Automated Trimethyl Sulfonium Hydroxide Derivatization Method for High-Throughput Fatty Acid Profiling by Gas Chromatography–Mass Spectrometry
Abstract
:1. Introduction
2. Results and Discussion
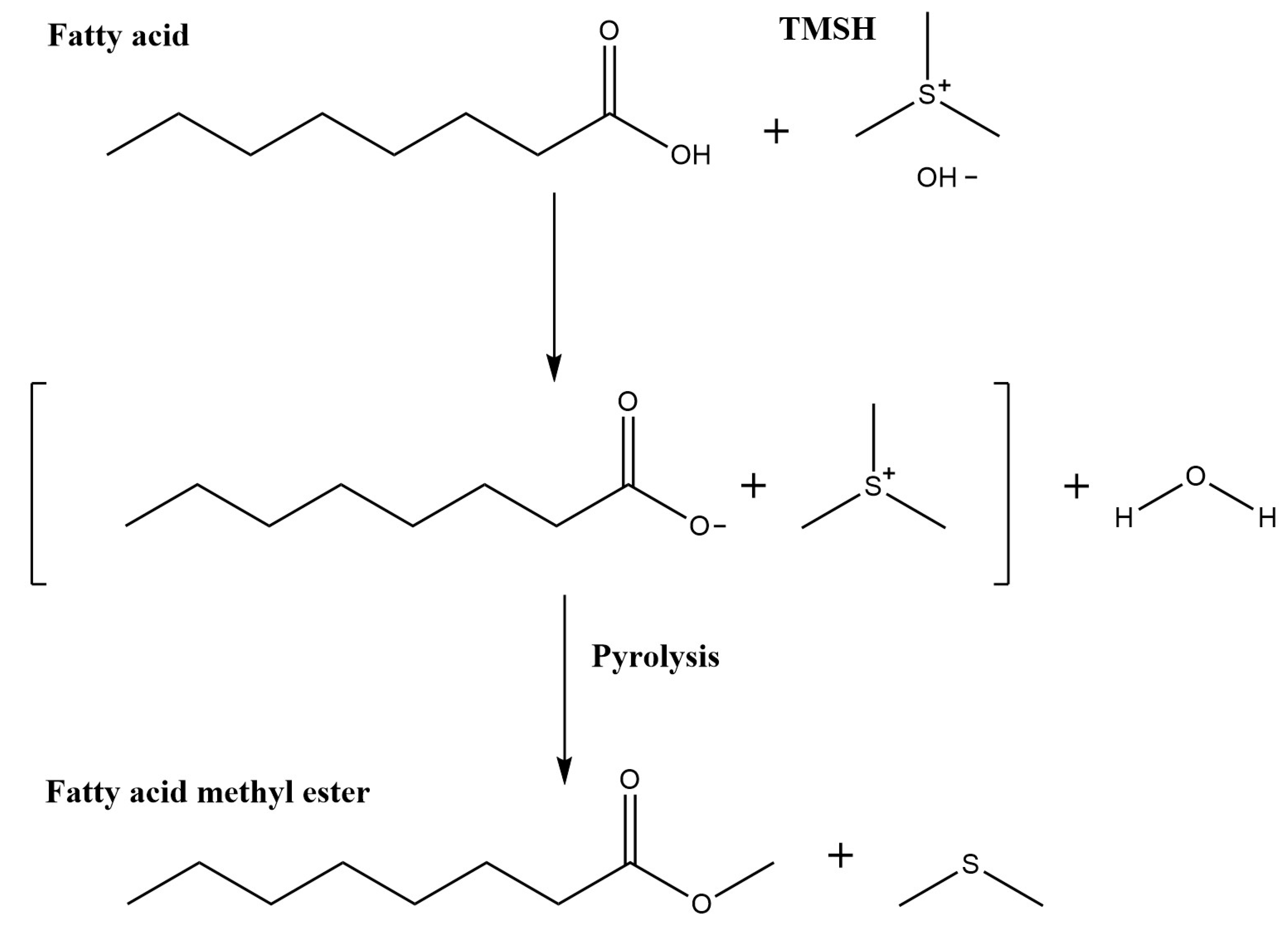
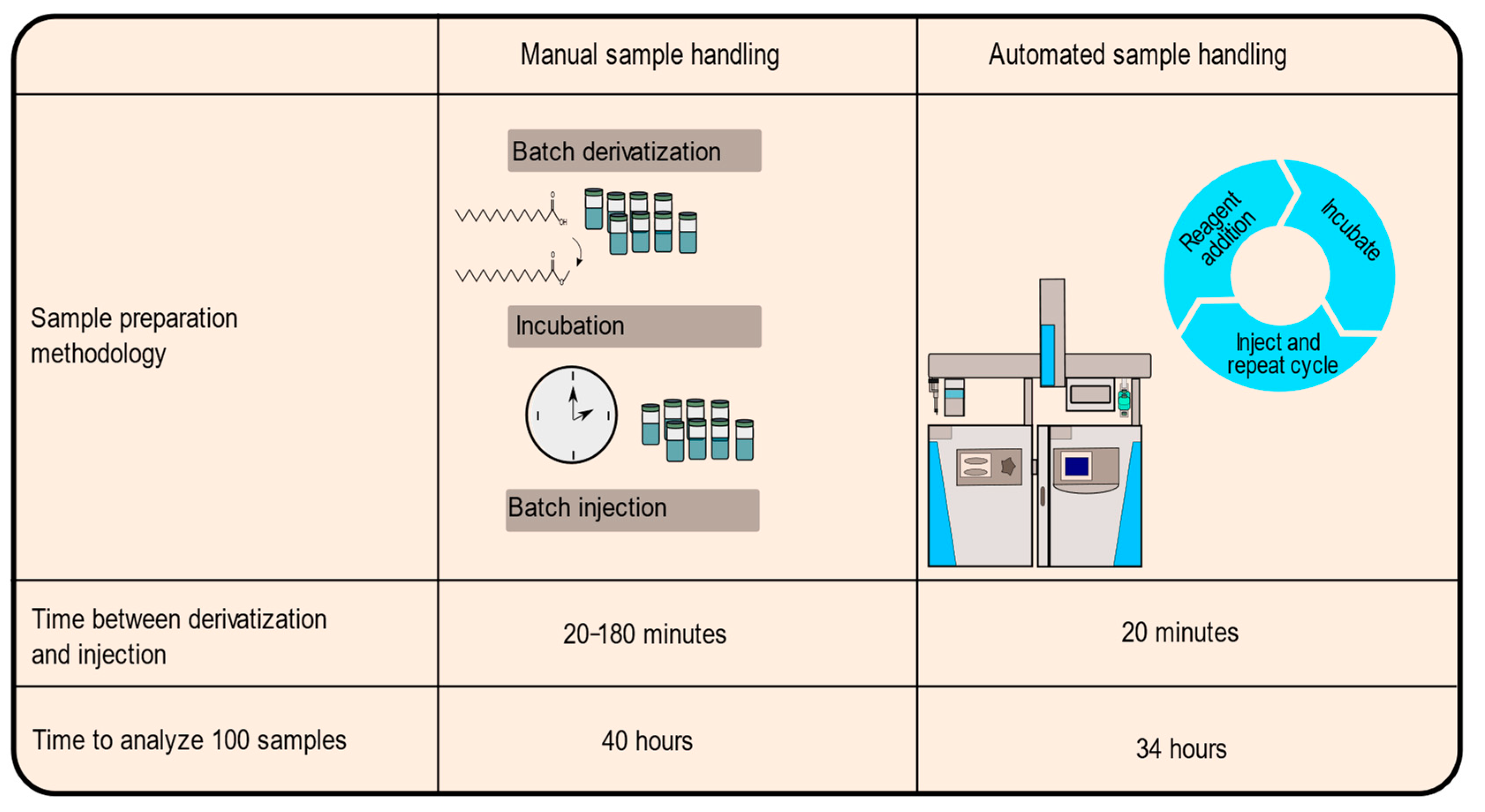
2.1. Adaptation of Method to Automation
2.2. Automated TMSH Derivatization Test on Fatty Acid Standard Mixture
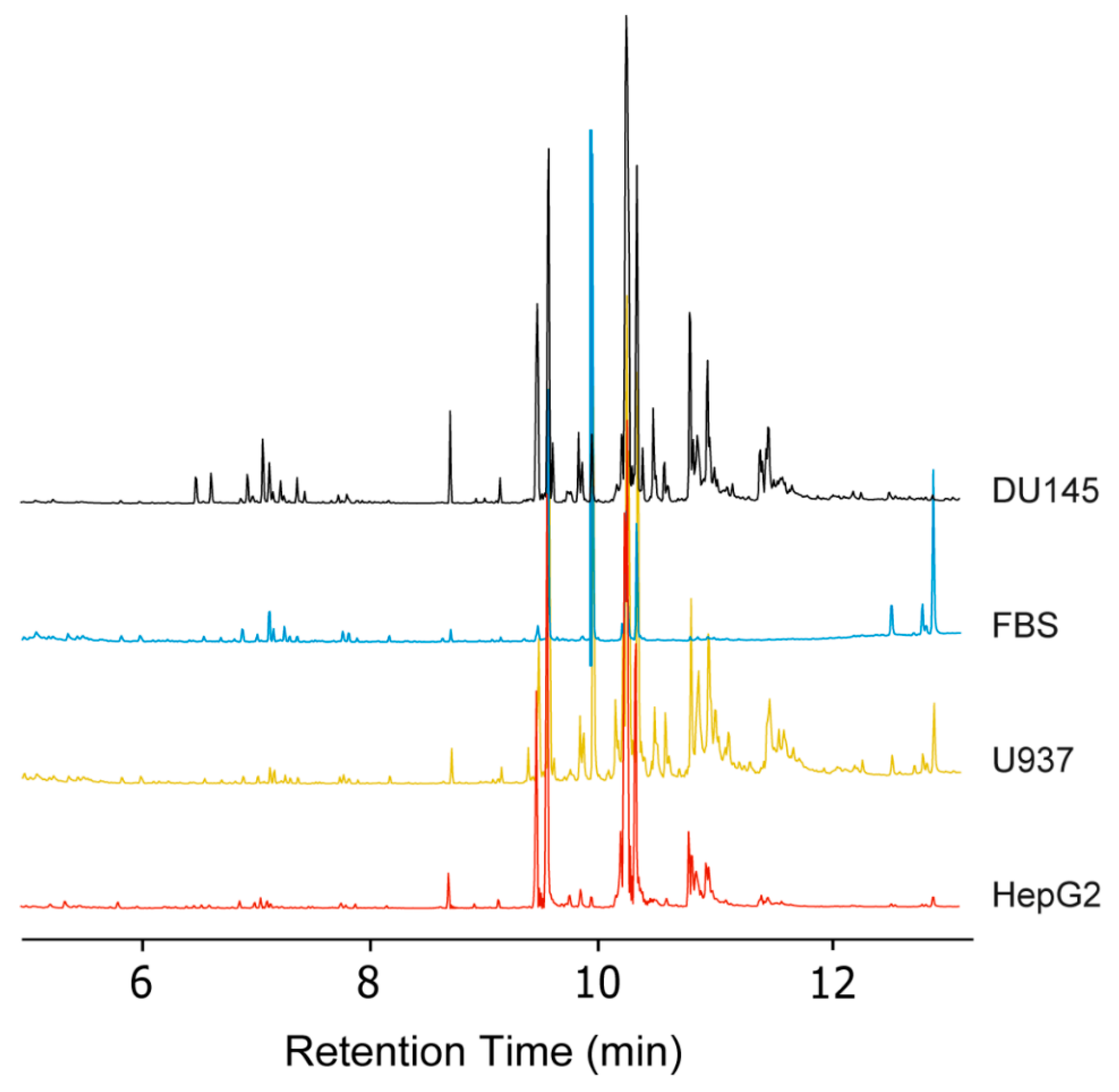
2.3. Validation of the Automated TMSH Derivatization with Biological Samples
3. Materials and Methods
3.1. Chemicals
3.2. Biological Samples
3.3. Lipid Extraction
3.4. Manual Derivatization
3.5. Automated Derivatization
3.6. GC–MS Setup and Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Albergamo, A.; Rigano, F.; Purcaro, G.; Mauceri, A.; Fasulo, S.; Mondello, L. Free fatty acid profiling of marine sentinels by nanoLC-EI-MS for the assessment of environmental pollution effects. Sci. Total Environ. 2016, 571, 955–962. [Google Scholar] [CrossRef] [PubMed]
- Song, E.M.; Byeon, J.-S.; Lee, S.M.; Yoo, H.J.; Kim, S.J.; Lee, S.-H.; Chang, K.; Hwang, S.W.; Yang, D.-H.; Jeong, J.-Y. Fecal fatty acid profiling as a potential new screening biomarker in patients with colorectal cancer. Dig. Dis. Sci. 2018, 63, 1229–1236. [Google Scholar] [CrossRef] [PubMed]
- Abdelmagid, S.A.; Clarke, S.E.; Nielsen, D.E.; Badawi, A.; El-Sohemy, A.; Mutch, D.M.; Ma, D.W. Comprehensive profiling of plasma fatty acid concentrations in young healthy Canadian adults. PLoS ONE 2015, 10, e0116195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.C.; Sun, C.H.; Liu, L.Y.; Sun, X.H.; Jin, X.W.; Song, W.L.; Liu, X.Q.; Wan, X.L. Serum fatty acid profiles using GC–MS and multivariate statistical analysis: Potential biomarkers of Alzheimer’s disease. Neurobiol. Aging 2012, 33, 1057–1066. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Nunez, B.; Dijck-Brouwer, D.A.; Muskiet, F.A. The relation of saturated fatty acids with low-grade inflammation and cardiovascular disease. J. Nutr. Biochem. 2016, 36, 1–20. [Google Scholar] [CrossRef]
- Morris, G.; Berk, M.; Carvalho, A.; Caso, J.R.; Sanz, Y.; Walder, K.; Maes, M. The Role of the Microbial Metabolites Including Tryptophan Catabolites and Short Chain Fatty Acids in the Pathophysiology of Immune-Inflammatory and Neuroimmune Disease. Mol. Neurobiol. 2017, 54, 4432–4451. [Google Scholar] [CrossRef]
- Manson, J.E.; Cook, N.R.; Lee, I.M.; Christen, W.; Bassuk, S.S.; Mora, S.; Gibson, H.; Albert, C.M.; Gordon, D.; Copeland, T.; et al. Marine n-3 Fatty Acids and Prevention of Cardiovascular Disease and Cancer. N Engl J. Med. 2019, 380, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Tiziani, S.; Kang, Y.; Harjanto, R.; Axelrod, J.; Piermarocchi, C.; Roberts, W.; Paternostro, G. Metabolomics of the tumor microenvironment in pediatric acute lymphoblastic leukemia. PLoS ONE 2013, 8, e82859. [Google Scholar] [CrossRef] [Green Version]
- Zini, C.A.; Lord, H.; Christensen, E.; de, A.T.; Caramao, E.B.; Pawliszyn, J. Automation of solid-phase microextraction-gas chromatography-mass spectrometry extraction of eucalyptus volatiles. J. Chromatogr. Sci. 2002, 40, 140–146. [Google Scholar] [CrossRef] [Green Version]
- Parkinson, D.R.; Bruheim, I.; Christ, I.; Pawliszyn, J. Full automation of derivatization--solid-phase microextraction-gas chromatography-mass spectrometry with a dual-arm system for the determination of organometallic compounds in aqueous samples. J. Chromatogr. A 2004, 1025, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Lan, C.; Liu, H.; Dong, J.; Luan, T. Full automation of solid-phase microextraction/on-fiber derivatization for simultaneous determination of endocrine-disrupting chemicals and steroid hormones by gas chromatography-mass spectrometry. Anal. Bioanal. Chem. 2006, 386, 391–397. [Google Scholar] [CrossRef]
- Hutchinson, J.P.; Setkova, L.; Pawliszyn, J. Automation of solid-phase microextraction on a 96-well plate format. J. Chromatogr. A 2007, 1149, 127–137. [Google Scholar] [CrossRef]
- Zarate, E.; Boyle, V.; Rupprecht, U.; Green, S.; Villas-Boas, S.G.; Baker, P.; Pinu, F.R. Fully Automated Trimethylsilyl (TMS) Derivatisation Protocol for Metabolite Profiling by GC–MS. Metabolites 2016, 7, 1. [Google Scholar] [CrossRef] [Green Version]
- Muller, Z.; Stelzl, E.; Bozic, M.; Haas, J.; Marth, E.; Kessler, H.H. Evaluation of automated sample preparation and quantitative PCR LCx assay for determination of human immunodeficiency virus type 1 RNA. J. Clin. Microbiol. 2004, 42, 1439–1443. [Google Scholar] [CrossRef] [Green Version]
- Gu, Q.; David, F.; Lynen, F.; Rumpel, K.; Dugardeyn, J.; Van Der Straeten, D.; Xu, G.; Sandra, P. Evaluation of automated sample preparation, retention time locked gas chromatography-mass spectrometry and data analysis methods for the metabolomic study of Arabidopsis species. J. Chromatogr. A 2011, 1218, 3247–3254. [Google Scholar] [CrossRef]
- Lerch, O.; Temme, O.; Daldrup, T. Comprehensive automation of the solid phase extraction gas chromatographic mass spectrometric analysis (SPE-GC/MS) of opioids, cocaine, and metabolites from serum and other matrices. Anal. Bioanal. Chem. 2014, 406, 4443–4451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiehn, O. Extending the breadth of metabolite profiling by gas chromatography coupled to mass spectrometry. Trends Anal. Chem 2008, 27, 261–269. [Google Scholar] [CrossRef] [Green Version]
- Tsugawa, H.; Bamba, T.; Shinohara, M.; Nishiumi, S.; Yoshida, M.; Fukusaki, E. Practical non-targeted gas chromatography/mass spectrometry-based metabolomics platform for metabolic phenotype analysis. J. Biosci. Bioeng. 2011, 112, 292–298. [Google Scholar] [CrossRef]
- Li, M.; Zhou, Z.; Nie, H.; Bai, Y.; Liu, H. Recent advances of chromatography and mass spectrometry in lipidomics. Anal. Bioanal. Chem. 2011, 399, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Dunn, W.B.; Broadhurst, D.; Begley, P.; Zelena, E.; Francis-McIntyre, S.; Anderson, N.; Brown, M.; Knowles, J.D.; Halsall, A.; Haselden, J.N.; et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2011, 6, 1060–1083. [Google Scholar] [CrossRef] [PubMed]
- Mateo-Castro, R.; Gimeno-Adelantado, J.V.; Bosch-Reig, F.; Doménech-Carbó, A.; Casas-Catalán, M.J.; Osete-Cortina, L.; De la Cruz-Cañizares, J.; Doménech-Carbó, M.T. Identification by GC-FID and GC–MS of amino acids, fatty and bile acids in binding media used in works of art. Fresenius J. Anal. Chem. 2001, 369, 642–646. [Google Scholar] [CrossRef] [PubMed]
- Abbiss, H.; Rawlinson, C.; Maker, G.L.; Trengove, R. Assessment of automated trimethylsilyl derivatization protocols for GC–MS-based untargeted metabolomic analysis of urine. Metabolomics 2015, 11, 1908–1921. [Google Scholar] [CrossRef]
- Giovane, A.; Balestrieri, A.; Napoli, C. New insights into cardiovascular and lipid metabolomics. J. Cell. Biochem. 2008, 105, 648–654. [Google Scholar] [CrossRef]
- Avula, S.G.C.; Belovich, J.M.; Xu, Y. Determination of fatty acid methyl esters derived from algae Scenedesmus dimorphus biomass by GC–MS with one-step esterification of free fatty acids and transesterification of glycerolipids. J. Sep. Sci. 2017, 40, 2214–2227. [Google Scholar] [CrossRef] [Green Version]
- Cui, Y.; Chen, X.; Liu, L.; Xie, W.; Wu, Y.; Wu, Q.; Wang, D. Gas chromatography-mass spectrometry analysis of the free fatty acids in serum obtained from patients with Alzheimer’s disease. Biomed. Mater. Eng 2015, 26, S2165–S2177. [Google Scholar] [CrossRef] [Green Version]
- Han, S.; Karlowicz-Bodalska, K.; Potaczek, P.; Wojcik, A.; Ozimek, L.; Szura, D.; Musial, W. Identification of unknown impurity of azelaic acid in liposomal formulation assessed by HPLC-ELSD, GC-FID, and GC–MS. AAPS PharmSciTech 2014, 15, 111–120. [Google Scholar] [CrossRef] [Green Version]
- Han, X.; Guo, J.; You, Y.; Yin, M.; Ren, C.; Zhan, J.; Huang, W. A fast and accurate way to determine short chain fatty acids in mouse feces based on GC–MS. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2018, 1099, 73–82. [Google Scholar] [CrossRef]
- Liu, Z.; Weng, R.; Feng, Y.; Li, Z.; Wang, L.; Su, X.; Yu, C. Fatty acid profiling of blood cell membranes by gas chromatography with mass spectrometry. J. Sep. Sci. 2016, 39, 3964–3972. [Google Scholar] [CrossRef] [PubMed]
- Quehenberger, O.; Armando, A.M.; Dennis, E.A. High sensitivity quantitative lipidomics analysis of fatty acids in biological samples by gas chromatography-mass spectrometry. Biochim. Biophys. Acta 2011, 1811, 648–656. [Google Scholar] [CrossRef] [Green Version]
- Argus, J.P.; Yu, A.K.; Wang, E.S.; Williams, K.J.; Bensinger, S.J. An optimized method for measuring fatty acids and cholesterol in stable isotope-labeled cells. J. Lipid Res. 2017, 58, 460–468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halket, J.M.; Waterman, D.; Przyborowska, A.M.; Patel, R.K.; Fraser, P.D.; Bramley, P.M. Chemical derivatization and mass spectral libraries in metabolic profiling by GC/MS and LC/MS/MS. J. Exp. Bot. 2005, 56, 219–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furuhashi, T.; Weckwerth, W. Introduction to Lipid (FAME) Analysis in Algae Using Gas Chromatography–Mass Spectrometry. Handb. Plant. Metab. 2013, 215–225. [Google Scholar]
- Fang, M.; Ivanisevic, J.; Benton, H.P.; Johnson, C.H.; Patti, G.J.; Hoang, L.T.; Uritboonthai, W.; Kurczy, M.E.; Siuzdak, G. Thermal Degradation of Small Molecules: A Global Metabolomic Investigation. Anal. Chem. 2015, 87, 10935–10941. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.X.; Wang, J. A simplified method for analysis of polyunsaturated fatty acids. BMC Biochem. 2005, 6, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Wang, H.; Zhu, M.J. A sensitive GC/MS detection method for analyzing microbial metabolites short chain fatty acids in fecal and serum samples. Talanta 2019, 196, 249–254. [Google Scholar] [CrossRef]
- Lepage, G.; Roy, C.C. Direct transesterification of all classes of lipids in a one-step reaction. J. Lipid Res. 1986, 27, 114–120. [Google Scholar] [CrossRef]
- Shin, H.-Y.; Lim, S.-M.; Bae, S.-Y.; Oh, S.C. Thermal decomposition and stability of fatty acid methyl esters in supercritical methanol. J. Anal. Appl. Pyrolysis 2011, 92, 332–338. [Google Scholar] [CrossRef]
- Olivares-Carrillo, P.; Quesada-Medina, J. Thermal decomposition of fatty acid chains during the supercritical methanol transesterification of soybean oil to biodiesel. J. Supercrit. Fluids 2012, 72, 52–58. [Google Scholar] [CrossRef]
- Firl, N.; Kienberger, H.; Hauser, T.; Rychlik, M. Determination of the fatty acid profile of neutral lipids, free fatty acids and phospholipids in human plasma. Clin. Chem. Lab. Med. 2013, 51, 799–810. [Google Scholar] [CrossRef]
- Sutherland, K. Derivatisation using m-(trifluoromethyl)phenyltrimethylammonium hydroxide of organic materials in artworks for analysis by gas chromatography-mass spectrometry: Unusual reaction products with alcohols. J. Chromatogr. A 2007, 1149, 30–37. [Google Scholar] [CrossRef]
- Khakimov, B.; Motawia, M.S.; Bak, S.; Engelsen, S.B. The use of trimethylsilyl cyanide derivatization for robust and broad-spectrum high-throughput gas chromatography-mass spectrometry based metabolomics. Anal. Bioanal. Chem. 2013, 405, 9193–9205. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, K.; Tanabe, T.; Kinoshita, M. Trimethylsulfonium hydroxide: A new methylating agent. J. Org. Chem. 1979, 44, 638–639. [Google Scholar] [CrossRef]
- Müller, K.-D.; Husmann, H.; Nalik, H.; Schomburg, G. Trans-esterification of fatty acids from microorganisms and human blood serum by trimethylsulfonium hydroxide (TMSH) for GC analysis. Chromatographia 1990, 30, 245–248. [Google Scholar] [CrossRef]
- Müller, K.D.; Nalik, H.P.; Schmid, E.N.; Husmann, H.; Schomburg, G. Fast identification of mycobacterium species by GC analysis with trimethylsulfonium hydroxide (TMSH) for transesterification. J. High. Resolut. Chromatogr. 1993, 16, 161–165. [Google Scholar] [CrossRef]
- Dron, J.; Linke, R.; Rosenberg, E.; Schreiner, M. Trimethylsulfonium hydroxide as derivatization reagent for the chemical investigation of drying oils in works of art by gas chromatography. J. Chromatogr. A 2004, 1047, 111–116. [Google Scholar] [CrossRef]
- Reinnicke, S.; Bernstein, A.; Elsner, M. Small and reproducible isotope effects during methylation with trimethylsulfonium hydroxide (TMSH): A convenient derivatization method for isotope analysis of negatively charged molecules. Anal. Chem. 2010, 82, 2013–2019. [Google Scholar] [CrossRef]
- Butte, W.; Eilers, J.; Kirsch, M. Trialkylsulfonium- and Trialkylselenoniumhydroxides for the Pyrolytic Alkylation of Acidic Compounds. Anal. Lett. 1982, 15, 841–850. [Google Scholar] [CrossRef]
- Ishida, Y.; Wakamatsu, S.; Yokoi, H.; Ohtani, H.; Tsuge, S. Compositional analysis of polyunsaturated fatty acid oil by one-step thermally assisted hydrolysis and methylation in the presence of trimethylsulfonium hydroxide. J. Anal. Appl. Pyrolysis 1999, 49, 267–276. [Google Scholar] [CrossRef]
- Vosmann, K.; Klein, E.; Weber, N. Reaction of fatty acid amides and ethanolamides with trimethylsulfonium hydroxide. J. Chromatogr. A 1997, 773, 239–247. [Google Scholar] [CrossRef]
- Koek, M.M.; Jellema, R.H.; van der Greef, J.; Tas, A.C.; Hankemeier, T. Quantitative metabolomics based on gas chromatography mass spectrometry: Status and perspectives. Metabolomics 2011, 7, 307–328. [Google Scholar] [CrossRef] [Green Version]
- Chiu, H.H.; Tsai, S.J.; Tseng, Y.J.; Wu, M.S.; Liao, W.C.; Huang, C.S.; Kuo, C.H. An efficient and robust fatty acid profiling method for plasma metabolomic studies by gas chromatography-mass spectrometry. Clin. Chim. Acta 2015, 451, 183–190. [Google Scholar] [CrossRef]
- Tan, B.; Zhang, Y.; Zhang, T.; He, J.; Luo, X.; Bian, X.; Wu, J.; Zou, C.; Wang, Y.; Fu, L. Identifying potential serum biomarkers of breast cancer through targeted free fatty acid profiles screening based on a GC–MS platform. Biomed. Chromatogr. 2020, 34, e4922. [Google Scholar] [CrossRef] [PubMed]
- Folch, J.; Lees, M.; Stanley, G.S. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef]
- Li, Y.; Watkins, B.A. Analysis of Fatty Acids in Food Lipids. Curr. Protoc. Food Anal. Chem. 2001, 00, D1.2.1–D1.2.15. [Google Scholar] [CrossRef]
- Barthet, V.; Chornick, T.; Daun, J. Comparison of Methods to Measure the Oil Contents in Oilseeds. J. Oleo Sci. 2002, 51, 589–597. [Google Scholar] [CrossRef]
- Hosli, R.; Tobler, A.; Konig, S.; Muhlebach, S. A quantitative phenytoin GC–MS method and its validation for samples from human ex situ brain microdialysis, blood and saliva using solid-phase extraction. J. Anal. Toxicol 2013, 37, 102–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lerch, O.; Zinn, P.; Gotze, H.J. Automation of the GC/MS analysis of mineral oil contaminations in water. Fresenius J. Anal. Chem 2000, 367, 195–200. [Google Scholar] [CrossRef]
- Shimbo, K.; Kubo, S.; Harada, Y.; Oonuki, T.; Yokokura, T.; Yoshida, H.; Amao, M.; Nakamura, M.; Kageyama, N.; Yamazaki, J.; et al. Automated precolumn derivatization system for analyzing physiological amino acids by liquid chromatography/mass spectrometry. Biomed. Chromatogr 2010, 24, 683–691. [Google Scholar] [CrossRef]

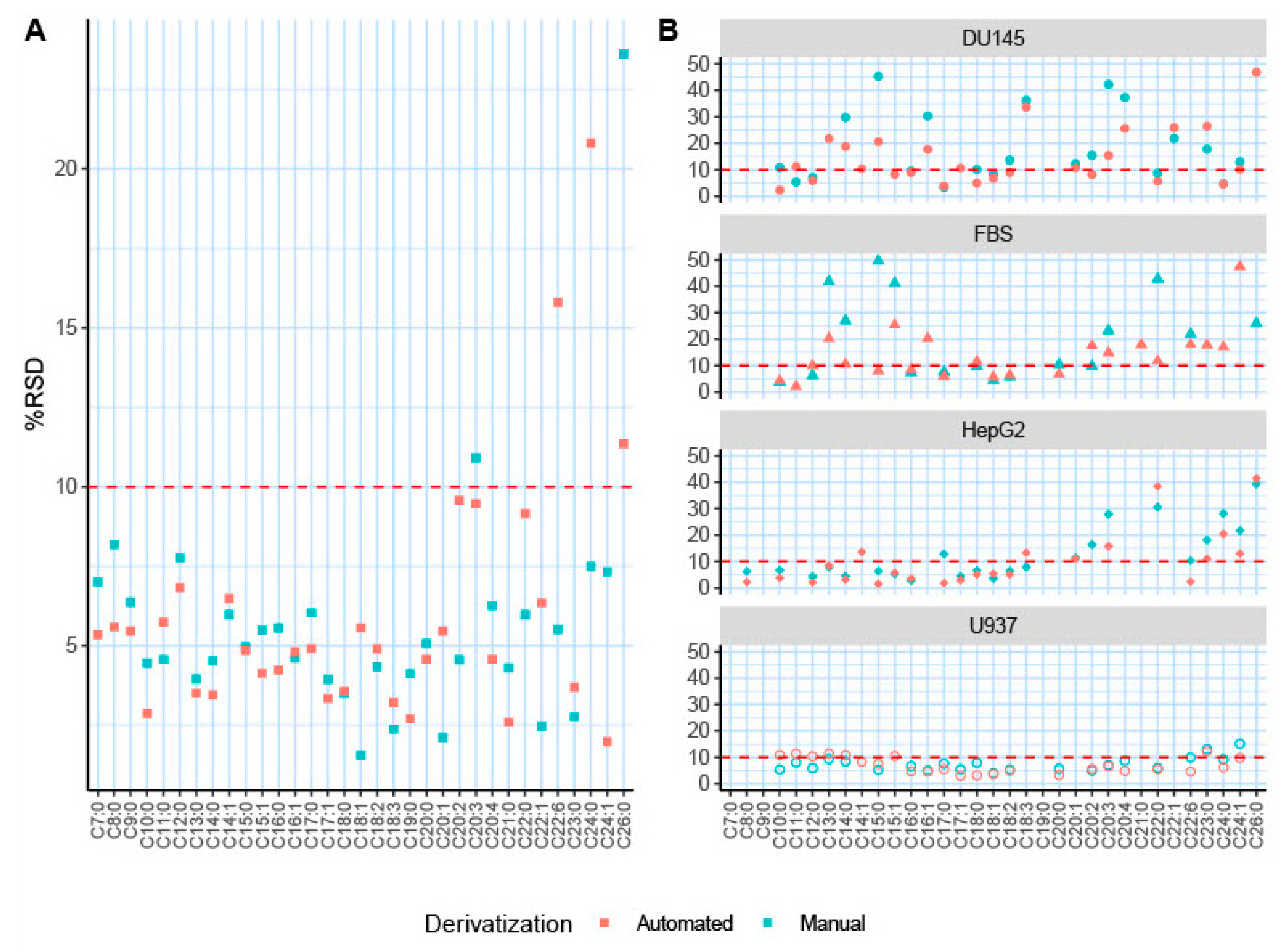
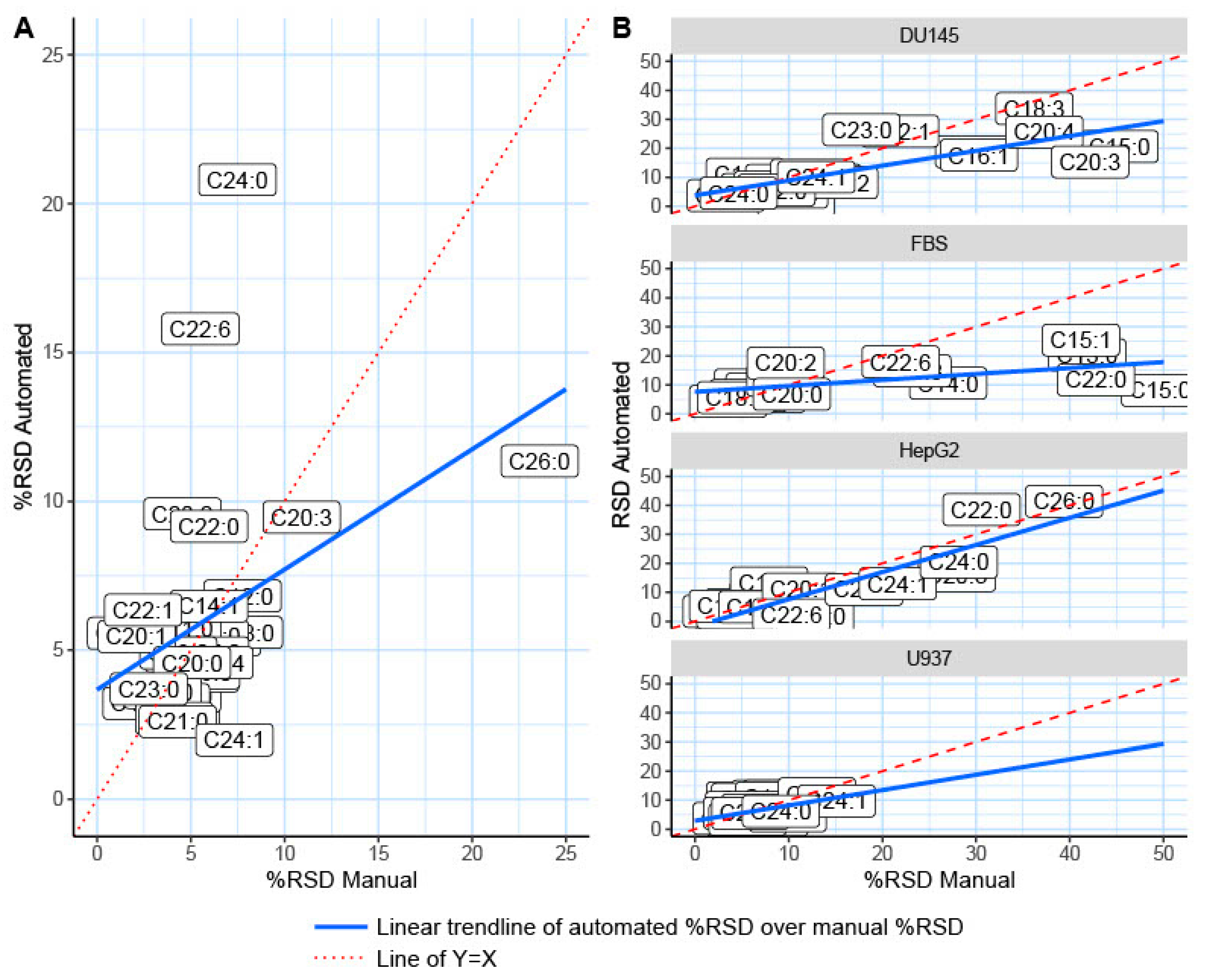
| Fatty Acid | HMDB ID | Carbons: Double Bonds | FAME Molecular Mass | Quantifier Ion (m/z) | Standards n = 6 | |
|---|---|---|---|---|---|---|
| Manual | Automatic | |||||
| Heptanoic acid | HMDB0000666 | C7:0 | 144.21 | 73.98 | 7.00 | 5.35 |
| Octanoic acid | HMDB0000482 | C8:0 | 158.24 | 73.98 | 8.18 | 5.59 |
| Nonanoic acid | HMDB0000847 | C9:0 | 172.26 | 73.98 | 6.36 | 5.46 |
| Decanoic acid | HMDB0000511 | C10:0 | 186.27 | 73.98 | 4.45 | 2.88 |
| Undecanoic acid | HMDB0000947 | C11:0 | 200.29 | 73.95 | 4.57 | 5.73 |
| Dodecanoic acid | HMDB0000638 | C12:0 | 214.32 | 73.93 | 7.76 | 6.82 |
| Tridecanoic acid | HMDB0000910 | C13:0 | 228.35 | 73.94 | 3.96 | 3.51 |
| 9(Z)-Tetradecenoic acid | HMDB0002000 | C14:1 | 240.36 | 55.05 | 5.97 | 6.48 |
| Tetradecanoic acid | HMDB0000806 | C14:0 | 242.38 | 73.99 | 4.53 | 3.45 |
| 9(Z)-Pentedecenoic acid | HMDB0029765 | C15:1 | 254.41 | 55.05 | 5.49 | 4.13 |
| Pentadecanoic acid | HMDB0000826 | C15:0 | 256.42 | 73.96 | 4.97 | 4.85 |
| 9(Z)-Hexadecenoic acid | HMDB0003229 | C16:1 | 268.41 | 55.05 | 4.61 | 4.81 |
| Hexadecanoic acid | HMDB0000220 | C16:0 | 270.45 | 73.95 | 5.56 | 4.24 |
| 10(Z)-Heptadecenoic acid | HMDB0060038 | C17:1 | 282.46 | 55.05 | 3.94 | 3.33 |
| Heptadecanoic acid | HMDB0002259 | C17:0 | 284.46 | 73.96 | 6.03 | 4.91 |
| (6Z, 9Z, 12Z)-Octadecatrienoic acid | HMDB0003073 | C18:3 | 292.44 | 292.18 | 2.37 | 3.21 |
| (9Z, 12Z)-octadecadienoic acid | HMDB0000673 | C18:2 | 294.45 | 294.18 | 4.33 | 4.90 |
| 9(Z)-octadecenoic acid | HMDB0000207 | C18:1 | 296.68 | 296.20 | 1.55 | 5.57 |
| Octadecanoic acid | HMDB0000827 | C18:0 | 298.48 | 73.98 | 3.50 | 3.57 |
| Nonadecanoic acid | HMDB0000772 | C19:0 | 312.53 | 73.96 | 4.11 | 2.71 |
| (5Z, 8Z, 11Z, 14Z)-Icosatetraenoic acid | HMDB0001043 | C20:4 | 318.49 | 318.20 | 6.26 | 4.57 |
| (5Z, 8Z,11Z )-Eicosatrienoic acid | HMDB0010378 | C20:3 | 320.51 | 320.20 | 10.91 | 9.47 |
| (11Z, 14Z)-Eicosadienoic acid | HMDB0005060 | C20:2 | 322.51 | 322.25 | 4.56 | 9.58 |
| 13(Z)-Eicosenoic acid | HMDB0035159 | C20:1 | 324.50 | 324.19 | 2.10 | 5.45 |
| Eicosanoic acid | HMDB0002212 | C20:0 | 326.56 | 73.99 | 5.07 | 4.57 |
| Heinecosanoic acid | HMDB0002345 | C21:0 | 340.58 | 73.97 | 4.30 | 2.60 |
| (4Z, 7Z, 10Z, 13Z, 16Z, 19Z)-Docosahexaenoic acid | HMDB0002183 | C22:6 | 342.52 | 79.06 | 5.51 | 13.65 |
| (13Z)-Docosenoic acid | HMDB0002068 | C22:1 | 352.57 | 55.05 | 2.46 | 6.35 |
| Docosanoic acid | HMDB0000944 | C22:0 | 354.59 | 74.00 | 5.97 | 9.16 |
| Tricosanoic acid | HMDB0001160 | C23:0 | 368.62 | 74.00 | 2.76 | 3.70 |
| (15Z)-Tetracosenoic acid | HMDB0002368 | C24:1 | 379.62 | 55.06 | 7.33 | 1.99 |
| Tetracosanoic acid | HMDB0002003 | C24:0 | 381.36 | 73.99 | 7.49 | 12.58 |
| Hexacosanoic acid | HMDB0002356 | C26:0 | 410.69 | 73.96 | 23.61 | 4.27 |
| Fatty Acid | Carbons:Double Bonds | HepG2 n = 6 | DU145 n = 6 | U937 n = 6 | FBS n = 6 | ||||
|---|---|---|---|---|---|---|---|---|---|
| Manual | Automatic | Manual | Automatic | Manual | Automatic | Manual | Automatic | ||
| Heptanoic acid | C7:0 | NF | NF | NF | NF | NF | NF | NF | NF |
| Octanoic acid | C8:0 | 12.29 | 6.74 | 10.80 | 5.16 | 7.88 | 5.33 | 24.24 | 18.95 |
| Nonanoic acid | C9:0 | NF | NF | NF | NF | NF | NF | NF | NF |
| Decanoic acid | C10:0 | 4.77 | 4.79 | 10.36 | 2.23 | 3.86 | 4.76 | 7.32 | 4.87 |
| Undecanoic acid | C11:0 | 4.13 | 3.68 | NF | NF | NF | NF | 6.52 | 5.87 |
| Dodecanoic acid | C12:0 | 3.33 | 2.55 | 5.29 | 2.94 | 3.15 | 4.48 | 7.18 | 5.63 |
| Tridecanoic acid | C13:0 | 5.44 | 7.24 | NF | NF | 14.27 | 11.59 | 21.36 | 23.31 |
| 9(Z)-Tetradecenoic acid | C14:1 | NF | NF | NF | NF | NF | NF | NF | NF |
| Tetradecanoic acid | C14:0 | 5.23 | 5.43 | NF | NF | 3.02 | 4.06 | 8.70 | 5.09 |
| 9(Z)-Pentedecenoic acid | C15:1 | 3.81 | 8.69 | 51.44 | 5.88 | 6.97 | 3.00 | 5.50 | 12.39 |
| Pentadecanoic acid | C15:0 | 6.17 | 7.41 | 41.89 | 21.60 | 3.13 | 4.66 | 11.34 | 7.35 |
| 9(Z)-Hexadecenoic acid | C16:1 | 3.37 | 3.19 | 30.09 | 17.32 | 2.26 | 1.82 | 19.98 | 7.27 |
| Hexadecanoic acid | C16:0 | 3.25 | 5.00 | 7.93 | 11.17 | 2.19 | 3.28 | 6.83 | 5.04 |
| 10(Z)-Heptadecenoic acid | C17:1 | 4.79 | 14.33 | NF | NF | 4.51 | 3.04 | 9.08 | 9.14 |
| Heptadecanoic acid | C17:0 | 6.49 | 3.79 | 19.65 | 7.05 | 1.00 | 1.03 | 4.69 | 2.69 |
| (6Z, 9Z, 12Z)-Octadecatrienoic acid | C18:3 | NF | NF | NF | NF | 14.21 | 19.39 | NF | NF |
| (9Z, 12Z)-octadecadienoic acid | C18:2 | NF | NF | NF | NF | 5.42 | 2.74 | 20.74 | 5.43 |
| 9(Z)-octadecenoic acid | C18:1 | 1.26 | 6.18 | 8.15 | 5.88 | 1.25 | 1.28 | 15.88 | 4.60 |
| Octadecanoic acid | C18:0 | 9.03 | 2.78 | 11.31 | 7.59 | 1.79 | 2.04 | 5.28 | 4.33 |
| (5Z, 8Z, 11Z, 14Z)-Icosatetraenoic acid | C20:4 | 14.34 | 15.46 | 71.96 | 28.56 | 9.21 | 3.77 | NF | NF |
| (5Z, 8Z, 11Z)-Eicosatrienoic acid | C20:3 | 11.10 | 15.08 | 40.73 | 20.70 | 10.79 | 5.57 | NF | NF |
| (11Z, 14Z)-Eicosadienoic acid | C20:2 | 7.70 | 12.23 | 18.63 | 12.29 | NF | NF | NF | NF |
| 13(Z)-Eicosenoic acid | C20:1 | 6.19 | 12.72 | 11.22 | 4.70 | 7.81 | 4.96 | NF | NF |
| Eicosanoic acid | C20:0 | 27.20 | 5.50 | 10.44 | 4.78 | 5.07 | 3.46 | 44.30 | 29.24 |
| Heinecosanoic acid | C21:0 | NF | NF | NF | NF | NF | NF | NF | NF |
| (4Z, 7Z, 10Z, 13Z, 16Z, 19Z)-Docosahexaenoic acid | C22:6 | 22.28 | 14.46 | NF | NF | 6.48 | 5.94 | NF | NF |
| (13Z)-Docosenoic acid | C22:1 | NF | NF | 11.85 | 9.28 | NF | NF | NF | NF |
| Docosanoic acid | C22:0 | 8.70 | 20.28 | 11.27 | 3.11 | 5.10 | 6.38 | 28.19 | 17.57 |
| Tricosanoic acid | C23:0 | NF | NF | 47.77 | 14.83 | 8.81 | 6.36 | NF | NF |
| (15Z)-Tetracosenoic acid | C24:1 | 8.30 | 14.30 | 12.74 | 6.92 | 8.66 | 3.37 | 42.72 | 6.93 |
| Tetracosanoic acid | C24:0 | 6.59 | 33.25 | 4.77 | 4.48 | 8.20 | 2.44 | 44.78 | 7.33 |
| Hexacosanoic acid | C26:0 | 10.07 | 7.00 | 8.50 | 7.60 | 6.25 | 3.08 | 11.95 | 7.41 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gries, P.; Rathore, A.S.; Lu, X.; Chiou, J.; Huynh, Y.B.; Lodi, A.; Tiziani, S. Automated Trimethyl Sulfonium Hydroxide Derivatization Method for High-Throughput Fatty Acid Profiling by Gas Chromatography–Mass Spectrometry. Molecules 2021, 26, 6246. https://doi.org/10.3390/molecules26206246
Gries P, Rathore AS, Lu X, Chiou J, Huynh YB, Lodi A, Tiziani S. Automated Trimethyl Sulfonium Hydroxide Derivatization Method for High-Throughput Fatty Acid Profiling by Gas Chromatography–Mass Spectrometry. Molecules. 2021; 26(20):6246. https://doi.org/10.3390/molecules26206246
Chicago/Turabian StyleGries, Paul, Atul Singh Rathore, Xiyuan Lu, Jennifer Chiou, Yen Bao Huynh, Alessia Lodi, and Stefano Tiziani. 2021. "Automated Trimethyl Sulfonium Hydroxide Derivatization Method for High-Throughput Fatty Acid Profiling by Gas Chromatography–Mass Spectrometry" Molecules 26, no. 20: 6246. https://doi.org/10.3390/molecules26206246
APA StyleGries, P., Rathore, A. S., Lu, X., Chiou, J., Huynh, Y. B., Lodi, A., & Tiziani, S. (2021). Automated Trimethyl Sulfonium Hydroxide Derivatization Method for High-Throughput Fatty Acid Profiling by Gas Chromatography–Mass Spectrometry. Molecules, 26(20), 6246. https://doi.org/10.3390/molecules26206246







