The Sesquiterpene Synthase PtTPS5 Produces (1S,5S,7R,10R)-Guaia-4(15)-en-11-ol and (1S,7R,10R)-Guaia-4-en-11-ol in Oomycete-Infected Poplar Roots
Abstract
:1. Introduction
2. Results
2.1. P. cactorum Infection Induces the Accumulation of Terpenes, Aromatic Compounds, and Fatty Acids in P. trichocarpa Roots
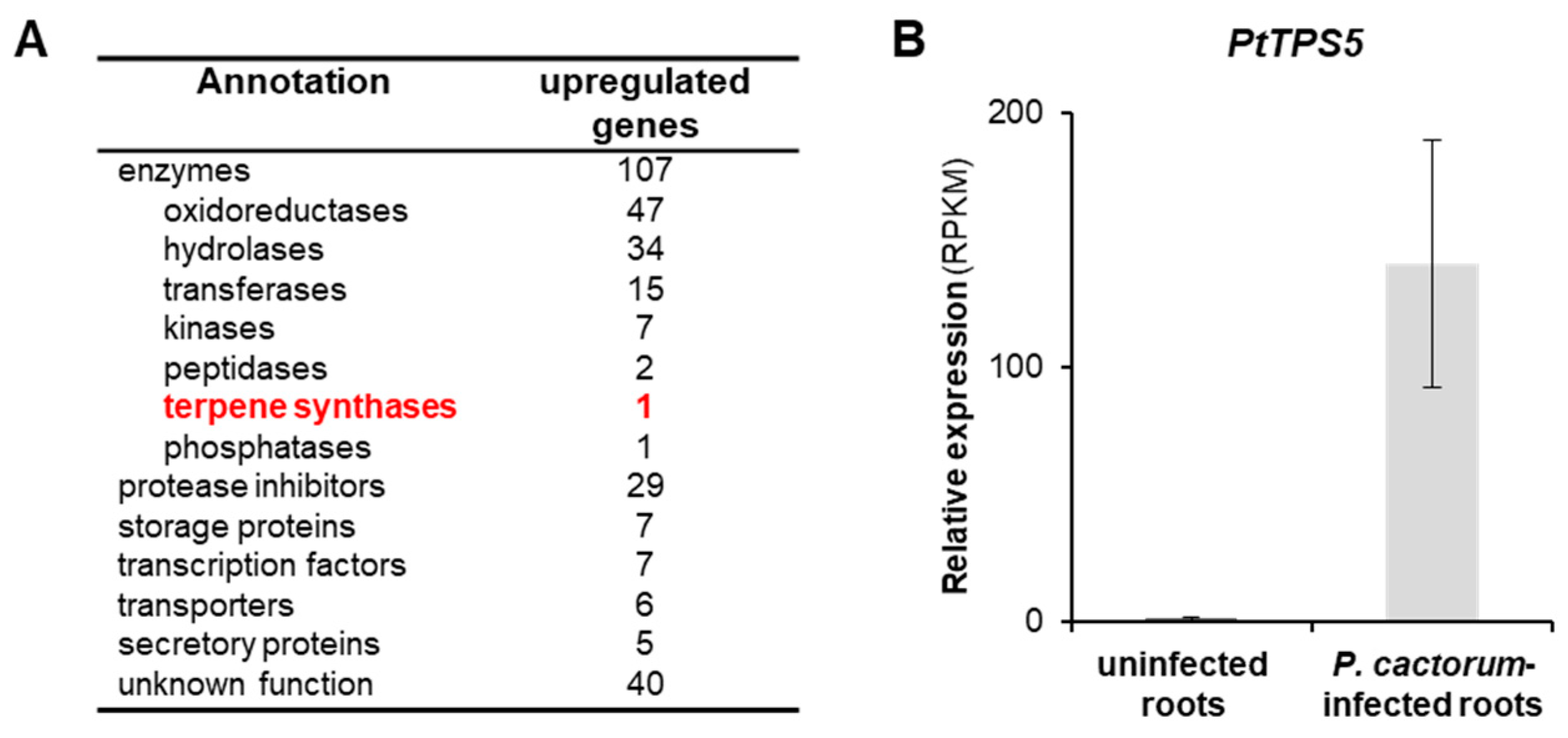
2.2. Transcriptome Analysis of Infected and Non-Infected Poplar Roots Revealed a Sesquiterpene Synthase Gene PtTPS5 that Is highly Induced upon P. cactorum Infection
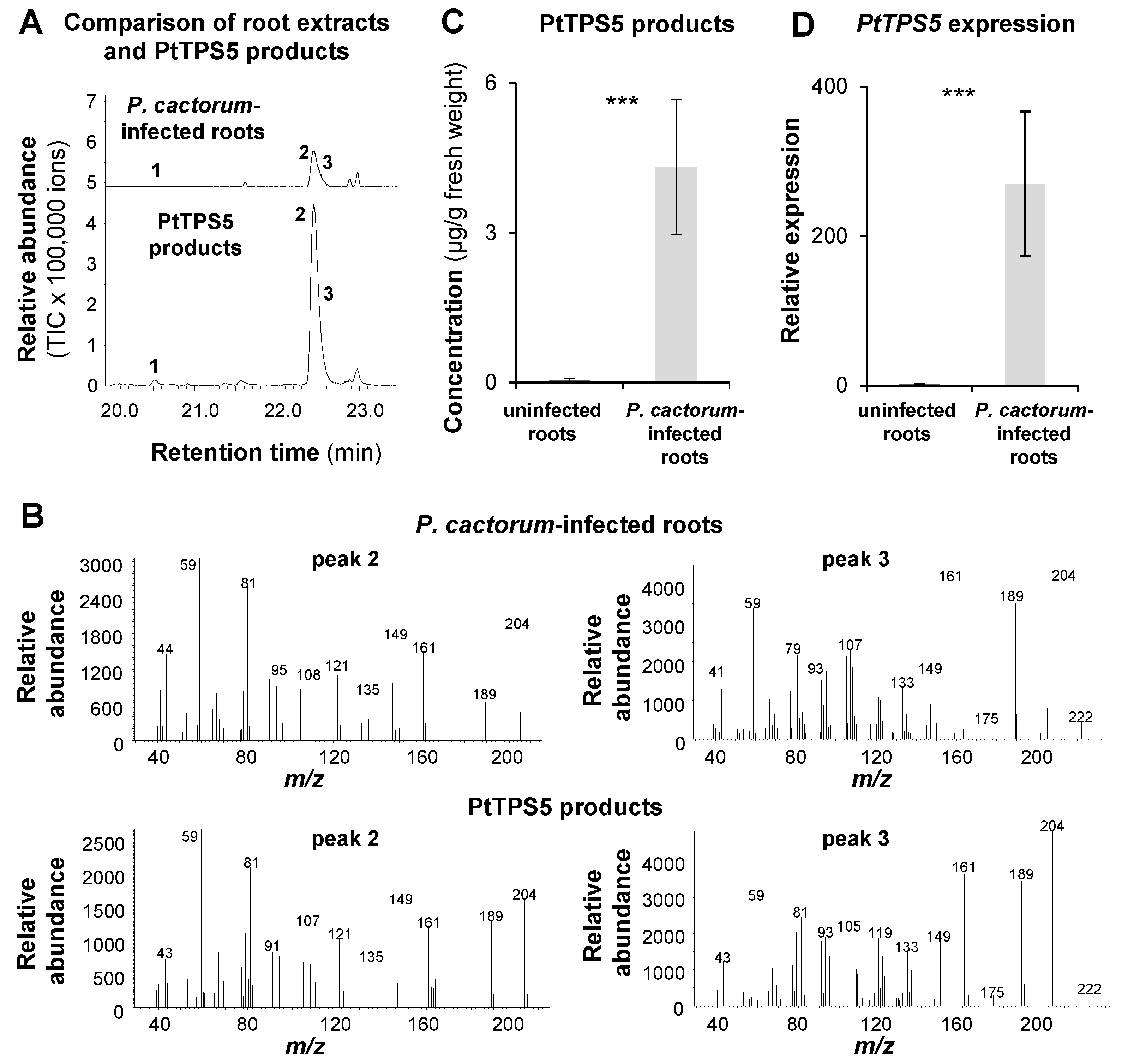
2.3. PtTPS5 Produces (1S,5S,7R,10R)-guaia-4(15)-en-11-ol and (1S,7R,10R)-guaia-4-en-11-ol as Major Products
2.4. The Accumulation of (1S,5S,7R,10R)-guaia-4(15)-en-11-ol, (1S,7R,10R)-guaia-4-en-11-ol, and Hedycaryol in P. cactorum-Infected and Non-Infected Roots Matches the Expression of PtTPS5
3. Discussion
4. Materials and Methods
4.1. Biological Material
4.2. Phytophthora Cactorum Treatment
4.3. Hexane Extraction of Root Tissue and GC-MS/GC-FID Analysis
4.4. Methanol Extraction of Root Tissue and HPLC-UV, LC-MS/MS Analysis of Methanol Extracts
4.5. RNA Extraction and Reverse Transcription
4.6. Heterologous Expression of PtTPS5 and Enzyme Assays
4.7. RNA Sequencing and RT-qPCR Analysis
4.8. Compound Isolation and Structure Elucidation
 /cm–1 = 2953 (m), 2923 (s), 2854 (m), 1714 (w), 1650 (w), 1456 (m), 1376 (m), 1260 (m), 1094 (s), 1020 (s), 873 (m), 800 (s).
/cm–1 = 2953 (m), 2923 (s), 2854 (m), 1714 (w), 1650 (w), 1456 (m), 1376 (m), 1260 (m), 1094 (s), 1020 (s), 873 (m), 800 (s). /cm–1 = 2954 (s), 2923 (s), 2854 (s), 1723 (w), 1670 (w), 1459 (m), 1376 (m), 1260 (w), 1096 (w), 1025 (w), 800 (w).
/cm–1 = 2954 (s), 2923 (s), 2854 (s), 1723 (w), 1670 (w), 1459 (m), 1376 (m), 1260 (w), 1096 (w), 1025 (w), 800 (w).4.9. Statistical Analysis
4.10. Accession Numbers
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Tholl, D. Biosynthesis and biological functions of terpenoids in plants. In Biotechnology of Isoprenoids; Schrader, J., Bohlmann, J., Eds.; Springer: Cham, Switzerland, 2015; pp. 63–106. [Google Scholar]
- Unsicker, S.B.; Kunert, G.; Gershenzon, J. Protective perfumes: The role of vegetative volatiles in plant defense against herbivores. Curr. Opin. Plant Biol. 2009, 12, 479–485. [Google Scholar] [CrossRef]
- Junker, R.R.; Gershenzon, J.; Unsicker, S.B. Floral odor bouquet loses its ant repellent properties after inhibition of terpene biosynthesis. J. Chem. Ecol. 2011, 37, 1323–1331. [Google Scholar] [CrossRef]
- Zhou, W.; Kügler, A.; McGale, E.; Haverkamp, A.; Knaden, M.; Guo, H.; Beran, F.; Yon, F.; Li, R.; Lackus, N.; et al. Tissue-specific emission of (E)-α-bergamotene helps resolve the dilemma when pollinators are also herbivores. Curr. Biol. 2017, 27, 1336–1341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brooks, C.J.; Watson, D.G. Terpenoid phytoalexins. Nat. Prod. Rep. 1991, 8, 367–389. [Google Scholar] [CrossRef] [PubMed]
- Schmelz, E.A.; Kaplan, F.; Huffaker, A.; Dafoe, N.J.; Vaughan, M.M.; Ni, X.Z. Identity, regulation, and activity of inducible diterpenoid phytoalexins in maize. Proc. Natl. Acad. Sci. USA 2011, 108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huffaker, A.; Kaplan, F.; Vaughan, M.M.; Dafoe, N.J.; Ni, X.; Rocca, J.R.; Alborn, H.T.; Teal, P.E.; Schmelz, E.A. Novel acidic sesquiterpenoids constitute a dominant class of pathogen-induced phytoalexins in maize. Plant Physiol. 2011, 156, 2082–2097. [Google Scholar] [CrossRef] [Green Version]
- Degenhardt, J.; Köllner, T.G.; Gershenzon, J. Monoterpene and sesquiterpene synthases and the origin of terpene skeletal diversity in plants. Phytochemistry 2009, 70, 1621–1637. [Google Scholar] [CrossRef] [PubMed]
- Dudareva, N.; Pichersky, E.; Gershenzon, J. Biochemistry of plant volatiles. Plant Physiol. 2004, 135, 1893–1902. [Google Scholar] [CrossRef] [Green Version]
- Bathe, U.; Tissier, A. Cytochrome P450 enzymes: A driving force of plant diterpene diversity. Phytochemistry 2019, 161, 149–162. [Google Scholar] [CrossRef]
- Danner, H.; Böckler, G.A.; Irmisch, S.; Yuan, J.S.; Chen, F.; Gershenzon, J.; Unsicker, S.B.; Köllner, T.G. Four terpene synthases produce major compounds of the gypsy moth feeding-induced volatile blend of Populus trichocarpa. Phytochemistry 2011, 72. [Google Scholar] [CrossRef]
- Irmisch, S.; Jiang, Y.; Chen, F.; Gershenzon, J.; Köllner, T.G. Terpene synthases and their contribution to herbivore-induced volatile emission in western balsam poplar (Populus trichocarpa). BMC Plant Biol. 2014, 14, 270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irmisch, S.; Müller, A.T.; Schmidt, L.; Günther, J.; Gershenzon, J.; Köllner, T.G. One amino acid makes the difference: The formation of ent-kaurene and 16α-hydroxy-ent-kaurane by diterpene synthases in poplar. BMC Plant Biol. 2015, 15, 262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lackus, N.D.; Lackner, S.; Gershenzon, J.; Unsicker, S.B.; Köllner, T.G. The occurrence and formation of monoterpenes in herbivore-damaged poplar roots. Sci. Rep. 2018, 8, 17936. [Google Scholar] [CrossRef] [PubMed]
- Lackus, N.D.; Petersen, N.P.; Nagel, R.; Schmidt, A.; Irmisch, S.; Gershenzon, J.; Kollner, T.G. Identification and characterization of trans-isopentenyl diphosphate synthases involved in herbivory-induced volatile terpene formation in Populus trichocarpa. Molecules 2019, 24, 2408. [Google Scholar] [CrossRef] [Green Version]
- Jones, R.; Sutherland, M. Hedycaryol, the precursor of elemol. Chem. Commun. 1968, 20, 1229–1230. [Google Scholar] [CrossRef]
- Böckler, G.A.; Gershenzon, J.; Unsicker, S.B. Phenolic glycosides of the Salicaceae and their role as anti-herbivore defenses. Phytochemistry 2011, 72, 1497–1509. [Google Scholar] [CrossRef]
- Schena, L.; Duncan, J.M.; Cooke, D.E.L. Development and application of a PCR-based ‘molecular tool box’ for the identification of Phytophthora species damaging forests and natural ecosystems. Plant Pathol. 2008, 57, 64–75. [Google Scholar] [CrossRef]
- Tissandie, L.; Viciana, S.; Brevard, H.; Meierhenrich, U.J.; Filippi, J.J. Towards a complete characterisation of guaiacwood oil. Phytochemistry 2018, 149, 64–81. [Google Scholar] [CrossRef]
- Xu, H.; Dickschat, J.S. Germacrene A–A central intermediate in sesquiterpene biosynthesis. Chemistry 2020. [Google Scholar] [CrossRef]
- Rinkel, J.; Dickschat, J.S. Addressing the chemistry of germacrene A by isotope labeling experiments. Org. Lett. 2019, 21, 2426–2429. [Google Scholar] [CrossRef]
- Rabe, P.; Rinkel, J.; Nubbemeyer, B.; Köllner, T.G.; Chen, F.; Dickschat, J.S. Terpene cyclases from social amoebae. Angew. Chem. Int. Ed. 2016, 55, 15420–15423. [Google Scholar] [CrossRef] [PubMed]
- Cornforth, J.W.; Cornforth, R.H.; Popják, G.; Yengoyan, L. Studies on the biosynthesis of cholesterol. XX. Steric course of decarboxylation of 5-pyrophosphomevalonate and of the carbon to carbon bond formation in the biosynthesis of farnesyl pyrophosphate. J. Biol. Chem. 1966, 241, 3970–3987. [Google Scholar] [CrossRef]
- Lauterbach, L.; Rinkel, J.; Dickschat, J.S. Two bacterial diterpene synthases from Allokutzneria albata produce bonnadiene, phomopsene, and allokutznerene. Angew. Chem. Int. Ed. 2018, 57, 8280–8283. [Google Scholar] [CrossRef] [PubMed]
- Toljamo, A.; Blande, D.; Karenlampi, S.; Kokko, H. Reprogramming of strawberry (Fragaria vesca) root transcriptome in response to Phytophthora cactorum. PLoS ONE 2016, 11, e0161078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yadav, H.; Dreher, D.; Athmer, B.; Porzel, A.; Gavrin, A.; Baldermann, S.; Tissier, A.; Hause, B. Medicago terpene synthase 10 is involved in defense against an oomycete root pathogen. Plant Physiol. 2019, 180, 1598–1613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Köllner, T.G.; O’Maille, P.E.; Gatto, N.; Boland, W.; Gershenzon, J.; Degenhardt, J. Two pockets in the active site of maize sesquiterpene synthase TPS4 carry out sequential parts of the reaction scheme resulting in multiple products. Arch. Biochem. Biophys. 2006, 448, 83–92. [Google Scholar] [CrossRef]
- Davis, G.D.; Essenberg, M. (+)-δ-cadinene is a product of sesquiterpene cyclase activity in cotton. Phytochemistry 1995, 39, 553–567. [Google Scholar] [CrossRef]
- Chen, X.-Y.; Chen, Y.; Heinstein, P.; Davisson, V.J. Cloning, expression, and characterization of (+)-δ-cadinene synthase: A catalyst for cotton phytoalexin biosynthesis. Arch. Biochem. Biophys. 1995, 324, 255–266. [Google Scholar] [CrossRef]
- Mao, H.; Liu, J.; Ren, F.; Peters, R.J.; Wang, Q. Characterization of CYP71Z18 indicates a role in maize zealexin biosynthesis. Phytochemistry 2016, 121, 4–10. [Google Scholar] [CrossRef]
- Tian, X.; Ruan, J.-X.; Huang, J.-Q.; Yang, C.-Q.; Fang, X.; Chen, Z.-W.; Hong, H.; Wang, L.-J.; Mao, Y.-B.; Lu, S.; et al. Characterization of gossypol biosynthetic pathway. Proc. Natl. Acad. Sci. USA 2018, 115, E5410. [Google Scholar] [CrossRef] [Green Version]
- Upchurch, R.G. Fatty acid unsaturation, mobilization, and regulation in the response of plants to stress. Biotechnol. Lett. 2008, 30, 967–977. [Google Scholar] [CrossRef] [PubMed]
- Rojas, C.M.; Senthil-Kumar, M.; Tzin, V.; Mysore, K.S. Regulation of primary plant metabolism during plant-pathogen interactions and its contribution to plant defense. Front. Plant Sci. 2014, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pohl, C.H.; Kock, J.L.; Thibane, V.S. Antifungal free fatty acids: A review. Sci. Microb. Pathog. Commun. Curr. Res. Technol. Adv. 2011, 3, 61–71. [Google Scholar]
- Kachroo, A.; Kachroo, P. Fatty acid-derived signals in plant defense. Annu. Rev. Phytopathol. 2009, 47, 153–176. [Google Scholar] [CrossRef] [PubMed]
- Kalemba, D.; Kunicka, A. Antibacterial and antifungal properties of essential oils. Curr. Med. Chem. 2003, 10, 813–829. [Google Scholar] [CrossRef]
- Böckler, G.A.; Gershenzon, J.; Unsicker, S.B. Gypsy moth caterpillar feeding has only a marginal impact on phenolic compounds in old-growth black poplar. J. Chem. Ecol. 2013, 39, 1301–1312. [Google Scholar] [CrossRef]
- Lackner, S.; Lackus, N.D.; Paetz, C.; Köllner, T.G.; Unsicker, S.B. Aboveground phytochemical responses to belowground herbivory in poplar trees and the consequence for leaf herbivore preference. Plant Cell Environ. 2019. [Google Scholar] [CrossRef] [Green Version]
- Lackus, N.D.; Müller, A.; Kröber, T.D.U.; Reichelt, M.; Schmidt, A.; Nakamura, Y.; Paetz, C.; Luck, K.; Lindroth, R.L.; Constabel, C.P.; et al. The occurrence of sulfated salicinoids in poplar and their formation by sulfotransferase 1. Plant Physiol. 2020, 183, 137–151. [Google Scholar] [CrossRef] [Green Version]
- Ramírez-Carvajal, G.A.; Morse, A.M.; Davis, J.M. Transcript profiles of the cytokinin response regulator gene family in Populus imply diverse roles in plant development. New Phytol. 2008, 177, 77–89. [Google Scholar] [CrossRef]
- Xu, M.; Zhang, B.; Su, X.; Zhang, S.; Huang, M. Reference gene selection for quantitative real-time polymerase chain reaction in Populus. Anal. Biochem. 2011, 408, 337–339. [Google Scholar] [CrossRef]
- Wang, H.L.; Chen, J.; Tian, Q.; Wang, S.; Xia, X.; Yin, W. Identification and validation of reference genes for Populus euphratica gene expression analysis during abiotic stresses by quantitative real-time pcr. Physiol. Plant 2014, 152, 529–545. [Google Scholar] [CrossRef] [PubMed]

| Compound | Uninfected Roots | Infected Roots | t-Value/T-Value | p-Value |
|---|---|---|---|---|
| Aromatic compounds | ||||
| Benzylalcohol # | 0.22 ± 0.20 | 0.77 ± 0.20 | 48.00 (WR) | 0.038 * |
| Salicylaldehyde # | 4.02 ± 1.00 | 4.93 ± 1.79 | 0.02 (ST) | 0.988 |
| 2-Phenylethanol # | 0.11 ± 0.04 | 0.74 ± 0.23 | 39.00 (WR) | <0.001 *** |
| Benzyl salicylate | n.q. | n.q. | - | - |
| Terpenes | ||||
| Limonene # | n.q | n.q | - | - |
| 1,8-Cineole # | n.q | n.q | - | - |
| α-Terpineol # | 0.10 ± 0.03 | 0.44 ± 0.05 | 5.26 (ST) | <0.001 *** |
| Elemol | 0.01 ± 0.01 | 0.27 ± 0.09 | 40.50 (WR) | 0.002 ** |
| Guaia-4(15)-en-11-ol # + Guaia-4-en-11-ol # | 0.04 ± 0.04 | 4.31 ± 1.35 | 36.00 (WR) | <0.001 *** |
| Fatty acids/aldehydes | ||||
| (E)-4-Nonenal | n.q. | n.q. | - | - |
| Myristaldehyde | 3.16 ± 0.34 | 8.53 ± 2.17 | 3.13 (ST) | 0.007 ** |
| Myristic acid # | 0.40 ± 0.08 | 6.13 ± 1.59 | 8.19 (ST) | <0.001 *** |
| Pentadecanoic acid # | 1.51 ± 0.30 | 4.49 ± 0.51 | 38.00 (WR) | <0.001 *** |
| Palmitic acid # | 24.53 ± 3.57 | 72.04 ± 6.13 | 36.00 (WR) | <0.001 *** |
| Oleic acid # | 5.04 ± 0.92 | 7.34 ± 1.09 | 1.51 (ST) | 0.154 |
| Stearic acid # | 1.65 ±0.19 | 5.29 ± 0.71 | 6.39 (ST) | <0.001 *** |
| Others | ||||
| 1-Hexanol | 0.14 ± 0.01 | 0.17 ± 0.01 | 12.00 (WR) | 0.038 * |
| Unidentified compound | traces | 1.64 ± 0.88 | 40.00 (WR) | 0.002 ** |
| Compound | Uninfected Roots | Infected Roots | t-Value/T-Value | p-Value |
|---|---|---|---|---|
| Salicin | 41.87 ± 16.03 | 77.22 ± 34.06 | 19.00 (WR) | 0.336 |
| Salicin-7-sulfate | 2.08 ± 0.26 | 3.39 ± 0.39 | 2.52 (ST) | 0.026 * |
| Salirepin | 14.50 ± 2.39 | 20.62 ± 2.79 | 1.53 (ST) | 0.151 |
| Salirepin-7-sulfate | 0.30 ± 0.03 | 0.42 ± 0.05 | 1.77 (ST) | 0.1 |
| Salicortin | 368.31 ± 172.48 | 204.06 ± 70.32 | 2.25 (ST) | 0.056 |
| Tremulacin | 3.16 ± 1.27 | 2.49 ± 1.98 | 0.28 (ST) | 0.785 |
| Homaloside D | 16.77 ± 9.28 | 7.78 ± 3.04 | 33.00 (WR) | 0.596 |
| C | Guaia-4(15)-en-11-ol (1) | Guaia-4-en-11-ol (2) | ||
|---|---|---|---|---|
| 13C | 1H | 13C | 1H | |
| 1 | 52.34 (CH) | 1.18 (m) | 55.60 (CH) | 2.28 (m) |
| 2 | 32.42 (CH2) | 1.76 (m, Hα) 1.00 (m, Hβ) | 30.42 (CH2) | 1.92 (m) 1.51 (m) |
| 3 | 32.96 (CH2) | 2.32 (ddm, J = 15.9, 7.8, Hβ) 2.19 (m, Hα) | 36.60 (CH2) | 2.23 (m) 2.13 (m) |
| 4 | 159.08 (Cq) | – | 131.74 (Cq) | – |
| 5 | 46.33 (CH) | 2.15 (m) | 138.62 (Cq) | – |
| 6 | 34.56 (CH2) | 1.88 (ddd, J = 13.9, 9.2, 6.1, Hβ) 1.54 (ddd, J = 13.9, 11.3, 8.0, Hα) | 30.37 (CH2) | 2.67 (d, J = 15.2) 2.00 (m) |
| 7 | 49.56 (CH) | 1.41 (m) | 49.30 (CH) | 1.36 (m) |
| 8 | 26.71 (CH2) | 1.74 (m, Hα) 1.10 (m, Hβ) | 31.51 (CH2) | 1.90 (m) 1.00 (m) |
| 9 | 40.29 (CH2) | 1.77 (m, Hβ) 0.96 (m, Hα) | 40.26 (CH2) | 1.78 (m) 1.10 (m) |
| 10 | 42.32 (CH) | 1.06 (m) | 39.37 (CH) | 1.33 (m) |
| 11 | 73.26 (Cq) | – | 72.82 (Cq) | – |
| 12 | 27.75 (CH3) | 1.03 (s) | 26.89 (CH3) | 1.03 (s) |
| 13 | 25.78 (CH3) | 1.01 (s) | 26.66 (CH3) | 1.02 (s) |
| 14 | 21.58 (CH3) | 0.87 (d, J = 6.5) | 22.01 (CH3) | 0.93 (d, J = 6.6) |
| 15 | 104.24 (CH2) | 5.00 (m, HE) 4.91 (br, HZ) | 14.60 (CH3) | 1.63 (br s) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lackus, N.D.; Morawetz, J.; Xu, H.; Gershenzon, J.; Dickschat, J.S.; Köllner, T.G. The Sesquiterpene Synthase PtTPS5 Produces (1S,5S,7R,10R)-Guaia-4(15)-en-11-ol and (1S,7R,10R)-Guaia-4-en-11-ol in Oomycete-Infected Poplar Roots. Molecules 2021, 26, 555. https://doi.org/10.3390/molecules26030555
Lackus ND, Morawetz J, Xu H, Gershenzon J, Dickschat JS, Köllner TG. The Sesquiterpene Synthase PtTPS5 Produces (1S,5S,7R,10R)-Guaia-4(15)-en-11-ol and (1S,7R,10R)-Guaia-4-en-11-ol in Oomycete-Infected Poplar Roots. Molecules. 2021; 26(3):555. https://doi.org/10.3390/molecules26030555
Chicago/Turabian StyleLackus, Nathalie D., Jennifer Morawetz, Houchao Xu, Jonathan Gershenzon, Jeroen S. Dickschat, and Tobias G. Köllner. 2021. "The Sesquiterpene Synthase PtTPS5 Produces (1S,5S,7R,10R)-Guaia-4(15)-en-11-ol and (1S,7R,10R)-Guaia-4-en-11-ol in Oomycete-Infected Poplar Roots" Molecules 26, no. 3: 555. https://doi.org/10.3390/molecules26030555






