Towards Rational Biosurfactant Design—Predicting Solubilization in Rhamnolipid Solutions
Abstract
:1. Introduction
2. Results and Discussion
2.1. Solubilizing Properties of Rhamnolipid Biocomplex
2.2. Predicting MSR with EA and KSVM
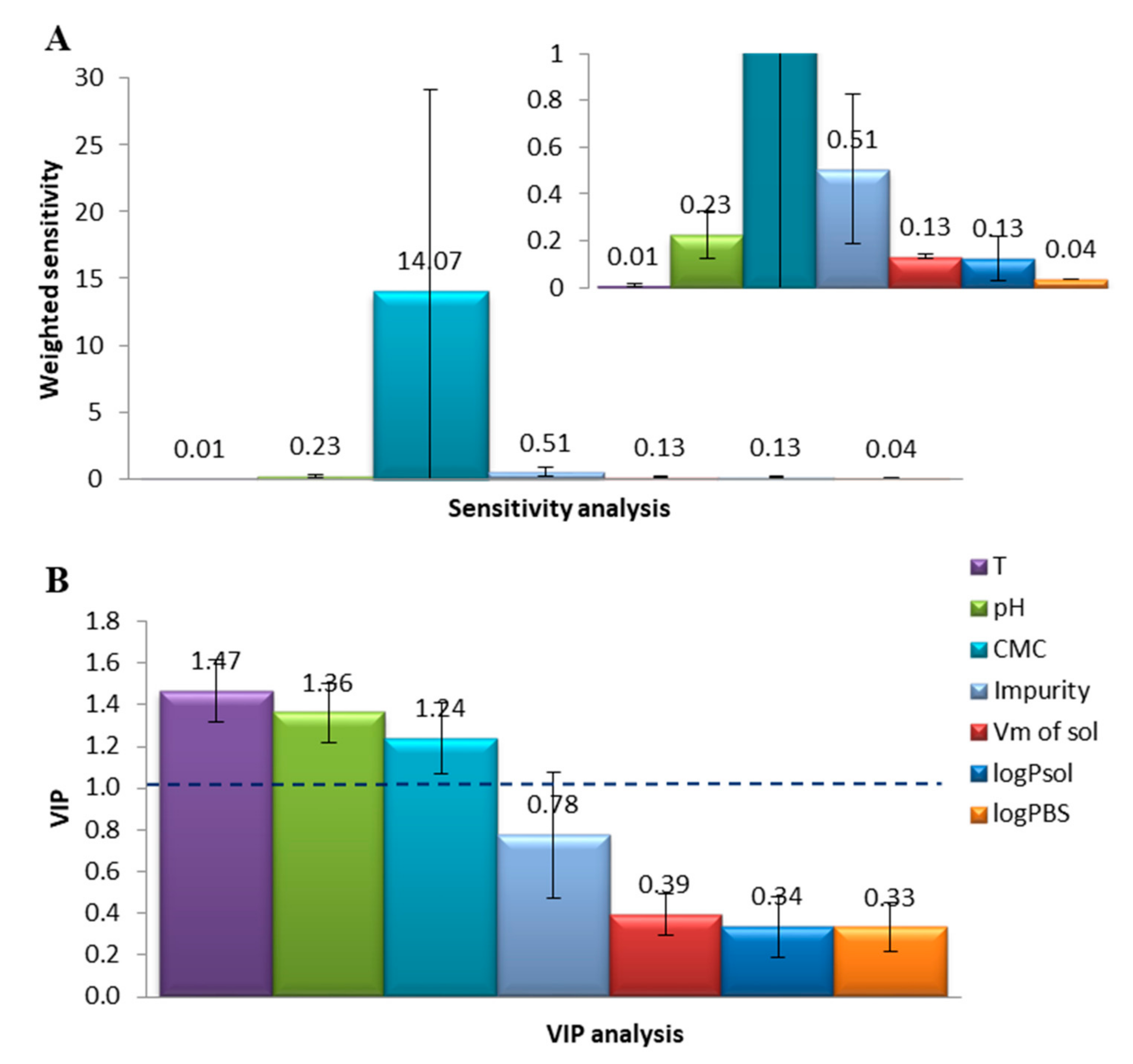
2.3. Sensitivity Analysis
2.4. PCA and Descriptor Overlap
3. Materials and Methods
3.1. Materials
3.2. Dodecane Solubilization
3.3. Solubilization Efficiency
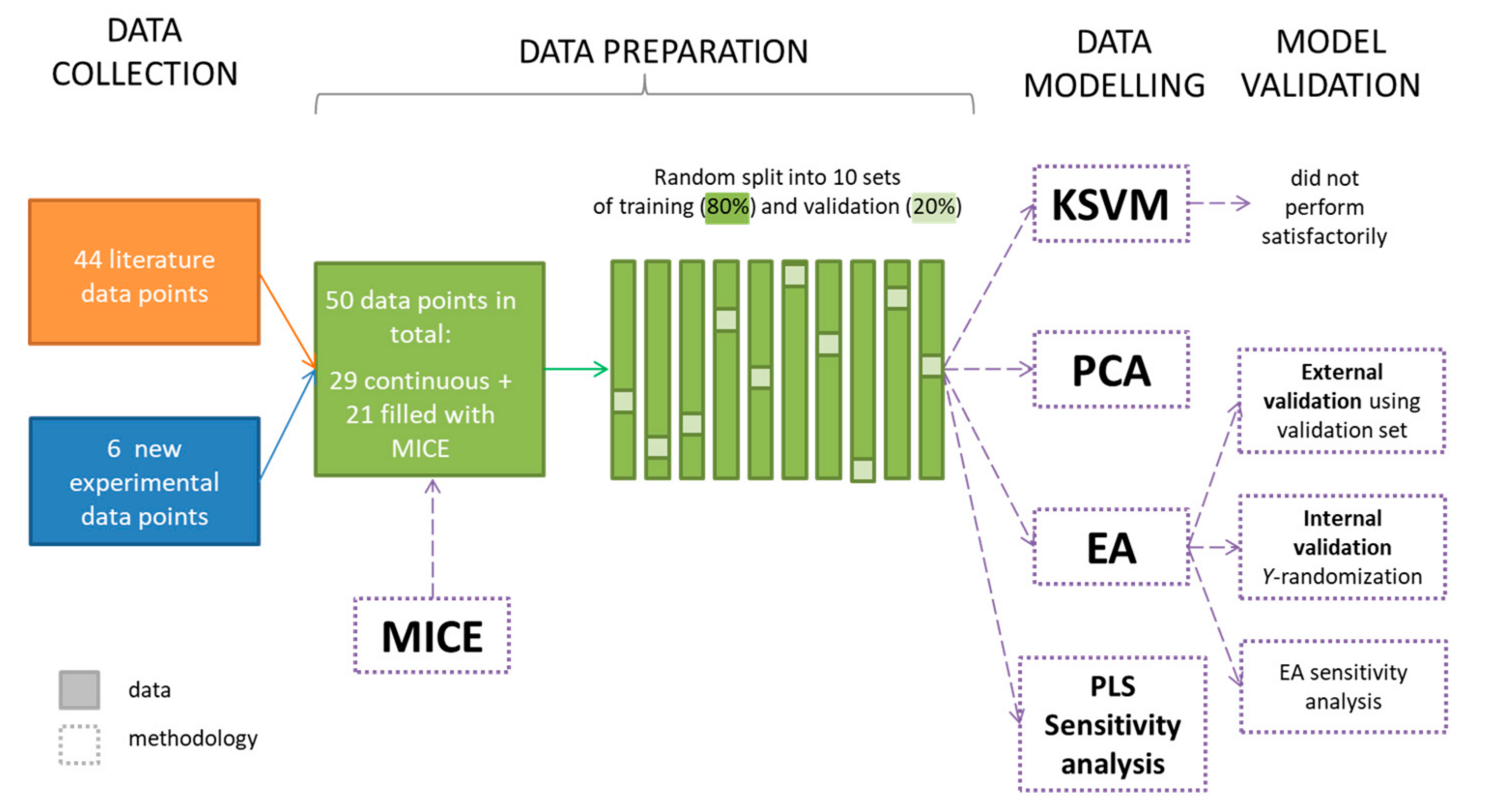
3.4. Data Collection
3.5. MICE
3.6. Computational Modelling
3.7. EA Generations
3.8. Model Validation—Y-Randomization
3.9. SVM Regression Methodology
3.10. Sensitivity for EA and KSVM
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
Abbreviations
References
- Clifford, J.S.; Ioannidis, M.A.; Legge, R.L. Enhanced aqueous solubilization of tetrachloroethylene by a rhamnolipid biosurfactant. J. Colloid Interface Sci. 2007, 305, 361–365. [Google Scholar] [CrossRef] [PubMed]
- Whang, L.-M.; Liu, P.-W.G.; Ma, C.-C.; Cheng, S.-S. Application of biosurfactants, rhamnolipid, and surfactin, for enhanced biodegradation of diesel-contaminated water and soil. J. Hazard. Mater. 2008, 151, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Mulligan, C.N.; Yong, R.; Gibbs, B. Surfactant-enhanced remediation of contaminated soil: A review. Eng. Geol. 2001, 60, 371–380. [Google Scholar] [CrossRef]
- Liu, Y.; Zeng, G.; Zhong, H.; Wang, Z.; Liu, Z.; Cheng, M.; Liu, G.; Yang, X.; Liu, S. Effect of rhamnolipid solubilization on hexadecane bioavailability: Enhancement or reduction? J. Hazard. Mater. 2017, 322, 394–401. [Google Scholar] [CrossRef] [PubMed]
- Nasongkla, N.; Bey, E.; Ren, J.; Ai, H.; Khemtong, C.; Guthi, J.S.; Chin, S.-F.; Sherry, A.D.; Boothman, D.A.; Gao, J. Multifunctional polymeric micelles as cancer-targeted, MRI-ultrasensitive drug delivery systems. Nano Lett. 2006, 6, 2427–2430. [Google Scholar] [CrossRef] [PubMed]
- Kaur, P.; Garg, T.; Rath, G.; Murthy, R.; Goyal, A.K. Surfactant-based drug delivery systems for treating drug-resistant lung cancer. Drug Deliv. 2016, 23, 717–728. [Google Scholar] [CrossRef]
- Maldonado-Valderrama, J.; Wilde, P.; Macierzanka, A.; Mackie, A. The role of bile salts in digestion. Adv. Colloid Interface Sci. 2011, 165, 36–46. [Google Scholar] [CrossRef]
- Scott, M.J.; Jones, M.N. The biodegradation of surfactants in the environment. Biochim. Biophys. Acta Biomembr. 2000, 1508, 235–251. [Google Scholar] [CrossRef] [Green Version]
- Tehrani-Bagha, A.R.; Oskarsson, H.; Van Ginkel, C.; Holmberg, K. Cationic ester-containing gemini surfactants: Chemical hydrolysis and biodegradation. J. Colloid Interface Sci. 2007, 312, 444–452. [Google Scholar] [CrossRef]
- Toribio, J.; Escalante, A.E.; Soberón-Chávez, G. Rhamnolipids: Production in bacteria other than Pseudomonas aeruginosa. Eur. J. Lipid Sci. Technol. 2010, 112, 1082–1087. [Google Scholar] [CrossRef]
- Nitschke, M.; Costa, S. Biosurfactants in food industry. Trends Food Sci. Technol. 2007, 18, 252–259. [Google Scholar] [CrossRef]
- Chrzanowski, Ł.; Ławniczak, Ł.; Czaczyk, K. Why do microorganisms produce rhamnolipids? World J. Microbiol. Biotechnol. 2012, 28, 401–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdel-Mawgoud, A.M.; Lépine, F.; Déziel, E. Rhamnolipids: Diversity of structures, microbial origins and roles. Appl. Microbiol. Biotechnol. 2010, 86, 1323–1336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hošková, M.; Schreiberová, O.; Ježdík, R.; Chudoba, J.; Masák, J.; Sigler, K.; Řezanka, T. Characterization of rhamnolipids produced by non-pathogenic Acinetobacter and Enterobacter bacteria. Bioresour. Technol. 2013, 130, 510–516. [Google Scholar] [CrossRef] [PubMed]
- Elshikh, M.; Funston, S.; Chebbi, A.; Ahmed, S.; Marchant, R.; Banat, I.M. Rhamnolipids from non-pathogenic Burkholderia thailandensis E264: Physicochemical characterization, antimicrobial and antibiofilm efficacy against oral hygiene related pathogens. New Biotechnol. 2017, 36, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Wigneswaran, V.; Nielsen, K.F.; Sternberg, C.; Jensen, P.R.; Folkesson, A.; Jelsbak, L. Biofilm as a production platform for heterologous production of rhamnolipids by the non-pathogenic strain Pseudomonas putida KT2440. Microb. Cell Fact. 2016, 15, 181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Twigg, M.S.; Tripathi, L.; Zompra, A.; Salek, K.; Irorere, V.; Gutierrez, T.; Spyroulias, G.; Marchant, R.; Banat, I.M. Identification and characterisation of short chain rhamnolipid production in a previously uninvestigated, non-pathogenic marine pseudomonad. Appl. Microbiol. Biotechnol. 2018, 102, 8537–8549. [Google Scholar] [CrossRef] [Green Version]
- Özdemir, G.; Peker, S.; Helvaci, S. Effect of pH on the surface and interfacial behavior of rhamnolipids R1 and R2. Colloids Surf. A 2004, 234, 135–143. [Google Scholar] [CrossRef]
- Jemil, N.; Ayed, H.B.; Hmidet, N.; Nasri, M. Characterization and properties of biosurfactants produced by a newly isolated strain Bacillus methylotrophicus DCS1 and their applications in enhancing solubility of hydrocarbon. World J. Microbiol. Biotechnol. 2016, 32, 175. [Google Scholar] [CrossRef]
- Kłosowska-Chomiczewska, I.E.; Medrzycka, K.; Karpenko, E. Biosurfactants—Biodegradability, toxicity, efficiency in comparison with synthetic surfactants. In Proceedings of the Research and Application of New Technologies in Wastewater Treatment and Municipal Solid Waste Disposal in Ukraine, Sweden, and Poland, Cracow, Poland, 17–19 October 2011; pp. 1–9. [Google Scholar]
- Li, G.; Lan, G.; Liu, Y.; Chen, C.; Lei, L.; Du, J.; Lu, Y.; Li, Q.; Du, G.; Zhang, J. Evaluation of biodegradability and biotoxicity of surfactants in soil. RSC Adv. 2017, 7, 31018–31026. [Google Scholar] [CrossRef] [Green Version]
- Özdemir, G.; Sezgin, Ö. Keratin–rhamnolipids and keratin–sodium dodecyl sulfate interactions at the air/water interface. Colloids Surf. B 2006, 52, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Ju, L.-K.; Dashtbozorg, S.; Vongpanish, N. Wound Dressings with Enhanced Gas Permeation and Other Beneficial Properties. U.S. Patent 9,468,700, 18 October 2016. [Google Scholar]
- Ławniczak, Ł.; Marecik, R.; Chrzanowski, Ł. Contributions of biosurfactants to natural or induced bioremediation. Appl. Microbiol. Biotechnol. 2013, 97, 2327–2339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chrzanowski, Ł.; Dziadas, M.; Ławniczak, Ł.; Cyplik, P.; Białas, W.; Szulc, A.; Lisiecki, P.; Jeleń, H. Biodegradation of rhamnolipids in liquid cultures: Effect of biosurfactant dissipation on diesel fuel/B20 blend biodegradation efficiency and bacterial community composition. Bioresour. Technol. 2012, 111, 328–335. [Google Scholar] [CrossRef] [PubMed]
- Marecik, R.; Wojtera-Kwiczor, J.; Ławniczak, Ł.; Cyplik, P.; Szulc, A.; Piotrowska-Cyplik, A.; Chrzanowski, Ł. Rhamnolipids increase the phytotoxicity of diesel oil towards four common plant species in a terrestrial environment. Water Air Soil Pollut. 2012, 223, 4275–4282. [Google Scholar] [CrossRef] [Green Version]
- Zhong, H.; Yang, X.; Tan, F.; Brusseau, M.L.; Yang, L.; Liu, Z.; Zeng, G.; Yuan, X. Aggregate-based sub-CMC solubilization of n-alkanes by monorhamnolipid biosurfactant. New J. Chem. 2016, 40, 2028–2035. [Google Scholar] [CrossRef] [Green Version]
- Tiso, T.; Ihling, N.; Kubicki, S.; Biselli, A.; Schonhoff, A.; Bator, I.; Thies, S.; Karmainski, T.; Kruth, S.; Willenbrink, A.-L. Integration of genetic and process engineering for optimized rhamnolipid production using Pseudomonas putida. Front. Bioeng. Biotechnol. 2020, 8, 976. [Google Scholar] [CrossRef]
- He, N.; Wu, T.; Jiang, J.; Long, X.; Shao, B.; Meng, Q. Toward high-efficiency production of biosurfactant rhamnolipids using sequential fed-batch fermentation based on a fill-and-draw strategy. Colloids Surf. B 2017, 157, 317–324. [Google Scholar] [CrossRef]
- Jiang, J.; Zu, Y.; Li, X.; Meng, Q.; Long, X. Recent progress towards industrial rhamnolipids fermentation: Process optimization and foam control. Bioresour. Technol. 2020, 298, 122394. [Google Scholar] [CrossRef]
- Moutinho, L.F.; Moura, F.R.; Silvestre, R.C.; Romão-Dumaresq, A.S. Microbial biosurfactants: A broad analysis of properties, applications, biosynthesis and techno-economical assessment of rhamnolipid production. Biotechnol. Prog. 2020, e3093. [Google Scholar] [CrossRef]
- Kłosowska-Chomiczewska, I. Physicochemical Properties and Possibilities of Application of Biosurfactant Preparation Produced by Pseudomonas PS-17 (pol.). Ph.D. Thesis, Gdańsk University of Technology, Gdańsk, Poland, 2014. [Google Scholar]
- Zhang, Y.; Miller, R.M. Effect of rhamnolipid (biosurfactant) structure on solubilization and biodegradation of n-alkanes. Appl. Environ. Microbiol. 1995, 61, 2247–2251. [Google Scholar] [CrossRef] [Green Version]
- Hansch, C.; Hoekman, D.; Leo, A.; Weininger, D.; Selassie, C.D. Chem-bioinformatics: Comparative QSAR at the interface between chemistry and biology. Chem. Rev. 2002, 102, 783–812. [Google Scholar] [CrossRef]
- Cherkasov, A.; Muratov, E.N.; Fourches, D.; Varnek, A.; Baskin, I.I.; Cronin, M.; Dearden, J.; Gramatica, P.; Martin, Y.C.; Todeschini, R. QSAR modeling: Where have you been? Where are you going to? J. Med. Chem. 2014, 57, 4977–5010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kłosowska-Chomiczewska, I.; Mędrzycka, K.; Hallmann, E.; Karpenko, E.; Pokynbroda, T.; Macierzanka, A.; Jungnickel, C. Rhamnolipid CMC prediction. J. Colloid Interface Sci. 2017, 488, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Kłosowska-Chomiczewska, I.E.; Artichowicz, W.; Preiss, U.; Jungnickel, C. Multicomponent ionic liquid CMC prediction. Phys. Chem. Chem. Phys. 2017, 19, 25309–25318. [Google Scholar] [CrossRef] [PubMed]
- Masrat, R.; Maswal, M.; Dar, A.A. Competitive solubilization of naphthalene and pyrene in various micellar systems. J. Hazard. Mater. 2013, 244, 662–670. [Google Scholar] [CrossRef] [PubMed]
- Goklen, K.E.; Hatton, T.A. Liquid-liquid extraction of low molecular-weight proteins by selective solubilization in reversed micelles. Sep. Sci. Technol. 1987, 22, 831–841. [Google Scholar] [CrossRef]
- Hill, A.J.; Ghoshal, S. Micellar solubilization of naphthalene and phenanthrene from nonaqueous-phase liquids. Environ. Sci. Technol. 2002, 36, 3901–3907. [Google Scholar] [CrossRef]
- Chaiko, M.A.; Nagarajan, R.; Ruckenstein, E. Solubilization of single-component and binary mixtures of hydrocarbons in aqueous micellar solutions. J. Colloid Interface Sci. 1984, 99, 168–182. [Google Scholar] [CrossRef]
- Bernardez, L.A.; Ghoshal, S. Selective solubilization of polycyclic aromatic hydrocarbons from multicomponent nonaqueous-phase liquids into nonionic surfactant micelles. Environ. Sci. Technol. 2004, 38, 5878–5887. [Google Scholar] [CrossRef]
- Yu, J.; Fischman, D.A.; Steck, T.L. Selective solubilization of proteins and phospholipids from red blood cell membranes by nonionic detergents. J. Cell Biochem. 1973, 1, 233–248. [Google Scholar] [CrossRef]
- Gaudin, T.; Rotureau, P.; Pezron, I.; Fayet, G. New QSPR Models to Predict the Critical Micelle Concentration of Sugar-Based Surfactants. Ind. Eng. Chem. Res. 2016, 55, 11716–11726. [Google Scholar] [CrossRef]
- Salager, J.-L.; Antón, R.; Anderez, J.M.; Aubry, J.-M. Formulation des micro-émulsions par la méthode HLD. Techniques de l’Ingénieur 2001, 157, 2001. [Google Scholar]
- Acosta, E.; Szekeres, E.; Sabatini, D.A.; Harwell, J.H. Net-average curvature model for solubilization and supersolubilization in surfactant microemulsions. Langmuir 2003, 19, 186–195. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Edelen, A.; Neighbors, B.; Sabatini, D.A. Biocompatible lecithin-based microemulsions with rhamnolipid and sophorolipid biosurfactants: Formulation and potential applications. J. Colloid Interface Sci. 2010, 348, 498–504. [Google Scholar] [CrossRef] [PubMed]
- Rongsayamanont, W.; Soonglerdsongpha, S.; Khondee, N.; Pinyakong, O.; Tongcumpou, C.; Sabatini, D.A.; Luepromchai, E. Formulation of crude oil spill dispersants based on the HLD concept and using a lipopeptide biosurfactant. J. Hazard. Mater. 2017, 334, 168–177. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Morgan, C.; Poindexter, L.; Fernandez, J. Application of the Hydrophilic–Lipophilic Deviation Concept to Surfactant Characterization and Surfactant Selection for Enhanced Oil Recovery. J. Surfactants Deterg. 2019. [Google Scholar] [CrossRef]
- Sunwoo, C.K.; Wade, W.H. Optimal surfactant structures for cosurfactant-free microemulston systems I. C16 and C14 Guerbet alcohol hydrophobes. J. Dispers. Sci. Technol. 1992, 13, 491–514. [Google Scholar] [CrossRef]
- Zhang, G.; Yu, J.; Du, C.; Lee, R. Formulation of surfactants for very low/high salinity surfactant flooding without alkali. In Proceedings of the SPE International Symposium on Oilfield Chemistry, The Woodlands, TX, USA, 13–15 April 2015. [Google Scholar]
- Sahigara, F.; Mansouri, K.; Ballabio, D.; Mauri, A.; Consonni, V.; Todeschini, R. Comparison of different approaches to define the applicability domain of QSAR models. Molecules 2012, 17, 4791–4810. [Google Scholar] [CrossRef] [Green Version]
- Wan, J.; Meng, D.; Long, T.; Ying, R.; Ye, M.; Zhang, S.; Li, Q.; Zhou, Y.; Lin, Y. Simultaneous Removal of Lindane, Lead and Cadmium from Soils by Rhamnolipids Combined with Citric Acid. PLoS ONE 2015, 10, e0129978. [Google Scholar] [CrossRef]
- Zhong, H.; Liu, Y.; Liu, Z.; Jiang, Y.; Tan, F.; Zeng, G.; Yuan, X.; Yan, M.; Niu, Q.; Liang, Y. Degradation of pseudo-solubilized and mass hexadecane by a Pseudomonas aeruginosa with treatment of rhamnolipid biosurfactant. Int. Biodeter. Biodegr. 2014, 94, 152–159. [Google Scholar] [CrossRef]
- Guo, Y.; Mulligan, C.N.; Nieh, M.P. An unusual morphological transformation of rhamnolipid aggregates induced by concentration and addition of styrene: A small angle neutron scattering (SANS) study. Colloids Surf. A 2011, 373, 42–50. [Google Scholar] [CrossRef]
- Wan, J.; Chai, L.; Lu, X.; Lin, Y.; Zhang, S. Remediation of hexachlorobenzene contaminated soils by rhamnolipid enhanced soil washing coupled with activated carbon selective adsorption. J. Hazard. Mater. 2011, 189, 458–464. [Google Scholar] [CrossRef] [PubMed]
- Shin, K.H.; Kim, J.Y.; Kim, K.W. Effect of Biosurfactant Addition on the Biodegradation of phenanthrene in soil-water system. Environ. Eng. Res. 2008, 13, 8–13. [Google Scholar] [CrossRef] [Green Version]
- Bai, G.; Brusseau, M.L.; Miller, R.M. Influence of cation type, ionic strength, and pH on solubilization and mobilization of residual hydrocarbon by a biosurfactant. J. Contam. Hydrol. 1998, 30, 265–279. [Google Scholar] [CrossRef]
- Zhang, Y.; Maier, W.J.; Miller, R.M. Effect of rhamnolipids on the dissolution, bioavailability, and biodegradation of phenanthrene. Environ. Sci. Technol. 1997, 31, 2211–2217. [Google Scholar] [CrossRef]
- Li, S.; Pi, Y.; Bao, M.; Zhang, C.; Zhao, D.; Li, Y.; Sun, P.; Lu, J. Effect of rhamnolipid biosurfactant on solubilization of polycyclic aromatic hydrocarbons. Mar. Pollut. Bull. 2015, 101, 219–225. [Google Scholar] [CrossRef]
- Zhang, X.; Guo, Q.; Hu, Y.; Lin, H. Effects of monorhamnolipid and dirhamnolipid on sorption and desorption of triclosan in sediment–water system. Chemosphere 2013, 90, 581–587. [Google Scholar] [CrossRef]
- Zhu, S.; Liang, S.; You, X. Effect of Rhamnolipid Biosurfactant on Solubilization and Biodegradation of Polycyclic Aromatic Hydrocarbons. In Proceedings of the 2013 Third International Conference on Intelligent System Design and Engineering Applications, Hong Kong, China, 16–18 January 2013; pp. 651–654. [Google Scholar]
- Yin, H.; Qiang, J.; Jia, Y.; Ye, J.; Peng, H.; Qin, H.; Zhang, N.; He, B. Characteristics of biosurfactant produced by Pseudomonas aeruginosa S6 isolated from oil-containing wastewater. Process. Biochem. 2009, 44, 302–308. [Google Scholar] [CrossRef]
- Harendra, S.; Vipulanandan, C. Degradation of high concentrations of PCE solubilized in SDS and biosurfactant with Fe/Ni bi-metallic particles. Colloids Surf. A 2008, 322, 6–13. [Google Scholar] [CrossRef]
- Bordas, F.; Lafrance, P.; Villemur, R. Conditions for effective removal of pyrene from an artificially contaminated soil using Pseudomonas aeruginosa 57SJ rhamnolipids. Environ. Pollut. 2005, 138, 69–76. [Google Scholar] [CrossRef]
- Bodour, A.A.; Guerrero-Barajas, C.; Jiorle, B.V.; Malcomson, M.E.; Paull, A.K.; Somogyi, A.; Trinh, L.N.; Bates, R.B.; Maier, R.M. Structure and characterization of flavolipids, a novel class of biosurfactants produced by Flavobacterium sp. strain MTN11. Appl. Environ. Microbiol. 2004, 70, 114–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barkay, T.; Navon-Venezia, S.; Ron, E.; Rosenberg, E. Enhancement of solubilization and biodegradation of polyaromatic hydrocarbons by the bioemulsifier alasan. Appl. Environ. Microbiol. 1999, 65, 2697–2702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thangamani, S.; Shreve, G.S. Effect of anionic biosurfactant on hexadecane partitioning in multiphase systems. Environ. Sci. Technol. 1994, 28, 1993–2000. [Google Scholar] [CrossRef] [PubMed]
- OECD. Guidance Document on the Validation of (Quantitative) Structure-Activity Relationships [(Q)SAR] Models; OECD Environment Health and Safety Publications: Paris, France, 2007; Volume 69. [Google Scholar]
- Aşçı, Y.; Nurbaş, M.; Açıkel, Y.S. Sorption of Cd (II) onto kaolin as a soil component and desorption of Cd (II) from kaolin using rhamnolipid biosurfactant. J. Hazard. Mater. 2007, 139, 50–56. [Google Scholar] [CrossRef]
- Cohen, R.; Exerowa, D. Surface forces and properties of foam films from rhamnolipid biosurfactants. Adv. Colloid Interface Sci. 2007, 134, 24–34. [Google Scholar] [CrossRef]
- Shin, K.-H.; Kim, K.-W.; Kim, J.-Y.; Lee, K.-E.; Han, S.-S. Rhamnolipid morphology and phenanthrene solubility at different pH values. J. Environ. Qual. 2008, 37, 509–514. [Google Scholar] [CrossRef]
- Santos, S.F.; Zanette, D.; Fischer, H.; Itri, R. A systematic study of bovine serum albumin (BSA) and sodium dodecyl sulfate (SDS) interactions by surface tension and small angle X-ray scattering. J. Colloid Interface Sci. 2003, 262, 400–408. [Google Scholar] [CrossRef]
- Edwards, A.L. An Introduction to Linear Regression and Correlation, 2nd ed.; W.H. Freeman: New York, NY, USA, 1984. [Google Scholar]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction, Springer Series in Statistics; Springer: New York, NY, USA, 2009. [Google Scholar]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Łozińska, N.; Głowacz-Różyńska, A.; Artichowicz, W.; Lu, Y.; Jungnickel, C. Microencapsulation of fish oil–determination of optimal wall material and encapsulation methodology. J. Food Eng. 2020, 268, 109730. [Google Scholar] [CrossRef]
- Myers, D. Surfaces, Interfaces, and Colloids: Principles and Applications, 2nd ed.; VCH: New York, NY, USA, 1999. [Google Scholar]
- Rangel-Yagui, C.O.; Pessoa, A., Jr.; Tavares, L.C. Micellar solubilization of drugs. J. Pharm. Pharm. Sci. 2005, 8, 147–163. [Google Scholar]
- Brophy, J.; Bawden, D. Is Google enough? Comparison of an internet search engine with academic library resources. Aslib Proc. 2005, 57, 498–512. [Google Scholar] [CrossRef] [Green Version]
- Molinspiration Chemoinformatics; Molinspiration: Brastislava, Slovak Republic, 2014.
- Addinsoft, Version 2019.1.2; XLSTAT Statistical and Data Analysis Solution: Boston, MA, USA, 2019.
- Jia, Q.; Zhao, Y.; Yan, F.; Wang, Q. QSAR model for predicting the toxicity of organic compounds to fathead minnow. Environ. Sci. Pollut. Res. 2018, 25, 35420–35428. [Google Scholar] [CrossRef] [PubMed]
- Veerasamy, R.; Rajak, H.; Jain, A.; Sivadasan, S.; Varghese, C.P.; Agrawal, R.K. Validation of QSAR models-strategies and importance. Int. J. Drug Des. Discov. 2011, 3, 511–519. [Google Scholar]
- Linden, L.; Goss, K.-U.; Endo, S. 3D-QSAR predictions for α-cyclodextrin binding constants using quantum mechanically based descriptors. Chemosphere 2017, 169, 693–699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cherkassky, V.; Ma, Y. Selection of Meta-Parameters for Support Vector Regression. In Proceedings of the Artificial Neural Networks—ICANN 2002, Madrid, Spain, 28–30 August 2002. [Google Scholar] [CrossRef]
- Cawley, G.C.; Talbot, N.L. On over-fitting in model selection and subsequent selection bias in performance evaluation. J. Mach. Learn. Res. 2010, 11, 2079–2107. [Google Scholar]
- Tobias, R.D. An Introduction to Partial Least Squares Regression. In Proceedings of the Twentieth Annual SAS Users Group International Conference; SAS Institute Inc: Cary, NC, USA, 1995; pp. 1250–1257. [Google Scholar]
- Vinzi, V.E.; Chin, W.W.; Henseler, J.; Wang, H. Handbook of Partial Least Squares; Springer: New York, NY, USA, 2010; Volume 201. [Google Scholar]
- Angrist, J.D.; Imbens, G.W.; Krueger, A.B. Jackknife instrumental variables estimation. J. Appl. Econom. 1999, 14, 57–67. [Google Scholar] [CrossRef]
- Farrés, M.; Platikanov, S.; Tsakovski, S.; Tauler, R. Comparison of the variable importance in projection (VIP) and of the selectivity ratio (SR) methods for variable selection and interpretation. J. Chemom. 2015, 29, 528–536. [Google Scholar] [CrossRef]
- Al-Bahry, S.; Al-Wahaibi, Y.; Elshafie, A.; Al-Bemani, A.; Joshi, S.; Al-Makhmari, H.; Al-Sulaimani, H. Biosurfactant production by Bacillus subtilis B20 using date molasses and its possible application in enhanced oil recovery. Int. Biodeterior. Biodegr. 2013, 81, 141–146. [Google Scholar] [CrossRef]
- Luna, J.M.; Sarubbo, L.; Campos-Takaki, G.M. A new biosurfactant produced by Candida glabrata UCP 1002: Characteristics of stability and application in oil recovery. Braz. Arch. Biol. Technol. 2009, 52, 785–793. [Google Scholar] [CrossRef]
- Thavasi, R.; Jayalakshmi, S.; Banat, I.M. Application of biosurfactant produced from peanut oil cake by Lactobacillus delbrueckii in biodegradation of crude oil. Bioresour. Technol. 2011, 102, 3366–3372. [Google Scholar] [CrossRef]
- Luna, J.M.; Rufino, R.D.; Sarubbo, L.A.; Campos-Takaki, G.M. Characterisation, surface properties and biological activity of a biosurfactant produced from industrial waste by Candida sphaerica UCP0995 for application in the petroleum industry. Colloids Surf. B 2013, 102, 202–209. [Google Scholar] [CrossRef] [PubMed]




| Process | Surfactant | Matrix | Compound Selectively Solubilized | Ref. |
|---|---|---|---|---|
| Separation of proteins with similar, low molecular size from their ternary mixture | Anionic surfactant: Aerosol OT/AOT (sodium di-2-ethylhexyl sulfosuccinate) | Ternary mixture of LMW proteins (ribonuclease-a, cytochrome-c and lysozyme) | Ribonuclease-a or cytochrome-c or lysozyme | [39] |
| Selective solubilization | Nonionic surfactants: Brij 30, C12E4Brij 35, C12E23 Tergitol NP10, C9PE10.5 Triton X-100, C8PE9.5Tween 80, C18S6E20 | Naphthalene, hexadecane and/or phenantrene | Naphtalene or phenantrene | [40] |
| Selective solubilization | Cationic surfactants: Cetylpyridinium chloride, C16PyrCl Dodecyl ammonium chloride, C12NH3Cl Dodecyl trimethyl ammonium chloride, N[C12,C,C,C]Cl Anionic surfactants: Sodium dodecyl sulfate, C12SO4Na Sodium diamyl sulfosuccinate (Aerosol AY) | Benzene-hexane mixtures | Benzene | [41] |
| Selective solubilization of PAHs from NAPL | Nonionic surfactants: Brij 30, C12E4 Brij 35, C12E23 Tergitol NP10, C9PE10.5 Triton X-100, C8PE9.5Tween 80, C18S6E20 | Ternary mixtures of naphtalene, phenantrene and hexadecane | Phenantrene 1 | [42] |
| Competitive solubilization of naphthalene and pyrene | Nonionic surfactants: Brij30, C12E4 Brij56, C16E10 Cationic surfactants: Dodecylethyldimethyl ammonium bromide, N[C12,C2,C,C]Br Cethyltrimethyl ammonium chloride, N[C16,C,C,C]Br Anionic surfactant: Sodium dodecyl sulfate, C12SO4Na | Binary mixtures of pyrene and naphthalene | Pyrene | [38] |
| Selective solubilization of glycerolipid and glycoprotein species from isolated human erythrocyte membranes | Nonionic surfactant: Triton X-100 | Isolated human erythrocyte membranes | Glycolipid, glycerolipid | [43] |
| Parameter | Impurity | logPBS | CMC [mg/L] | logPsol | Vm sol. [Å3] | pH | T [°C] | MSR |
|---|---|---|---|---|---|---|---|---|
| Min. value | 0 | 4.51 | 0.0010 | 2.69 | 94.19 | 4 | 20 | 1.35 × 10−5 |
| Max. value | 2 | 5.21 | 700.00 | 9.00 | 314.59 | 9 | 30 | 82.10 |
| Mean | 0.73 | 4.88 | 101.98 | 5.80 | 207.04 | 6.76 | 24.0 | 4.97 |
| SD | 0.80 | 0.22 | 118.20 | 1.99 | 59.42 | 0.78 | 2.7 | 16.06 |
| Parameter | Chemometric Tool | |
|---|---|---|
| EA | KSVM | |
| External validation: | ||
| R2val ± CI | 0.773 ± 0.052 | 0.166 ± 0.152 |
| MSEval ± CI | 0.296 ± 0.084 | 1.176 ± 0.483 |
| Y-randomization: | ||
| G | 2.613 | - |
| G (Critical value) | 2.355 | - |
| p-value | 0.006 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kłosowska-Chomiczewska, I.E.; Kotewicz-Siudowska, A.; Artichowicz, W.; Macierzanka, A.; Głowacz-Różyńska, A.; Szumała, P.; Mędrzycka, K.; Hallmann, E.; Karpenko, E.; Jungnickel, C. Towards Rational Biosurfactant Design—Predicting Solubilization in Rhamnolipid Solutions. Molecules 2021, 26, 534. https://doi.org/10.3390/molecules26030534
Kłosowska-Chomiczewska IE, Kotewicz-Siudowska A, Artichowicz W, Macierzanka A, Głowacz-Różyńska A, Szumała P, Mędrzycka K, Hallmann E, Karpenko E, Jungnickel C. Towards Rational Biosurfactant Design—Predicting Solubilization in Rhamnolipid Solutions. Molecules. 2021; 26(3):534. https://doi.org/10.3390/molecules26030534
Chicago/Turabian StyleKłosowska-Chomiczewska, Ilona E., Adrianna Kotewicz-Siudowska, Wojciech Artichowicz, Adam Macierzanka, Agnieszka Głowacz-Różyńska, Patrycja Szumała, Krystyna Mędrzycka, Elżbieta Hallmann, Elena Karpenko, and Christian Jungnickel. 2021. "Towards Rational Biosurfactant Design—Predicting Solubilization in Rhamnolipid Solutions" Molecules 26, no. 3: 534. https://doi.org/10.3390/molecules26030534
APA StyleKłosowska-Chomiczewska, I. E., Kotewicz-Siudowska, A., Artichowicz, W., Macierzanka, A., Głowacz-Różyńska, A., Szumała, P., Mędrzycka, K., Hallmann, E., Karpenko, E., & Jungnickel, C. (2021). Towards Rational Biosurfactant Design—Predicting Solubilization in Rhamnolipid Solutions. Molecules, 26(3), 534. https://doi.org/10.3390/molecules26030534








