Malate Transport and Metabolism in Nitrogen-Fixing Legume Nodules
Abstract
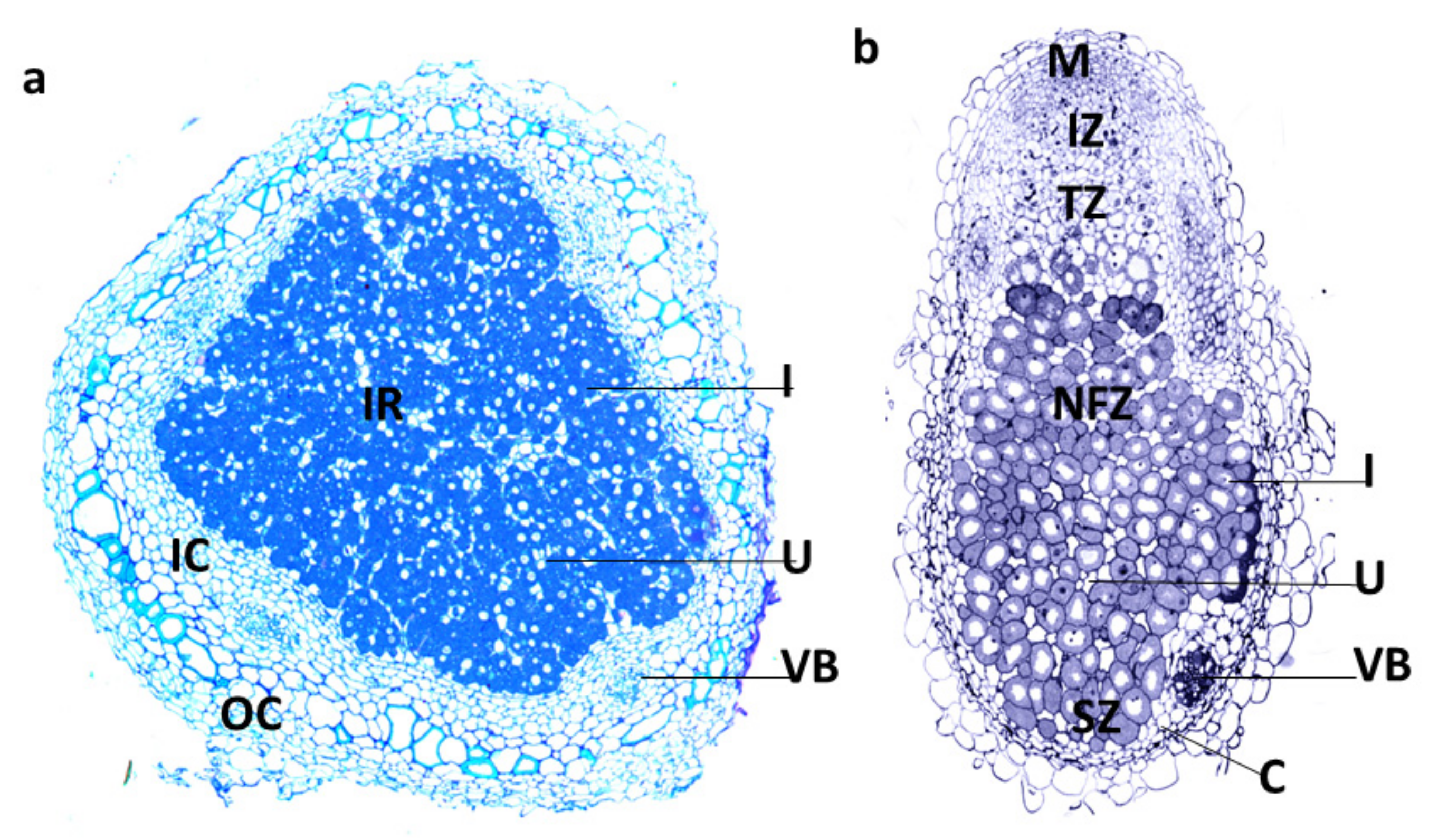
:1. Introduction
2. Nodule Metabolism and Provision of Carbon to the Bacteroids
3. The Role of Mitochondria in Nitrogen-Fixing Nodules
- Oxygen levels in infected cells are maintained at very low levels (sub-micromolar) to allow operation of the nitrogenase enzyme [3,11]. Mitochondria from these cells have the capacity to respire under very low oxygen levels to supply the quantities of ATP estimated to be required to support symbiosome function and ammonia assimilation in infected cells [31,42,43]. The apparent oxygen affinity of cytochrome oxidase in nodule mitochondria is significantly greater than in those from roots [42].
- They have very low alternative electron transport pathway activity, with virtually no alternative oxidase protein [47] and very low rates of cyanide-insensitive oxygen uptake. They also have very low rates of rotenone-insensitive malate uptake that is indicative of low internal alternative NADP(H) dehydrogenase (NDA) levels, although they oxidise external NADH rapidly [44]. This indicates that mitochondrial ATP production in infected cells is more efficient than in roots and other tissues.
- They have very low malic enzyme activity, but very high malate dehydrogenase activity [32,34] and export oxaloacetate rapidly [33]. Together with low activity of other TCAC enzymes [33], this suggests that mitochondria of infected cells operate a truncated TCAC, oxidising principally malate to produce ATP and OAA for ammonia assimilation (Figure 2).
4. Malate Metabolism in Bacteroids
5. Malate Transporters in Nodules
6. The Role of GABA in Nodule Metabolism and Transport
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Oldroyd, G.E.; Murray, J.D.; Poole, P.S.; Downie, J.A. The rules of engagement in the legume-rhizobial symbiosis. Annu. Rev. Gen. 2011, 45, 119–144. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, L.F.; Day, D.A. The peribacteroid membrane. Physiol. Plant. 1997, 100, 30–44. [Google Scholar] [CrossRef]
- Udvardi, M.; Poole, P.S. Transport and Metabolism in Legume-Rhizobia Symbioses. Annu. Rev. Plant Biol. 2013, 64, 781–805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gavrin, A.; Kaiser, B.N.; Geiger, D.; Tyerman, S.D.; Wen, Z.; Bisseling, T.; Fedorova, E.E. Adjustment of Host Cells for Accommodation of Symbiotic Bacteria: Vacuole Defunctionalization, HOPS Suppression, and TIP1g Retargeting in Medicago. Plant Cell 2014, 26, 3809. [Google Scholar] [CrossRef] [Green Version]
- Mohd Noor, S.; Day, D.; Smith, P. The Symbiosome Membrane. In Biological Nitrogen Fixation; Bruijn, F.J., Ed.; John Wiley & Sons: Hoboken, NJ, USA, 2015; pp. 683–694. [Google Scholar]
- Udvardi, M.K.; Day, D.A. Metabolite transport across symbiotic membranes of legume nodules. Annu. Rev. Plant Physiol. 1997, 48, 493–523. [Google Scholar] [CrossRef] [PubMed]
- Clarke, V.C.; Loughlin, P.C.; Gavrin, A.; Chen, C.; Brear, E.M.; Day, D.A.; Smith, P.M. Proteomic analysis of the soybean symbiosome identifies new symbiotic proteins. Mol. Cell. Proteom. 2015, 14, 1301–1322. [Google Scholar] [CrossRef] [Green Version]
- Rolfe, B.G.; Gresshoff, P.M. Genetic Analysis of Legume Nodule Initiation. Annu. Rev. Plant Physiol. 1988, 39, 297–319. [Google Scholar] [CrossRef]
- Gavrin, A.; Loughlin, P.C.; Brear, E.; Griffith, O.W.; Bedon, F.; Suter Grotemeyer, M.; Escudero, V.; Reguera, M.; Qu, Y.; Mohd-Noor, S.N.; et al. Soybean Yellow Stripe-like 7 is a symbiosome membrane peptide transporter important for nitrogen fixation. Plant Physiol. 2021, 186, 581–598. [Google Scholar] [CrossRef]
- Schubert, K. Enzymes of Purine Biosynthesis and Catabolism in Glycine max: I. Comparison of Activities with N₂ Fixation and Composition of Xylem Exudate during Nodule Development. Plant Physiol. 1981, 68, 1115–1122. [Google Scholar] [CrossRef] [Green Version]
- Vance, C.P. Carbon and nitrogen metabolism in legume nodules. In Nitrogen-Fixing Leguminous Symbioses, Nitrogen Fixation: Origins, Applications, and Research Progress; Dilworth, M.J., James, E.K., Sprent, J.I., Newton, W.E., Eds.; Springer: Dordrecht, The Netherlands, 2008; pp. 293–320. [Google Scholar]
- Gordon, A.J.; Ryle, G.J.A.; Mitchell, D.F.; Powell, C.E. The Flux of 14C-Labelled Photosynthate through Soyabean Root Nodules during N2 Fixation. J. Exp. Bot. 1985, 36, 756–769. [Google Scholar] [CrossRef]
- Rosendahl, L.; Vance, C.P.; Pedersen, W.B. Products of Dark CO2 Fixation in Pea Root Nodules Support Bacteroid Metabolism. Plant Physiol. 1990, 93, 12–19. [Google Scholar] [CrossRef] [Green Version]
- Vance, C.P.; Miller, S.S.; Driscoll, B.T.; Robinson, D.L.; Trepp, G.; Gantt, J.S.; Samas, D.A. Nodule Carbon Metabolism: Organic Acids for N2 Fixation. In Biological Nitrogen Fixation for the 21st Century; Elmerich, C., Kondorosi, A., Newton, W.E., Eds.; Springer: Dordrecht, The Netherlands, 1998; Volume 31, pp. 443–448. [Google Scholar]
- Smith, P.M.C.; Winter, H.; Storer, P.J.; Bussell, J.D.; Schuller, K.A.; Atkins, C.A. Effect of short-term N2 deficiency on expression of the ureide pathway in cowpea root nodules. Plant Physiol. 2002, 129, 1216–1221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kouchi, H.; Fukai, K.; Katagiri, H.; Minamisawa, K.; Tajima, S. Isolation and enzymological characterization of infected and uninfected protoplasts from root nodules of Glycine max. Physiol. Plant. 1988, 73, 327–334. [Google Scholar] [CrossRef]
- Day, D.A.; Copeland, L. Carbon metabolism and compartmentation in nitrogen-fixing legume nodules. Plant Physiol. Biochem. 1991, 29, 185–201. [Google Scholar]
- Fedorova, M.; Tikhonovich, I.A.; Vance, C.P. Expression of C-assimilating enzymes in pea (Pisum sativum L.) root nodules. In situ localization in effective nodules. Plant Cell Environ. 1999, 22, 1249–1262. [Google Scholar] [CrossRef]
- Takanashi, K.; Takahashi, H.; Sakurai, N.; Sugiyama, A.; Suzuki, H.; Shibata, D.; Nakazono, M.; Yazaki, K. Tissue-Specific Transcriptome Analysis in Nodules of Lotus japonicus. Mol. Plant Microbe Interact. 2012, 25, 869–876. [Google Scholar] [CrossRef] [Green Version]
- Brown, S.M.; Oparka, K.J.; Sprent, J.I.; Walsh, K.B. Symplastic transport in soybean root nodules. Soil Biol. Biochem. 1995, 27, 387–399. [Google Scholar] [CrossRef]
- Abd-Alla, M.H.; Koyro, H.-W.; Yan, F.; Schubert, S.; Peiter, E. Functional structure of the indeterminate Vicia faba L. root nodule: Implications for metabolite transport. J. Plant Physiol. 2000, 157, 335–343. [Google Scholar] [CrossRef]
- Complainville, A.; Brocard, L.; Roberts, I.; Dax, E.; Sever, N.; Sauer, N.; Kondorosi, A.; Wolf, S.; Oparka, K.; Crespi, M. Nodule initiation involves the creation of a new symplasmic field in specific root cells of medicago species. Plant Cell 2003, 15, 2778–2791. [Google Scholar] [CrossRef] [Green Version]
- Gaudioso-Pedraza, R.; Beck, M.; Frances, L.; Kirk, P.; Ripodas, C.; Niebel, A.; Oldroyd, G.E.D.; Benitez-Alfonso, Y.; de Carvalho-Niebel, F. Callose-Regulated Symplastic Communication Coordinates Symbiotic Root Nodule Development. Curr. Biol. 2018, 28, 3562–3577.e6. [Google Scholar] [CrossRef] [Green Version]
- Kryvoruchko, I.S.; Sinharoy, S.; Torres-Jerez, I.; Sosso, D.; Pislariu, C.I.; Guan, D.; Murray, J.; Benedito, V.A.; Frommer, W.B.; Udvardi, M.K. MtSWEET11, a Nodule-Specific Sucrose Transporter of Medicago truncatula. Plant Physiol. 2016, 171, 554–565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugiyama, A.; Saida, Y.; Yoshimizu, M.; Takanashi, K.; Sosso, D.; Frommer, W.B.; Yazaki, K. Molecular Characterization of LjSWEET3, a Sugar Transporter in Nodules of Lotus japonicus. Plant Cell Physiol. 2017, 58, 298–306. [Google Scholar] [PubMed]
- Takanashi, K.; Sasaki, T.; Kan, T.; Saida, Y.; Sugiyama, A.; Yamamoto, Y.; Yazaki, K. A Dicarboxylate Transporter, LjALMT4, Mainly Expressed in Nodules of Lotus japonicus. Mol. Plant Microbe Interact. 2016, 29, 584–592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Day, D.A. Permeability of Isolated Infected Cells from Soybean Nodules. J. Exp. Bot. 1991, 42, 1325–1329. [Google Scholar] [CrossRef]
- Udvardi, M.K.; Price, G.D.; Gresshoff, P.M.; Day, D.A. A dicarboxylate transporter on the peribacteroid membrane of soybean nodules. FEBS Lett. 1988, 231, 36–40. [Google Scholar] [CrossRef] [Green Version]
- Ronson, C.W.; Astwood, P.M.; Downie, J.A. Molecular cloning and genetic organization of C4-dicarboxylate transport genes from Rhizobium leguminosarum. J. Bacteriol. 1984, 160, 903–909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pessi, G.; Ahrens, C.H.; Rehrauer, H.; Lindemann, A.; Hauser, F.; Fischer, H.M.; Hennecke, H. Genome-wide transcript analysis of Bradyrhizobium japonicum bacteroids in soybean root nodules. Mol. Plant Microbe Interact. 2007, 20, 1353–1363. [Google Scholar] [CrossRef] [Green Version]
- Rawsthorne, S.; Larue, T. Metabolism under Microaerobic Conditions of Mitochondria from Cowpea Nodules. Plant Physiol. 1986, 81, 1097–1102. [Google Scholar] [CrossRef] [Green Version]
- Day, D.A.; Mannix, M. Malate oxidation by soybean nodule mitochondria and the possible consequences for nitrogen fixation. Plant Physiol. Biochem. 1988, 26, 567–573. [Google Scholar]
- Bryce, J.H.; Day, D.A. Tricarboxylic Acid Cycle Activity in Mitochondria from Soybean Nodules and Cotyledons. J. Exp. Bot. 1990, 41, 961–967. [Google Scholar] [CrossRef]
- Rawsthorne, S.; Larue, T.A. Preparation and properties of mitochondria from cowpea nodules. Plant Physiol. 1986, 81, 1092–1096. [Google Scholar] [CrossRef] [Green Version]
- Van Ghelue, M.; Ribeiro, A.; Solheim, B.; Akkermans, A.D.; Bisseling, T.; Pawlowski, K. Sucrose synthase and enolase expression in actinorhizal nodules of Alnus glutinosa: Comparison with legume nodules. Mol. Gen. Genet. 1996, 250, 437–446. [Google Scholar] [PubMed]
- Ronson, C.W.; Lyttleton, P.; Robertson, J.G. C4-dicarboxylate transport mutants of Rhizobium trifolii form ineffective nodules on Trifolium repens. Proc. Natl. Acad. Sci. USA 1981, 78, 4284–4288. [Google Scholar] [CrossRef] [Green Version]
- Bolton, E.; Higgisson, B.; Harrington, A.; O’Gara, F. Dicarboxylic acid transport in Rhizobium meliloti: Isolation of mutants and cloning of dicarboxylic acid transport genes. Arch. Microbiol. 1986, 144, 142–146. [Google Scholar] [CrossRef]
- Yurgel, S.N.; Kahn, M.L. Dicarboxylate transport by rhizobia. FEMS Microbiol. Rev. 2004, 28, 489–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Streeter, J.G. Carbohydrate, organic Acid, and amino Acid composition of bacteroids and cytosol from soybean nodules. Plant Physiol. 1987, 85, 768–773. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Copeland, L. Role of malonate in chickpeas. Phytochemistry 2000, 54, 585–589. [Google Scholar] [CrossRef]
- Mitsch, M.J.; diCenzo, G.C.; Cowie, A.; Finan, T.M. Succinate Transport Is Not Essential for Symbiotic Nitrogen Fixation by Sinorhizobium meliloti or Rhizobium leguminosarum. Appl. Environ. Microbiol. 2018, 84, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Millar, A.H.; Day, D.A.; Bergersen, F.J. Microaerobic respiration and oxidative phosphorylation by soybean nodule mitochondria: Implications for nitrogen fixation. Plant Cell Environ. 1995, 18, 715–726. [Google Scholar] [CrossRef]
- Puppo, A.; Dimitrijevic, L.; Rigaud, J. O2 consumption and superoxide dismutase content in purified mitochondria from soybean root nodules. Plant Sci. 1987, 50, 3–11. [Google Scholar] [CrossRef]
- Day, D.A.; Price, G.D.; Gresshoff, P.M. Isolation and oxidative properties of mitochondria and bacteroids from soybean root nodules. Protoplasma 1986, 134, 121–129. [Google Scholar] [CrossRef]
- Bergersen, F.J. Root Nodules of Legumes: Structure and Functions; John Wiley and Sons: Chichester, UK, 1982. [Google Scholar]
- Whitehead, L.F.; Day, D.A.; Hardham, A.R. Cytoskeletal arrays in the cells of soybean root nodules: The role of actin microfilaments in the organisation of symbiosomes. Protoplasma 1998, 203, 194–205. [Google Scholar] [CrossRef]
- Millar, A.; Finnegan, P.; Whelan, J.; Drevon, J.; Day, D.A. Expression and kinetics of the mitochondrial alternative oxidase in nitrogen-fixing nodules of soybean roots. Plant Cell Environ. 1997, 20, 1273–1282. [Google Scholar] [CrossRef]
- McKay, I.A.; Dilworth, M.J.; Glenn, A.R. C4-dicarboxylate metabolism in free-living and bacteroid forms of Rhizobium leguminosarum MNF3841. J. Microbiol. 1988, 134, 1433–1440. [Google Scholar] [CrossRef] [Green Version]
- Copeland, L.; Quinnell, R.G.; Day, D.A. Malic Enzyme Activity in Bacteroids from Soybean Nodules. J. Gen. Micrbiol. 1989, 135, 2005–2011. [Google Scholar] [CrossRef] [Green Version]
- Kimura, I.; Tajima, S. Presence and characteristics of NADP-malic enzyme in soybean nodule bacteroids. J. Soil Sci. Plant Nutr. 1989, 35, 271–279. [Google Scholar] [CrossRef]
- Driscoll, B.T.; Finan, T.M. NAD+-dependent malic enzyme of Rhizobium meliloti is required for symbiotic nitrogen fixation. Mol. Microbiol. 1993, 7, 865–873. [Google Scholar] [CrossRef]
- Driscoll, B.T.; Finan, T.M. Properties of NAD+- and NADP+-dependent malic enzymes of Rhizobium meliloti and differential expression of their genes in nitrogen-fixing bacteroids. Microbiology 1997, 143, 489–498. [Google Scholar] [CrossRef] [Green Version]
- Udvardi, M.K.; Lister, D.L.; Day, D.A. ATPase activity and anion transport across the peribacteroid membrane of isolated soybean symbiosomes. Arch. Microbiol. 1991, 156, 362–366. [Google Scholar] [CrossRef]
- Ou Yang, L.-J.; Whelan, J.; Weaver, C.D.; Roberts, D.M.; Day, D.A. Protein phosphorylation stimulates the rate of malate uptake across the peribacteroid membrane of soybean nodules. FEBS Lett. 1991, 293, 188–190. [Google Scholar] [CrossRef] [Green Version]
- Udvardi, M.K.; Day, D.A. Electrogenic ATPase Activity on the Peribacteroid Membrane of Soybean Glycine max Root Nodules. Plant Physiol. 1989, 90, 982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pierre, O.; Engler, G.; Hopkins, J.; Brau, F.; Boncompagni, E.; Hérouart, D. Peribacteroid space acidification: A marker of mature bacteroid functioning in Medicago truncatula nodules. Plant Cell Environ. 2013, 36, 2059–2070. [Google Scholar] [PubMed]
- Niemietz, C.M.; Tyerman, S.D. Channel-mediated permeation of ammonia gas through the peribacteroid membrane of soybean nodules. FEBS Lett. 2000, 465, 110–114. [Google Scholar] [CrossRef] [Green Version]
- Guenther, J.F.; Chanmanivone, N.; Galetovic, M.P.; Wallace, I.S.; Cobb, J.A.; Roberts, D.M. Phosphorylation of soybean nodulin 26 on serine 262 enhances water permeability and is regulated developmentally and by osmotic signals. Plant Cell 2003, 15, 981–991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rivers, R.L.; Dean, R.M.; Chandy, G.; Hall, J.E.; Roberts, D.M.; Zeidel, M.L. Functional analysis of nodulin 26, an aquaporin in soybean root nodule symbiosomes. J. Biol. Chem. 1997, 272, 16256–16261. [Google Scholar] [CrossRef] [Green Version]
- Wienkoop, S.; Saalbach, G. Proteome analysis. Novel proteins identified at the peribacteroid membrane from Lotus japonicus root nodules. Plant Physiol. 2003, 131, 1080–1090. [Google Scholar] [CrossRef]
- Saalbach, G.; Erik, P.; Wienkoop, S. Characterisation by proteomics of peribacteroid space and peribacteroid membrane preparations from pea (Pisum sativum) symbiosomes. Proteomics 2002, 2, 325–337. [Google Scholar] [CrossRef]
- Corratgé-Faillie, C.; Lacombe, B. Substrate(un)specificity of Arabidopsis NRT1/PTR FAMILY(NPF) proteins. J. Exp. Bot. 2017, 68, 3107–3113. [Google Scholar] [CrossRef]
- Jeong, J.; Suh, S.; Guan, C.; Tsay, Y.-F.; Moran, N.; Oh, C.J.; An, C.S.; Demchenko, K.N.; Pawlowski, K.; Lee, Y. A nodule-specific dicarboxylate transporter from alder is a member of the peptide transporter family. Plant Physiol. 2004, 134, 969–978. [Google Scholar] [CrossRef] [Green Version]
- Segonzac, C.; Boyer, J.-C.; Ipotesi, E.; Szponarski, W.; Tillard, P.; Touraine, B.; Sommerer, N.; Rossignol, M.; Gibrat, R. Nitrate Efflux at the Root Plasma Membrane: Identification of an Arabidopsis Excretion Transporter. Plant Cell 2007, 19, 3760. [Google Scholar] [CrossRef] [Green Version]
- Do, T.H.T.; Martinoia, E.; Lee, Y. Functions of ABC transporters in plant growth and development. Curr. Opin. Plant Biol. 2018, 41, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Choi, Y.; Burla, B.; Kim, Y.Y.; Jeon, B.; Maeshima, M.; Yoo, J.Y.; Martinoia, E.; Lee, Y. The ABC transporter AtABCB14 is a malate importer and modulates stomatal response to CO2. Nat. Cell Biol. 2008, 10, 1217–1223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henrichs, S.; Wang, B.; Fukao, Y.; Zhu, J.; Charrier, L.; Bailly, A.; Oehring, S.C.; Linnert, M.; Weiwad, M.; Endler, A.; et al. Regulation of ABCB1/PGP1-catalysed auxin transport by linker phosphorylation. EMBO J. 2012, 31, 2965–2980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wandrey, M.; Trevaskis, B.; Brewin, N.; Udvardi, M.K. Molecular and Cell Biology of a Family of Voltage-Dependent Anion Channel Porins in Lotus japonicus. Plant Physiol. 2004, 134, 182. [Google Scholar] [CrossRef] [Green Version]
- Roux, B.; Rodde, N.; Jardinaud, M.-F.; Timmers, T.; Sauviac, L.; Cottret, L.; Carrère, S.; Sallet, E.; Courcelle, E.; Moreau, S.; et al. An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant J. 2014, 77, 817–837. [Google Scholar] [CrossRef]
- Mun, T.; Bachmann, A.; Gupta, V.; Stougaard, J.; Andersen, S.U. Lotus Base: An integrated information portal for the model legume Lotus japonicus. Sci. Rep. 2016, 6, 39447. [Google Scholar] [CrossRef] [Green Version]
- Hurth, M.A.; Suh, S.J.; Kretzschmar, T.; Geis, T.; Bregante, M.; Gambale, F.; Martinoia, E.; Neuhaus, H.E. Impaired pH Homeostasis in Arabidopsis Lacking the Vacuolar Dicarboxylate Transporter and Analysis of Carboxylic Acid Transport across the Tonoplast. Plant Physiol. 2005, 137, 901. [Google Scholar] [CrossRef] [Green Version]
- Emmerlich, V.; Linka, N.; Reinhold, T.; Hurth, M.A.; Traub, M.; Martinoia, E.; Neuhaus, H.E. The plant homolog to the human sodium/dicarboxylic cotransporter is the vacuolar malate carrier. Proc. Natl. Acad. Sci. USA 2003, 100, 11122–11126. [Google Scholar] [CrossRef] [Green Version]
- Liu, R.; Li, B.; Qin, G.; Zhang, Z.; Tian, S. Identification and Functional Characterization of a Tonoplast Dicarboxylate Transporter in Tomato (Solanum lycopersicum). Front. Plant Sci. 2017, 8, 186. [Google Scholar] [CrossRef] [Green Version]
- Negi, J.; Matsuda, O.; Nagasawa, T.; Oba, Y.; Takahashi, H.; Kawai-Yamada, M.; Uchimiya, H.; Hashimoto, M.; Iba, K. CO2 regulator SLAC1 and its homologues are essential for anion homeostasis in plant cells. Nature 2008, 452, 483–486. [Google Scholar] [CrossRef]
- Maierhofer, T.; Diekmann, M.; Offenborn, J.N.; Lind, C.; Bauer, H.; Hashimoto, K.; Ka, S.A.-R.; Luan, S.; Kudla, J.; Geiger, D.; et al. Site- and kinase-specific phosphorylation-mediated activation of SLAC1, a guard cell anion channel stimulated by abscisic acid. Sci. Signal. 2014, 7, ra86. [Google Scholar] [CrossRef]
- Scherzer, S.; Maierhofer, T.; Al-Rasheid, K.A.; Geiger, D.; Hedrich, R. Multiple calcium-dependent kinases modulate ABA-activated guard cell anion channels. Mol. Plant 2012, 5, 1409–1412. [Google Scholar] [CrossRef] [Green Version]
- Sasaki, T.; Yamamoto, Y.; Ezaki, B.; Katsuhara, M.; Ahn, S.J.; Ryan, P.R.; Delhaize, E.; Matsumoto, H. A wheat gene encoding an aluminum-activated malate transporter. Plant J. 2004, 37, 645–653. [Google Scholar] [CrossRef] [PubMed]
- Kovermann, P.; Meyer, S.; Hörtensteiner, S.; Picco, C.; Scholz-Starke, J.; Ravera, S.; Lee, Y.; Martinoia, E. The Arabidopsis vacuolar malate channel is a member of the ALMT family. Plant J. 2007, 52, 1169–1180. [Google Scholar] [CrossRef]
- Meyer, S.; Scholz-Starke, J.; De Angeli, A.; Kovermann, P.; Burla, B.; Gambale, F.; Martinoia, E. Malate transport by the vacuolar AtALMT6 channel in guard cells is subject to multiple regulation. Plant J. 2011, 67, 247–257. [Google Scholar] [CrossRef] [Green Version]
- Ou Yang, L.-J.; Udvardi, M.K.; Day, D.A. Specificity and regulation of the dicarboxylate carrier on the peribacteroid membrane of soybean nodules. Planta 1990, 182, 437. [Google Scholar] [CrossRef]
- Li, H.; Jiang, F.; Wu, P.; Wang, K.; Cao, Y. A High-Quality Genome Sequence of Model Legume Lotus japonicus (MG-20) Provides Insights into the Evolution of Root Nodule Symbiosis. Genes 2020, 11, 5. [Google Scholar] [CrossRef]
- Libault, M.; Farmer, A.; Brechenmacher, L.; Drnevich, J.; Langley, R.J.; Bilgin, D.D.; Radwan, O.; Neece, D.J.; Clough, S.J.; May, G.D.; et al. Complete Transcriptome of the Soybean Root Hair Cell, a Single-Cell Model, and Its Alteration in Response to Bradyrhizobium japonicum Infection. Plant Physiol. 2010, 152, 541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Severin, A.J.; Woody, J.L.; Bolon, Y.-T.; Joseph, B.; Diers, B.W.; Farmer, A.D.; Muehlbauer, G.J.; Nelson, R.T.; Grant, D.; Specht, J.E.; et al. RNA-Seq Atlas of Glycine max: A guide to the soybean transcriptome. BMC Plant Biol. 2010, 10, 160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shelp, B.J.; Bown, A.W.; McLean, M.D. Metabolism and functions of gamma-aminobutyric acid. Trends Plant Sci. 1999, 4, 446–452. [Google Scholar] [CrossRef]
- Sunita, R.A.; Stephen, T.D.; Matthew, G.; Xu, B. Gamma-Aminobutyric acid(GABA) signalling in plants. Cell. Mol. Life Sci. 2017, 74, 1577–1603. [Google Scholar]
- Bown, A.W.; Shelp, B.J. Plant GABA: Not just a metabolite. Trends Plant Sci. 2016, 21, 811–813. [Google Scholar] [CrossRef] [PubMed]
- Kinnersley, A.M.; Turano, F.J. Gamma aminobutyric acid(GABA) and plant responses to stress. Crit. Rev. Plant Sci. 2000, 19, 479–509. [Google Scholar] [CrossRef]
- Sulieman, S. Does GABA increase the efficiency of symbiotic N2 fixation in legumes? Plant Signal. Behav. 2011, 6, 32–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sulieman, S.; Schulze, J. The efficiency of nitrogen fixation of the model legume Medicago truncatula (Jemalong A17) is low compared to Medicago sativa. Plant Physiol. 2010, 167, 683–692. [Google Scholar] [CrossRef] [PubMed]
- Vance, C.P.; Heichel, G.H. Carbon in N2 fixation: Limitation or exquisite adaptation. Annu. Rev. Plant Biol. 1991, 42, 373–390. [Google Scholar] [CrossRef]
- White, J.P.; Prell, J.; Ramachandran, V.K.; Poole, P.S. Characterization of a γ-aminobutyric acid transport system of Rhizobium leguminosarum bv. viciae 3841. J. Bacteriol. 2009, 191, 1547–1555. [Google Scholar] [CrossRef] [Green Version]
- Michaeli, S.; Fait, A.; Lagor, K.; Nunes-Nesi, A.; Grillich, N.; Yellin, A.; Bar, D.; Khan, M.; Fernie, A.R.; Turano, F.J. A mitochondrial GABA permease connects the GABA shunt and the TCA cycle, and is essential for normal carbon metabolism. Plant J. 2011, 67, 485–498. [Google Scholar] [CrossRef]
- Bown, A.W.; Shelp, B.J. The metabolism and physiological roles of 4-aminobutyric acid. Life Sci. Adv. 1989, 8, 21–25. [Google Scholar]
- Jin, H.; Dilworth, M.; Glenn, A. 4-Aminobutyrate is not available to bacteroids of cowpea Rhizobium MNF2030 in snake bean nodules. Arch. Microbiol. 1990, 153, 455–462. [Google Scholar] [CrossRef]
- Miller, R.; McRae, D.; Joy, K. Glutamate and γ-Aminobutyrate Metabolism. Mol. Plant Microbe Interact. 1991, 4, 37–45. [Google Scholar] [CrossRef]
- Hosie, A.H.F.; Allaway, D.; Galloway, C.S.; Dunsby, H.A.; Poole, P.S. Rhizobium leguminosarum has a second general amino acid permease with unusually broad substrate specificity and high similarity to branched-chain amino acid transporters (Bra/LIV) of the ABC family. J. Bacteriol 2002, 184, 4071–4080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sulieman, S.; Schulze, J. Phloem-derived gamma-aminobutyric acid (GABA) is involved in upregulating nodule N2 fixation efficiency in the model legume Medicago truncatula. Plant Cell Environ. 2010, 33, 2162–2172. [Google Scholar] [CrossRef] [PubMed]
- Girousse, C.; Bonnemain, J.L.; Delrot, S.; Bournoville, R. Sugar and amino acid composition of phloem sap of Medicago sativa: A comparative study of two collecting methods. Plant Physiol. Biochem. 1991, 29, 41–48. [Google Scholar]
- Atkins, C.A.; Pate, J.S.; Peoples, M.B.; Joy, K.W. Amino acid transport and metabolism in relation to the nitrogen economy of a legume leaf. Plant Physiol. 1983, 71, 841–848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pate, J.S.; Peoples, M.B.; Atkins, C.A. Spontaneous phloem bleeding from cryopunctured fruits of a ureide-producing legume. Plant Physiol. 1984, 74, 499–505. [Google Scholar] [CrossRef] [Green Version]
- Parsons, R.; Baker, A. Cycling of amino compounds in symbiotic lupin. J. Exp. Bot. 1996, 47, 421–429. [Google Scholar] [CrossRef] [Green Version]
- Layzell, D.B.; LaRue, T.A. Modeling C and N transport to developing soybean fruits. Plant Physiol. 1982, 70, 1290–1298. [Google Scholar] [CrossRef]
- Beuve, N.; Rispail, N.; Laine, P.; Cliquet, J.B.; Ourry, A.; Le Deunff, E. Putative role of γ-aminobutyric acid (GABA) as a long-distance signal in up-regulation of nitrate uptake in Brassica napus L. Plant Cell Environ. 2004, 27, 1035–1046. [Google Scholar] [CrossRef] [Green Version]
- Barbosa, J.M.; Singh, N.K.; Cherry, J.H.; Locy, R.D. Nitrate uptake and utilization is modulated by exogenous gamma-aminobutyric acid in Arabidopsis thaliana seedlings. Plant Physiol. Biochem. 2010, 48, 443–450. [Google Scholar] [CrossRef]
- Ramesh, S.A.; Tyerman, S.D.; Xu, B.; Bose, J.; Kaur, S.; Conn, V.; Domingos, P.; Ullah, S.; Wege, S.; Shabala, S.; et al. GABA signalling modulates plant growth by directly regulating the activity of plant-specific anion transporters. Nat. Commun. 2015, 6, 78–79. [Google Scholar]
- Serraj, R.; Shelp, B.J.; Sinclair, T.R. Accumulation of gamma-aminobutyric acid in nodulated soybean in response to drought stress. Physiol. Plant. 1998, 102, 79–86. [Google Scholar] [CrossRef]
- Giacomello, S. A new era for plant science: Spatial single-cell transcriptomics. Curr. Opin. Plant Biol. 2021, 60, 102041. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Booth, N.J.; Smith, P.M.C.; Ramesh, S.A.; Day, D.A. Malate Transport and Metabolism in Nitrogen-Fixing Legume Nodules. Molecules 2021, 26, 6876. https://doi.org/10.3390/molecules26226876
Booth NJ, Smith PMC, Ramesh SA, Day DA. Malate Transport and Metabolism in Nitrogen-Fixing Legume Nodules. Molecules. 2021; 26(22):6876. https://doi.org/10.3390/molecules26226876
Chicago/Turabian StyleBooth, Nicholas J., Penelope M. C. Smith, Sunita A. Ramesh, and David A. Day. 2021. "Malate Transport and Metabolism in Nitrogen-Fixing Legume Nodules" Molecules 26, no. 22: 6876. https://doi.org/10.3390/molecules26226876





