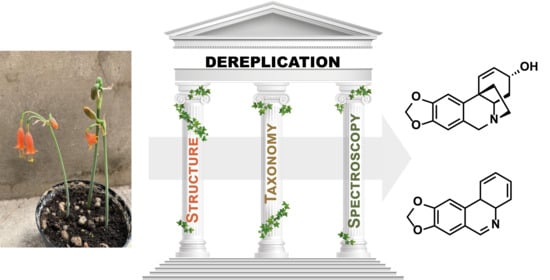
The Three Pillars of Natural Product Dereplication. Alkaloids from the Bulbs of Urceolina peruviana (C. Presl) J.F. Macbr. as a Preliminary Test Case
Abstract
:1. Introduction
1.1. General Considerations
1.2. The Three Pilars of Dereplication
1.2.1. Molecular Structures
1.2.2. Spectroscopy
1.2.3. Taxonomy
1.2.4. Databases
2. Results
2.1. KnapsakSearch
2.2. Predicted NMR Data for Natural Products (PNMRNP)
2.3. CSEARCH
2.4. Databases and Dereplication
2.5. Application to the Alkaloids of Urceolina peruviana (Amaryllidaceae)
2.5.1. Fraction A4, Major Compound
2.5.2. Fraction A7, Major Compound
2.5.3. Fraction A9, Major Compound
2.5.4. Fraction A11, Major Compound
2.5.5. Fraction A2, a Minor Compound
2.5.6. Database Searches
3. Materials
3.1. Chemicals
3.2. NMR
3.3. UPLC-HRMS
3.4. CPC
3.5. Plant Material
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, Y.; Kirchmair, J. Cheminformatics in natural product based drug discovery. Mol. Inform. 2020, 39, 2000171. [Google Scholar] [CrossRef]
- Hubert, J.; Nuzillard, J.-M.; Renault, J.-H. Dereplication strategies in natural product research: How many tools and methodologies behind the same concept? Phytochem. Rev. 2017, 16, 55–95. [Google Scholar] [CrossRef]
- Logan, D.C. Known knowns, known unknowns, unknown unknowns and the propagation of scientific enquiry. J. Exp. Bot. 2009, 60, 712–714. [Google Scholar] [CrossRef]
- Bakiri, A.B.; Plainchont, B.; Emerenciano, V.d.P.; Reynaud, R.; Hubert, J.; Renault, J.-H.; Nuzillard, J.-M. Computer-aided dereplication and structure elucidation of natural products at the university of Reims. Mol. Inform. 2017, 36, 1700027. [Google Scholar] [CrossRef] [Green Version]
- Rutz, A.; Dounoue-Kubo, M.; Ollivier, S.; Bisson, J.; Bagheri, M.; Saesong, T.; Ebrahimi, S.N.; Ingkaninan, K.; Wolfender, J.-L.; Allard, P.-M. Taxonomically informed scoring enhances confidence in natural products annotation. Front. Plant Sci. 2019, 10, 1329. [Google Scholar] [CrossRef] [Green Version]
- Heller, S.R.; McNaught, A.; Pletnev, I.; Stein, S.; Tchekhovskoi, D. Inchi, the IUPAC international chemical identifier. J. Cheminform. 2015, 7, 23. [Google Scholar] [CrossRef] [Green Version]
- Weininger, D. SMILES, a chemical language and information system, 1. Introduction to methodology and encoding rules. J. Chem. Inf. Model. 1988, 28, 31–36. [Google Scholar] [CrossRef]
- Dalby, A.; Nourse, J.G.; Hounshell, W.D.; Gushurst, A.K.I.; Grier, D.L.; Leland, B.A.; Laufer, J. Description of several chemical structure file formats used by computer programs developed at Molecular Design Limited. J. Chem. Inf. Model. 1992, 32, 244–255. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef] [Green Version]
- Blue Book—IUPAC. International Union of Pure and Applied Chemistry. Available online: https://iupac.org/what-we-do/books/bluebook/ (accessed on 13 December 2020).
- Empowering Innovation & Scientific Discoveries. CAS. Available online: https://www.cas.org (accessed on 13 December 2020).
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2019 update: Improved access to chemical data. Nucleic Acids Res. 2018, 47, D1102–D1109. [Google Scholar] [CrossRef] [Green Version]
- Pupier, M.; Nuzillard, J.-M.; Wist, J.; Schlörer, N.E.; Kuhn, S.; Erdelyi, M.; Steinbeck, C.; Williams, A.J.; Butts, C.; Claridge, T.D.W.; et al. NMReDATA, a standard to report the NMR assignment and parameters of organic compounds. Magn. Reson. Chem. 2018, 56, 703–715. [Google Scholar] [CrossRef] [Green Version]
- IUPAC CPEP Subcommittee on Electronic Data Standards. Available online: http://www.jcamp-dx.org/protocols.html (accessed on 13 December 2020).
- Solution. Allotrope Foundation. Available online: https://www.allotrope.org/solution (accessed on 13 December 2020).
- Wolfender, J.-L.; Nuzillard, J.-M.; van der Hooft, J.J.J.; Renault, J.-H.; Bertrand, S. Accelerating metabolite identification in natural product research: Toward an ideal combination of liquid chromatography–high-resolution tandem mass spectrometry and NMR profiling, in silico databases, and chemometrics. Anal. Chem. 2019, 91, 704–742. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.X.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef]
- Ulrich, E.L.; Akutsu, H.; Doreleijers, J.F.; Harano, Y.; Ioannidis, Y.E.; Lin, J.; Livny, M.; Mading, S.; Maziuk, D.; Miller, Z.; et al. BioMagResBank. Nucleic Acids Res. 2008, 36, D402–D408. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with global natural products social molecular networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [Green Version]
- Kuhn, S.; Colreavy-Donnelly, S.; Santana de Souza, J.; Borges, R.M. An integrated approach for mixture analysis using MS and NMR techniques. Faraday Discuss. 2019, 218, 339–353. [Google Scholar] [CrossRef]
- Hubert, J.; Nuzillard, J.-M.; Purson, S.; Hamzaoui, M.; Borie, N.; Reynaud, R.; Renault, J.-H. Identification of natural metabolites in mixture: A pattern recognition strategy based on 13C NMR. Anal. Chem. 2014, 86, 2955–2962. [Google Scholar] [CrossRef]
- Bruguière, A.; Derbré, S.; Coste, C.; Le Bot, M.; Siegler, B.; Leong, S.T.; Sulaiman, S.N.; Awang, K.; Richomme, P. 13C-NMR dereplication of Garcinia extracts: Predicted chemical shifts as reliable databases. Fitoterapia 2018, 131, 59–64. [Google Scholar] [CrossRef]
- Taxonomy Browser (Root). Available online: https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi (accessed on 13 December 2020).
- Tree of Life Web Project. Available online: http://tolweb.org/tree/ (accessed on 13 December 2020).
- Sorokina, M.; Steinbeck, C. Review on natural products databases: Where to find data in 2020. J. Cheminform. 2020, 12, 20. [Google Scholar] [CrossRef] [Green Version]
- Johnson, S.R.; Lange, B.M. Open-access metabolomics databases for natural product research: Present capabilities and future potential. Front. Bioeng. Biotechnol. 2015, 3, 22. [Google Scholar] [CrossRef] [Green Version]
- Fromanteau, D.L.G.; Gastmans, J.P.; Vestri, S.A.; Emerenciano, V.d.P.; Borges, J.H.G. A constraints generator in structural determination by microcomputer. Comput. Chem. 1993, 17, 369–378. [Google Scholar] [CrossRef]
- Scotti, M.T.; Herrera-Acevedo, C.; Oliveira, T.B.; Costa, R.P.O.; Santos, S.Y.K.O.; Rodrigues, R.P.; Scotti, L.; Da-Costa, F.B. SistematX, an online web-based cheminformatics tool for data management of secondary metabolites. Molecules 2018, 23, 103. [Google Scholar] [CrossRef] [Green Version]
- Afendi, F.M.; Okada, T.; Yamazaki, M.; Hirai-Morita, A.; Nakamura, Y.; Nakamura, K.; Ikeda, S.; Takahashi, H.; Altaf-Ul-Amin, M.; Darusman, L.K.; et al. KNApSAcK family databases: Integrated metabolite-plant species databases for multifaceted plant research. Plant Cell Physiol. 2012, 53, e1. [Google Scholar] [CrossRef] [Green Version]
- RDKit. Available online: https://www.rdkit.org/ (accessed on 13 December 2020).
- ISIDA/EdiSDF—Laboratoire de Chemoinformatique. Available online: http://infochim.u-strasbg.fr/spip.php?article83 (accessed on 13 December 2020).
- nuzillard/KnapsackSearck: Automated Data Search in the KNApSAcK Database. Available online: https://github.com/nuzillard/KnapsackSearch (accessed on 13 December 2020).
- KNApSAcK Core System. Available online: http://www.knapsackfamily.com/knapsack_core/top.php (accessed on 13 December 2020).
- Allard, P.-M.; Péresse, T.; Bisson, J.; Gindro, K.; Marcourt, L.; Pham, V.C.; Roussi, F.; Litaudon, M.; Wolfender, J.-L. Integration of molecular networking and in silico MS/MS fragmentation for natural products dereplication. Anal. Chem. 2016, 88, 3317–3323. [Google Scholar] [CrossRef]
- Daylight Theory: SMIRKS—A Reaction Transform Language. Available online: https://www.daylight.com/dayhtml/doc/theory/theory.smirks.html (accessed on 13 December 2020).
- Bento, A.P.; Hersey, A.; Félix, E.; Landrum, G.; Gaulton, A.; Atkinson, F.; Bellis, L.J.; De Veij, M.; Leach, A.R. An open source chemical structure curation pipeline using RDKit. J. Cheminform. 2020, 12, 51. [Google Scholar] [CrossRef]
- CIRpy—CIRpy 1.0.2 Documentation. Available online: https://cirpy.readthedocs.io/en/latest/ (accessed on 13 December 2020).
- Steinbeck, C.; Kuhn, S. NMRShiftDB—Compound identification and structure elucidation support through a free community-built web database. Phytochemistry 2004, 65, 2711–2717. [Google Scholar] [CrossRef]
- Vestri Alvarenga, S.A.; Gastmans, J.-P.; do Vale Rodrigues, G.; Moreno, P.R.H.; Emerenciano, V.d.P. A computer-assisted approach for chemotaxonomic studies—Diterpenes in Lamiaceae. Phytochemistry 2001, 56, 583–595. [Google Scholar] [CrossRef]
- Pelletier, S.W. The nature and definition of an alkaloid. In Alkaloids: Chemical and Biological Perspectives; Pelletier, S.W., Ed.; Wiley: New York, NY, USA, 1983; Volume 1, pp. 1–31. [Google Scholar]
- Djoumbou Feunang, Y.; Eisner, R.; Knox, C.; Chepelev, L.; Hastings, J.; Owen, G.; Fahy, E.; Steinbeck, C.; Subramanian, S.; Bolton, E.; et al. ClassyFire: Automated chemical classification with a comprehensive, computable taxonomy. J. Cheminform. 2016, 8, 61. [Google Scholar] [CrossRef] [Green Version]
- Daylight Theory: SMARTS—A Language for Describing Molecular Patterns. Available online: https://www.daylight.com/dayhtml/doc/theory/theory.smarts.html (accessed on 13 December 2020).
- Schaub, J.; Zielesny, A.; Steinbeck, C.; Sokorina, M. Too sweet: Cheminformatics for deglycosylation in natural products. J. Cheminform. 2020, 12, 67. [Google Scholar] [CrossRef]
- Bruneton, J. Pharmacognosie, 5th ed.; Lavoisier collection Tec&Doc: Cachan, France, 2016. [Google Scholar]
- Nuzillard, J.-M.; Emerenciano, V.d.P. Automatic structure elucidation through data base search and 2D NMR spectral analysis. Nat. Prod. Commun. 2006, 1, 57–64. [Google Scholar]
- Kim, H.W.; Wang, M.; Leber, C.A.; Nothias, L.-F.; Reher, R.; Kang, K.B.; van der Hooft, J.J.J.; Dorrestein, P.C.; Gerwick, W.H.; Cottrell, G.W. NPClassifier: A deep neural network-based structural classification tool for natural products. ChemRxiv. Prepr. 2020. [Google Scholar] [CrossRef]
- Predicted Carbon-13 NMR Data of Natural Products (PNMRNP). Zenodo. Available online: https://zenodo.org/record/3825257 (accessed on 13 December 2020).
- Sorokina, M.; Steinbeck, C. NaPLeS: A natural products likeness scorer—Web application and database. J. Cheminform. 2019, 11, 55. [Google Scholar] [CrossRef]
- Robien, W. Computer-assisted peer reviewing of spectral data: The CSEARCH protocol. Monatsh. Chem. 2019, 150, 927–932. [Google Scholar] [CrossRef] [Green Version]
- Boit, H.-G.; Döpke, W. Alkaloide aus Urceolina-, Hymenocallis-, Elisena-, Calostemma-, Eustephia- und Hippeastrum-Arten. Chem. Ber. 1957, 90, 1827–1830. [Google Scholar] [CrossRef]
- Giraud, L. Kallawaya: Guérisseurs Itinérants des Andes. Recherche sur les Pratiques Médicinales et Magiques; IRD Editions: Montpellier, France, 1984. [Google Scholar]
- Berkov, S.; Osorio, E.; Viladomat, F.; Bastida, J. Chapter Two—Chemodiversity, chemotaxonomy and chemoecology of Amaryllidaceae alkaloids. In The Alkaloids: Chemistry and Biology; Knölker, H.-J., Ed.; Academic Press: Cambridge, MA, USA, 2020; Volume 83, pp. 113–185. [Google Scholar] [CrossRef]
- Renault, J.-H.; Nuzillard, J.-M.; Maciuk, A.; Zèches-Hanrot, M. Use of Centrifugal Partition Chromatography for Purifying. Galanthamine. Patent WO 2006/064105, 22 June 2006. [Google Scholar]
- Renault, J.-H.; Le Crouerour, G.; Thépenier, P.; Nuzillard, J.-M.; Zèches-Hanrot, M.; Le Men-Olivier, L. Isolation of indole alkaloids from Catharanthus roseus by centrifugal partition chromatography in the pH-zone refining mode. J. Chromatogr. A 1999, 849, 421–431. [Google Scholar] [CrossRef]
- Martin, S.F. Chapter 3—The Amaryllidaceae alkaloids. In The Alkaloids: Chemistry and Pharmacology; Brossi, A., Ed.; Academic Press: Cambridge, MA, USA, 1987; Volume 30, pp. 251–376. [Google Scholar] [CrossRef]
- Haughwitz, R.D.; Jeffs, P.W.; Wenkert, E. 358. Proton magnetic resonance studies of some Amaryllidaceae alkaloids of the 5,10b-ethanophenanthridine series and of criwelline and tazettine. J. Chem. Soc. 1965, 2001–2009. [Google Scholar] [CrossRef]
- Das, M.K.; Kumar, N.; Bisai, A. Catalytic asymmetric total syntheses of naturally occurring Amarylidaceae alkaloids, (−)-crinine, (−)-epi-crinine, (−)-oxocrinine, (+)-epi-elwesine, (+)-vittatine, and (+)-epi-vittatine. Org. Lett. 2018, 20, 4421–4424. [Google Scholar] [CrossRef]




Sample Availability: Samples of compounds are not available from the authors. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lianza, M.; Leroy, R.; Machado Rodrigues, C.; Borie, N.; Sayagh, C.; Remy, S.; Kuhn, S.; Renault, J.-H.; Nuzillard, J.-M. The Three Pillars of Natural Product Dereplication. Alkaloids from the Bulbs of Urceolina peruviana (C. Presl) J.F. Macbr. as a Preliminary Test Case. Molecules 2021, 26, 637. https://doi.org/10.3390/molecules26030637
Lianza M, Leroy R, Machado Rodrigues C, Borie N, Sayagh C, Remy S, Kuhn S, Renault J-H, Nuzillard J-M. The Three Pillars of Natural Product Dereplication. Alkaloids from the Bulbs of Urceolina peruviana (C. Presl) J.F. Macbr. as a Preliminary Test Case. Molecules. 2021; 26(3):637. https://doi.org/10.3390/molecules26030637
Chicago/Turabian StyleLianza, Mariacaterina, Ritchy Leroy, Carine Machado Rodrigues, Nicolas Borie, Charlotte Sayagh, Simon Remy, Stefan Kuhn, Jean-Hugues Renault, and Jean-Marc Nuzillard. 2021. "The Three Pillars of Natural Product Dereplication. Alkaloids from the Bulbs of Urceolina peruviana (C. Presl) J.F. Macbr. as a Preliminary Test Case" Molecules 26, no. 3: 637. https://doi.org/10.3390/molecules26030637
APA StyleLianza, M., Leroy, R., Machado Rodrigues, C., Borie, N., Sayagh, C., Remy, S., Kuhn, S., Renault, J.-H., & Nuzillard, J.-M. (2021). The Three Pillars of Natural Product Dereplication. Alkaloids from the Bulbs of Urceolina peruviana (C. Presl) J.F. Macbr. as a Preliminary Test Case. Molecules, 26(3), 637. https://doi.org/10.3390/molecules26030637