrum resistance against insect and phytopathogens in transgenic tobacco. Plant Biotechnol. J. 2010, 8, 65–75. [Google Scholar] [CrossRef]
- Babu, M.R.; Sajeena, A.; Seetharaman, K.; Reddy, M.S. Advances in genetically engineered (transgenic) plants in pest management—An over view. Crop Prot. 2003, 22, 1071–1086. [Google Scholar] [CrossRef]
- Kos, M.; van Loon, J.J.A.; Dicke, M.; Vet, L.E.M. Transgenic plants as vital components of integrated pest management. Trends Biotechnol. 2009, 27, 621–627. [Google Scholar] [CrossRef]
- Parra, J.R.P. Técnicas de Criação de Insetos para Programas de Controle Biológico, 3rd ed.; ESALQ/FEALQ: Piracicaba, Brazil, 1996; p. 137. [Google Scholar]
- Waldbauer, G.P. The consumption and utilization of food by insects. In Advances in Insect Physiology; Beament, J.W.L., Treherne, J.E., Wigglesworth, V.B., Eds.; Academic Press: London, UK, 1968; Volume 5, pp. 229–288. [Google Scholar]
- Scriber, J.M.; Slansky, F., Jr. The nutritional ecology of immature insects. Annu. Rev. Entomol. 1981, 26, 183–211. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Oliveira, C.F.R.; Luz, L.A.; Paiva, P.M.G.; Coelho, L.C.B.B.; Marangoni, S.; Macedo, M.L.R. Evaluation of seed coagulant Moringa oleifera lectin (cMoL) as a bioinsecticidal tool with potential for the control of insects. Process Biochem. 2011, 46, 498–504. [Google Scholar] [CrossRef]
- Noelting, G.; Bernfeld, P. Sur les enzymes amylolytiques. III. La β-amylase: Dosage d’activité et controle de l’absence d’α-amylase. Helv. Chim. Acta 1948, 31, 286–290. [Google Scholar] [CrossRef] [PubMed]
- Erlanger, B.F.; Kokowsky, N.; Cohen, W. The preparation and properties of two new chromogenic substrates of trypsin. Arch. Biochem. Biophys. 1961, 95, 271–278. [Google Scholar] [CrossRef]
- Laemmli, U. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Shu, B.; Zhang, J.; Cui, G.; Sun, R.; Sethuraman, V.; Yi, X.; Zhong, G. Evaluation of reference genes for real-time quantitative PCR analysis in larvae of Spodoptera litura exposed to azadirachtin stress conditions. Front. Physiol. 2018, 9, 372. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29. [Google Scholar] [CrossRef]
- Käll, L.; Krogh, A.; Sonnhammer, E.L.L. Advantages of combined transmembrane topology and signal peptide prediction—the Phobius web server. Nucleic Acids Res. 2007, 35, W429–W432. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Eswar, N.; Webb, B.; Marti-Renom, M.A.; Madhusudhan, M.S.; Eramian, D.; Shen, M.-Y.; Pieper, U.; Sali, A. Comparative Protein Structure Modeling Using Modeller. Curr. Protoc. Bioinform. 2006, 15, 5.6.1–5.6.30. [Google Scholar] [CrossRef]
- Krauchenco, S.; Pando, S.C.; Marangoni, S.; Polikarpov, I. Crystal structure of the Kunitz (STI)-type inhibitor from Delonix regia seeds. Biochem. Biophys. Res. Commun. 2003, 312, 1303–1308. [Google Scholar] [CrossRef]
- Rypniewski, W.R.; Ostergaard, P.R.; Norregaard-Madsen, M.; Dauter, M.; Wilson, K.S. Fusarium oxysporum trypsin at atomic resolution at 100 and 283 K: A study of ligand binding. Acta Cryst. D 2001, 57, 8–19. [Google Scholar] [CrossRef]
- Broutin, I.; Arnoux, B.; Riche, C.; Lecroisey, A.; Keil, B.; Pascard, C.; Ducruix, A. 1.8 Å structure of Hypoderma lineatum collagenase: A member of the serine proteinase family. Acta Cryst. D 1996, 52, 380–392. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2009, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |







| Parameter | Control | 0.5% Talisin |
|---|---|---|
| Pupal mass (mg) | 275.08 ± 13.25 a | 272.86 ± 15.67 a |
| Larval stage (days) | 18.80 ± 0.97 a | 19.70 ± 0.78 b |
| Pupal stage (days) | 9.60 ± 0.91 a | 10.40 ± 1.01 a |
| Adult life span (days) | 9.66 ± 1.75 a | 8.33 ± 1.10 a |
| Total development time (days) | 35.70 ± 2.53 a | 39.00 ± 0.89 b |
| Survival to adulthood (%) | 100 a | 100 a |
| Parameter | Control | 0.5% Talisin |
|---|---|---|
| Relative Consumption Rate (g/g/day) | 0.7490 ± 0.0801 a | 0.7646 ± 0.2592 a |
| Relative Growth Ratio (g/g/day) | 0.0833 ± 0.0060 a | 0.0809 ± 0.0102 a |
| Relative Metabolic Ratio (g/g/day) | 0.3907 ± 0.0613 a | 0.4605 ± 0.2018 a |
| Approximate Digestibility (%) | 63.526 ± 4.0253 a | 76.8555 ± 7.6049 b |
| Efficiency of Conversion of Ingested Food (%) | 11.966 ± 1.8135 a | 11.2151 ± 3.4421 a |
| Efficiency of Conversion of Digested Food (%) | 17.861 ± 2.6050 a | 14.8380 ± 5.8188 a |
| Metabolic Cost (%) | 85.447 ± 6.5185 a | 92.5912 ± 6.5681 a |
| Predicted Structures | Sequence Length | Fold Quality (Z-Score) | Stereochemistry (G-Factors) | Ramachandran Most Favored (%) | Ramachandran Allowed (%) | Ramachandran Outliers (%) | Bad Bonds (%) | Bad Angles (%) |
|---|---|---|---|---|---|---|---|---|
| Talisin | 198 | −4.62 | −0.28 | 87.7 | 94.9 | 5.10 | 0.00 | 1.58 |
| Trypsin 6 | 233 | −5.82 | −0.21 | 94.8 | 98.3 | 1.73 | 0.06 | 2.14 |
| Trypsin 12 | 232 | −5.28 | −0.28 | 93.0 | 97.0 | 3.04 | 0.00 | 2.22 |
| Chymotrypsin 2 | 234 | −5.42 | −0.25 | 94.8 | 97.8 | 2.16 | 0.00 | 1.68 |
| Chymotrypsin 21 | 237 | −5.82 | −0.22 | 91.5 | 98.7 | 1.28 | 0.17 | 1.90 |
| Residues | Positions | Atom Names | Distances (Å) | Residues | Positions | Atom Names | Interactions |
|---|---|---|---|---|---|---|---|
| Trypsin 6 (XP_022821647.1 *) | Talisin (ACJ51124.1 *) | ||||||
| Asn | 37 | ND2 | 1.9 | Ser | 106 | OG | HB |
| Ser | 112 | OG | 3.1 | Ser | 50 | N | HB |
| Ile | 113 | O | 1.9 | Gln | 48 | NE2 | HB |
| Ile | 113 | N | 2.5 | Ser | 49 | OG | HB |
| Gly | 115 | N | 3.1 | Gln | 48 | NE2 | HB |
| Ala | 116 | CB | 3.6 | Leu | 34 | CD2 | H |
| Asn | 197 | O | 2.1 | Gln | 53 | NE2 | HB |
| Ile | 199 | N | 3.5 | Gln | 53 | OE1 | HB |
| Ile | 199 | N | 3.3 | Gln | 53 | NE2 | HB |
| Ser | 231 | OG | 1.9 | Pro | 35 | O | HB |
| Trypsin 12 (XP_022821658 *) | Talisin (ACJ51124.1 *) | ||||||
| Gly | 4 | O | 2.9 | Ile | 117 | O | HB |
| Thr | 133 | CG | 3.6 | Val | 116 | CG1 | H |
| Thr | 133 | OG1 | 3.6 | Ile | 117 | N | HB |
| Tyr | 134 | O | 3.2 | Tyr | 65 | OH | HB |
| Tyr | 134 | CE2 | 3.6 | Met | 126 | CE | H |
| Tyr | 135 | OH | 2.0 | Tyr | 65 | O | HB |
| Ala | 137 | O | 1.9 | Tyr | 65 | OH | HB |
| Pro | 138 | O | 3.3 | Val | 96 | N | HB |
| Thr | 139 | O | 1.7 | Ser | 94 | OG | HB |
| Thr | 139 | OG1 | 2.3 | Gln | 82 | OE1 | HB |
| Thr | 139 | OG1 | 3.2 | Val | 96 | N | HB |
| Ser | 141 | OG | 3.4 | Tyr | 93 | N | HB |
| Arg | 145 | NH2 | 2.3 | Ile | 117 | O | HB |
| Arg | 145 | NE | 2.7 | Ile | 117 | O | HB |
| Chymotrypsin 2 (ALO61082.1 *) | Distances (Å) | Talisin (ACJ51124.1 *) | Interactions | ||||
|---|---|---|---|---|---|---|---|
| Residues | Positions | Atom Names | Residues | Positions | Atom Names | ||
| Ile | 35 | O | 2.6 | Asn | 66 | OD1 | HB |
| Ile | 35 | O | 3.2 | Asn | 66 | ND2 | HB |
| His | 81 | CE1 | 3.5 | Val | 116 | CG1 | H |
| Ile | 110 | O | 2.8 | Asn | 66 | ND2 | HB |
| Leu | 112 | N | 3.2 | Tyr | 65 | O | HB |
| Asn | 119 | OD1 | 3.1 | Val | 149 | N | H |
| Asn | 119 | OD1 | 3.2 | Ser | 148 | OG | HB |
| Leu | 201 | CD2 | 3.6 | Tyr | 65 | CE1 | H |
| Ser | 229 | OG | 2.9 | Lys | 98 | NZ | HB |
| Gln | 232 | OE1 | 2.9 | Val | 63 | O | HB |
| Gln | 232 | OE1 | 3.2 | Asn | 80 | OD1 | HB |
| Ser | 233 | O | 3.4 | Ser | 94 | OG | HB |
| Ser | 233 | OG | 3.5 | Gln | 82 | OE1 | HB |
| Gln | 234 | O | 2.9 | Gln | 82 | ND2 | HB |
| Gln | 234 | OE1 | 2.9 | Tyr | 93 | OH | HB |
| Gln | 234 | NE2 | 3.0 | Tyr | 93 | OH | HB |
| Trypsin 21 (AIR09774.1 *) | Talisin (ACJ51124.1 *) | ||||||
| Leu | 9 | N | 3.0 | Asp | 70 | OD2 | HB |
| Glu | 11 | OE1 | 3.6 | Gly | 67 | N | HB |
| Gln | 102 | NE2 | 3.3 | Asp | 192 | OD2 | HB |
| Phe | 103 | O | 3.0 | Asp | 192 | N | HB |
| Ala | 111 | CB | 3.3 | Tyr | 93 | CD1 | H |
| Leu | 112 | O | 2.9 | Tyr | 93 | OH | HB |
| Ser | 116 | OG | 3.5 | Gln | 92 | OE1 | HB |
| Gln | 117 | OE1 | 2.9 | Tyr | 93 | N | HB |
| Tyr | 151 | OH | 3.6 | Asn | 66 | O | HB |
| Gln | 197 | NE2 | 3.1 | Asn | 66 | ND2 | HB |
| Gln | 197 | OE1 | 3.6 | Asn | 66 | ND2 | HB |
| Arg | 198 | NH2 | 3.4 | Tyr | 65 | O | HB |
| Arg | 198 | NH2 | 3.5 | Asn | 66 | O | HB |
| Gly | 200 | O | 3.6 | Tyr | 65 | OH | HB |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oliveira, C.T.; Machado, S.W.; Bezerra, C.d.S.; Cardoso, M.H.; Franco, O.L.; Silva, C.P.; Alves, D.G.; Rios, C.; Macedo, M.L.R. Effects of a Reserve Protein on Spodoptera frugiperda Development: A Biochemical and Molecular Approach to the Entomotoxic Mechanism. Molecules 2020, 25, 2195. https://doi.org/10.3390/molecules25092195
Oliveira CT, Machado SW, Bezerra CdS, Cardoso MH, Franco OL, Silva CP, Alves DG, Rios C, Macedo MLR. Effects of a Reserve Protein on Spodoptera frugiperda Development: A Biochemical and Molecular Approach to the Entomotoxic Mechanism. Molecules. 2020; 25(9):2195. https://doi.org/10.3390/molecules25092195
Chicago/Turabian StyleOliveira, Carolina Turatti, Suzy Wider Machado, Cézar da Silva Bezerra, Marlon Henrique Cardoso, Octávio Luiz Franco, Carlos Peres Silva, Demetrio Gomes Alves, Cristina Rios, and Maria Lígia R. Macedo. 2020. "Effects of a Reserve Protein on Spodoptera frugiperda Development: A Biochemical and Molecular Approach to the Entomotoxic Mechanism" Molecules 25, no. 9: 2195. https://doi.org/10.3390/molecules25092195
APA StyleOliveira, C. T., Machado, S. W., Bezerra, C. d. S., Cardoso, M. H., Franco, O. L., Silva, C. P., Alves, D. G., Rios, C., & Macedo, M. L. R. (2020). Effects of a Reserve Protein on Spodoptera frugiperda Development: A Biochemical and Molecular Approach to the Entomotoxic Mechanism. Molecules, 25(9), 2195. https://doi.org/10.3390/molecules25092195







