Effect of Concentrated Salts Solutions on the Stability of Immobilized Enzymes: Influence of Inactivation Conditions and Immobilization Protocol
Abstract
:1. Introduction
2. Results and Discussion
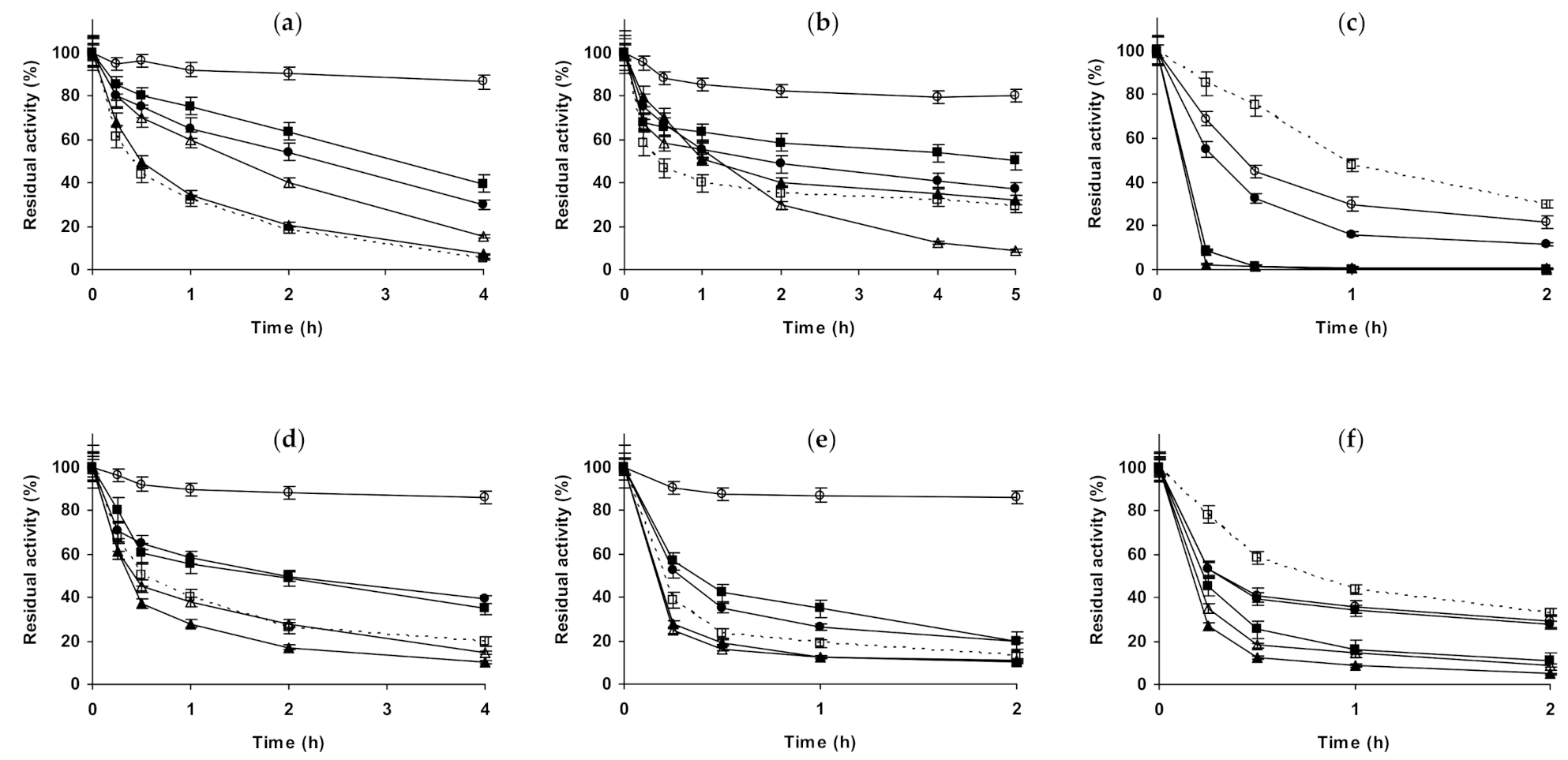
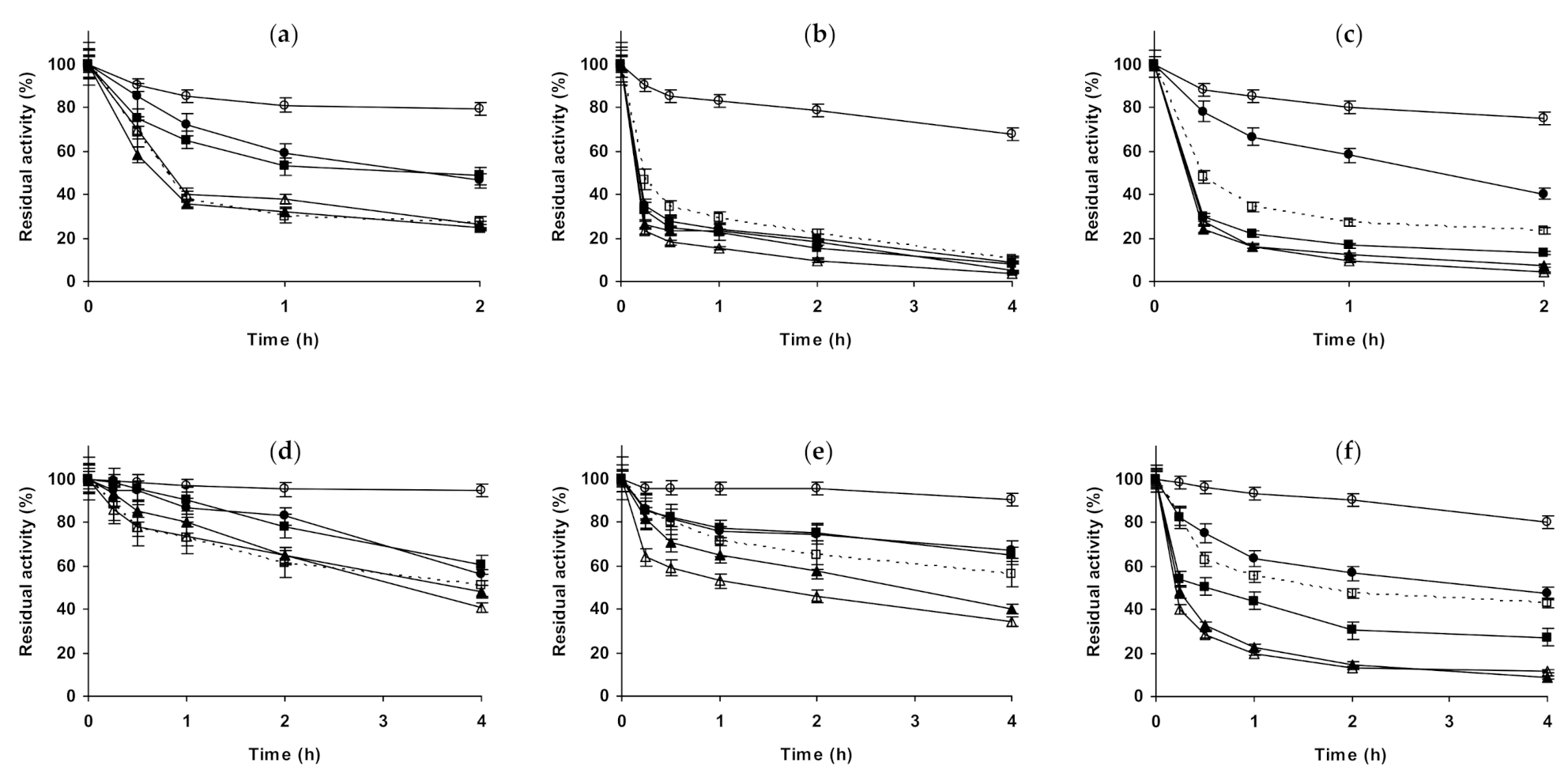
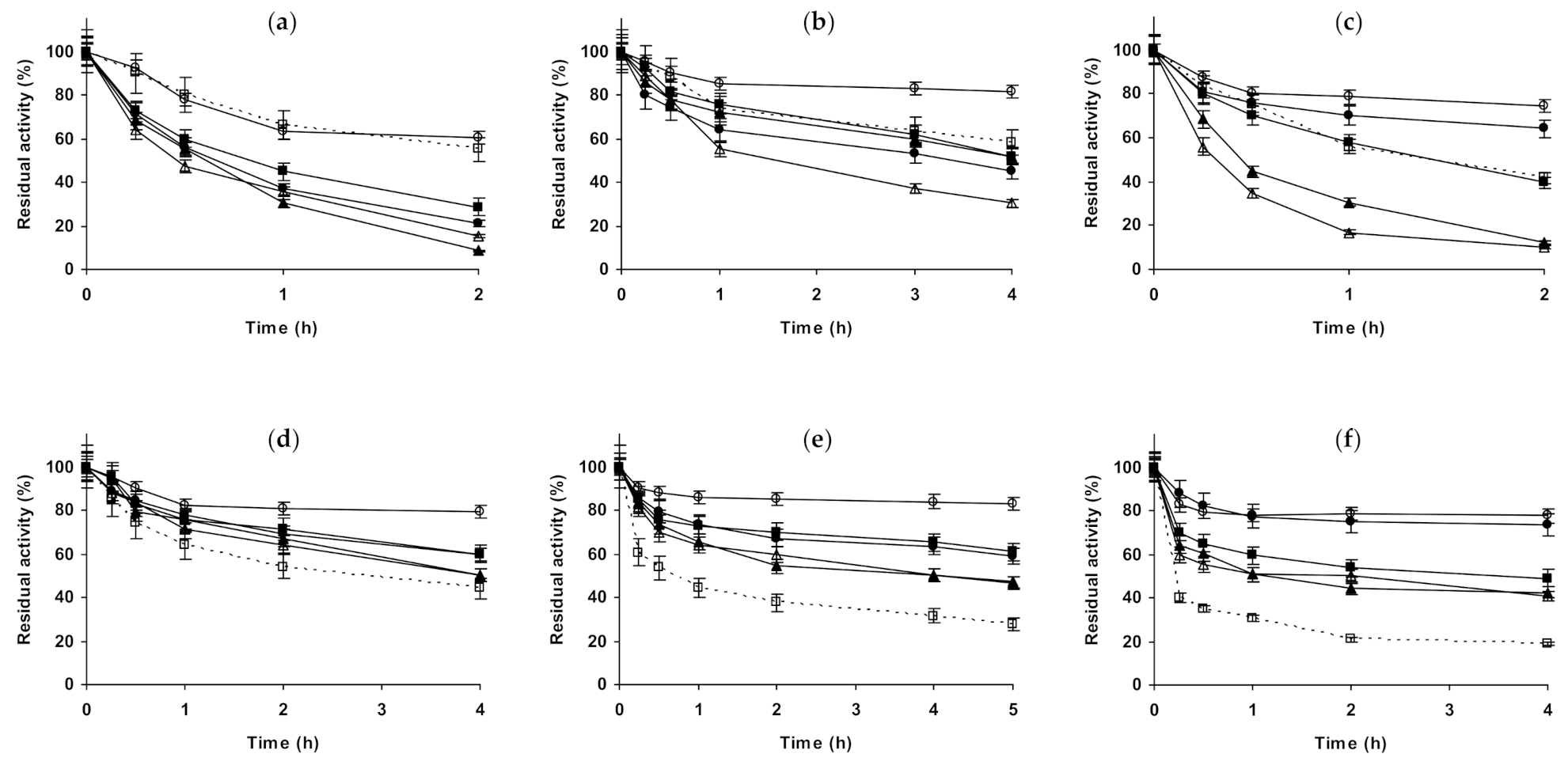
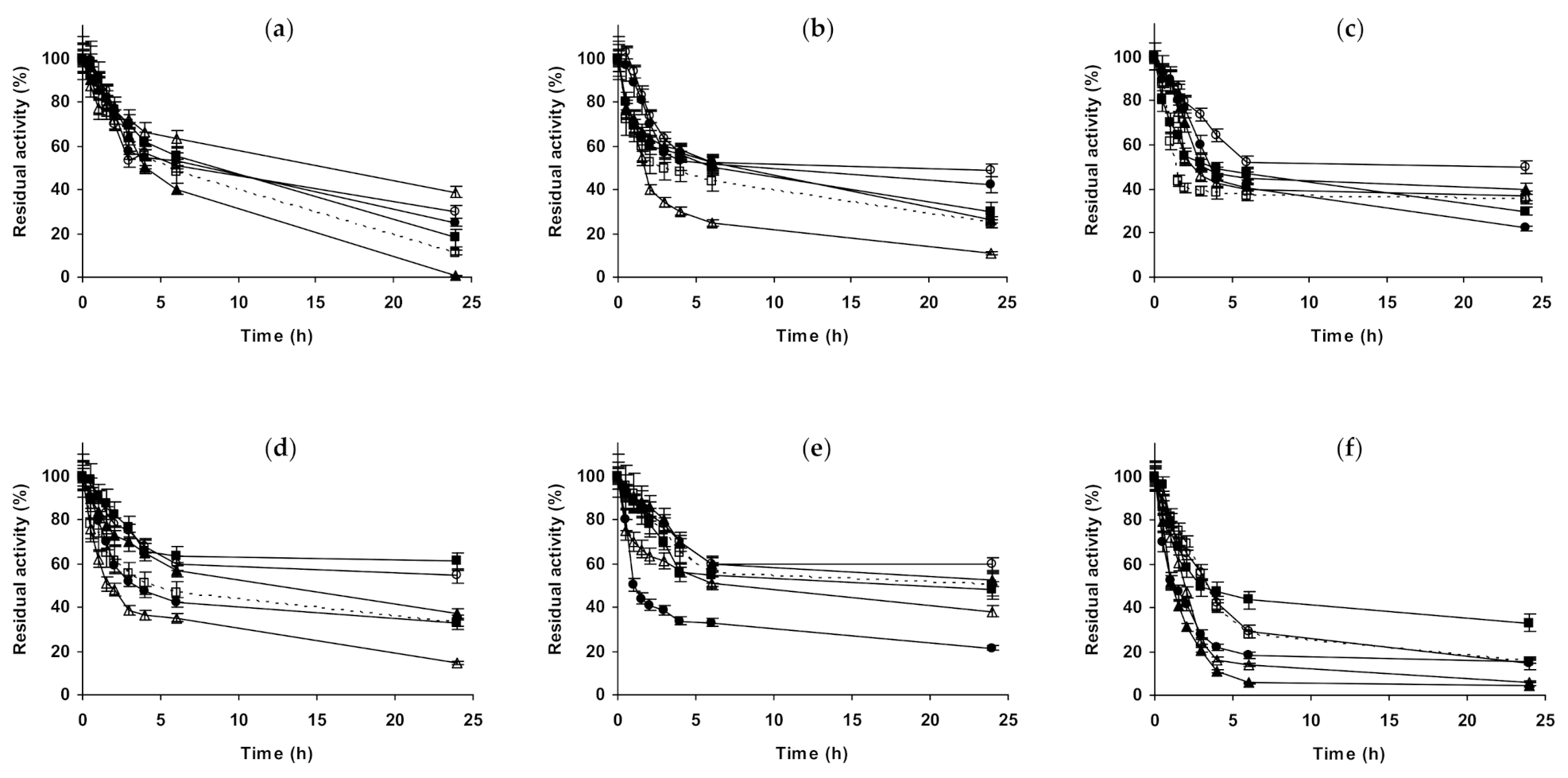
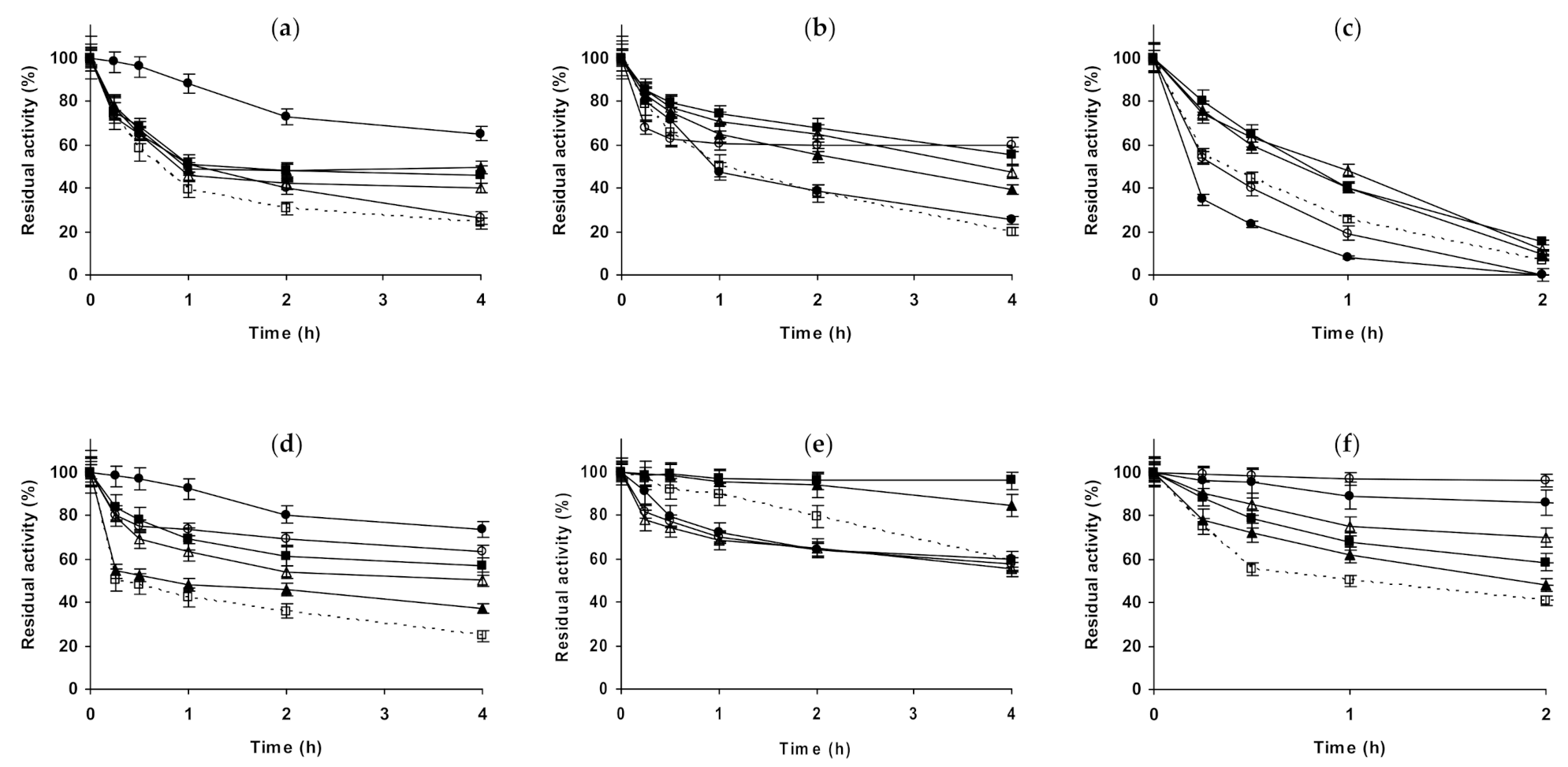
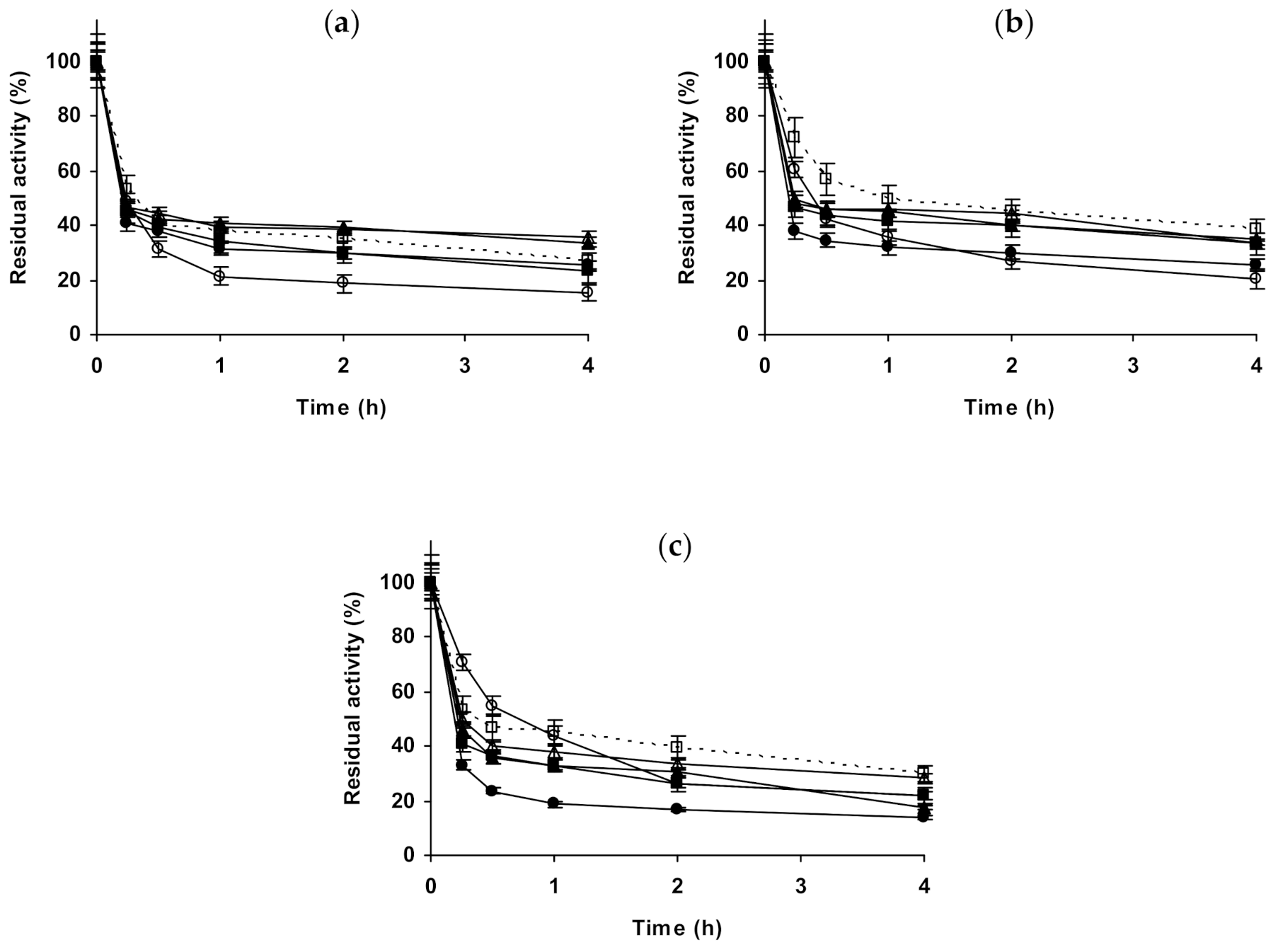
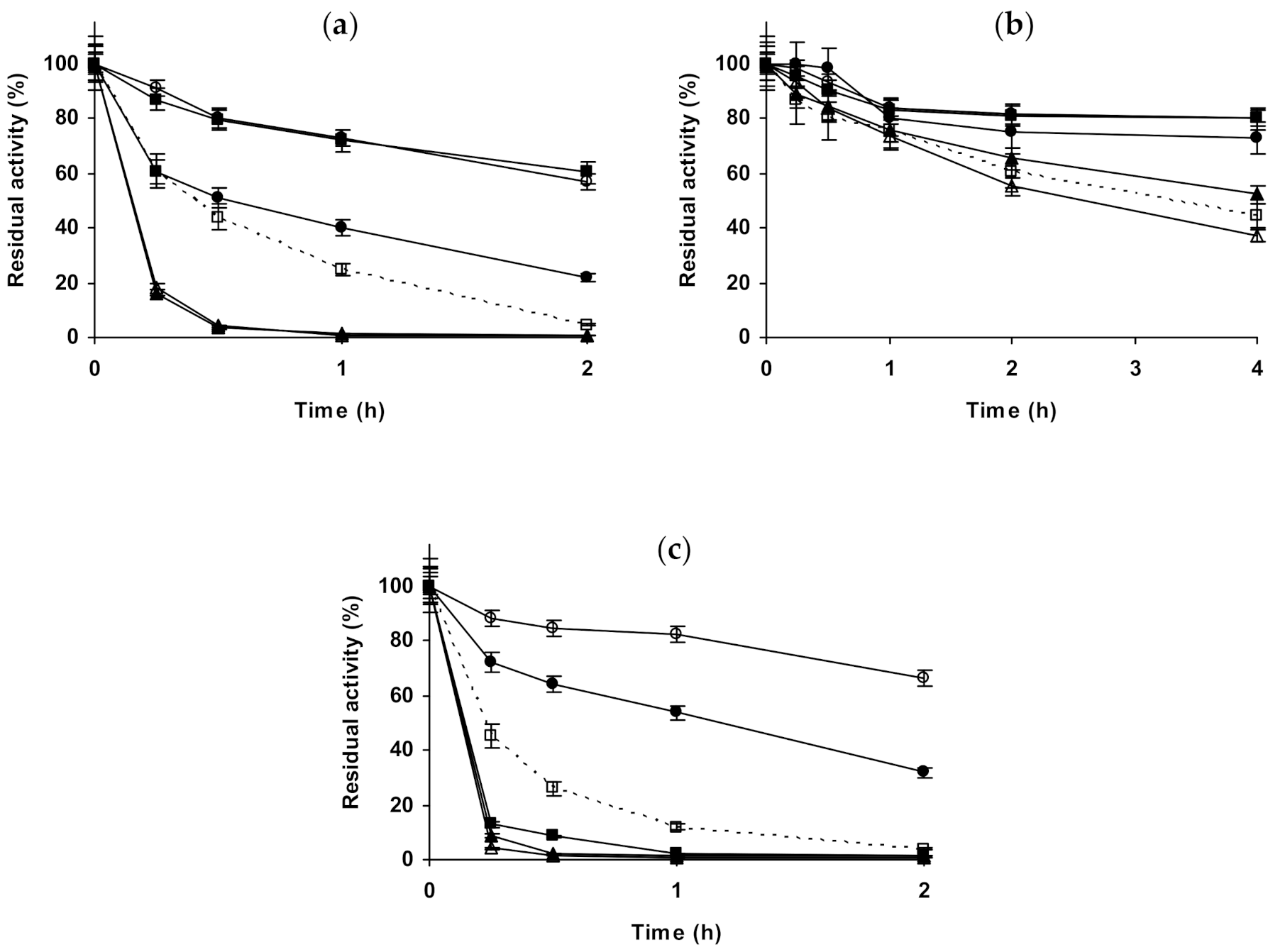
2.1. Effect of Different Salts on the Stability of Immobilized Lipases
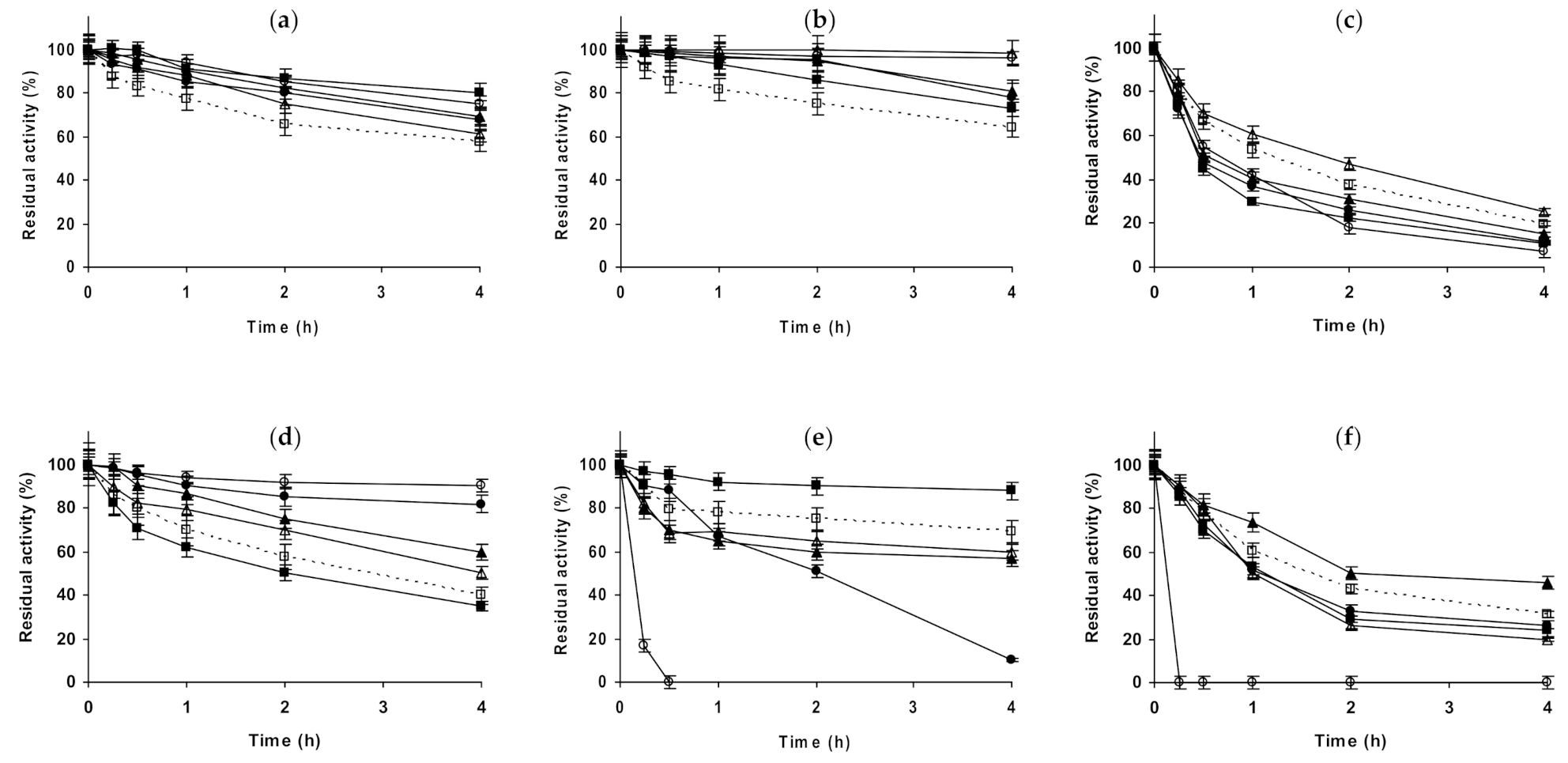
2.2. Effect of Different Salts on the Stability of Immobilized Proteases
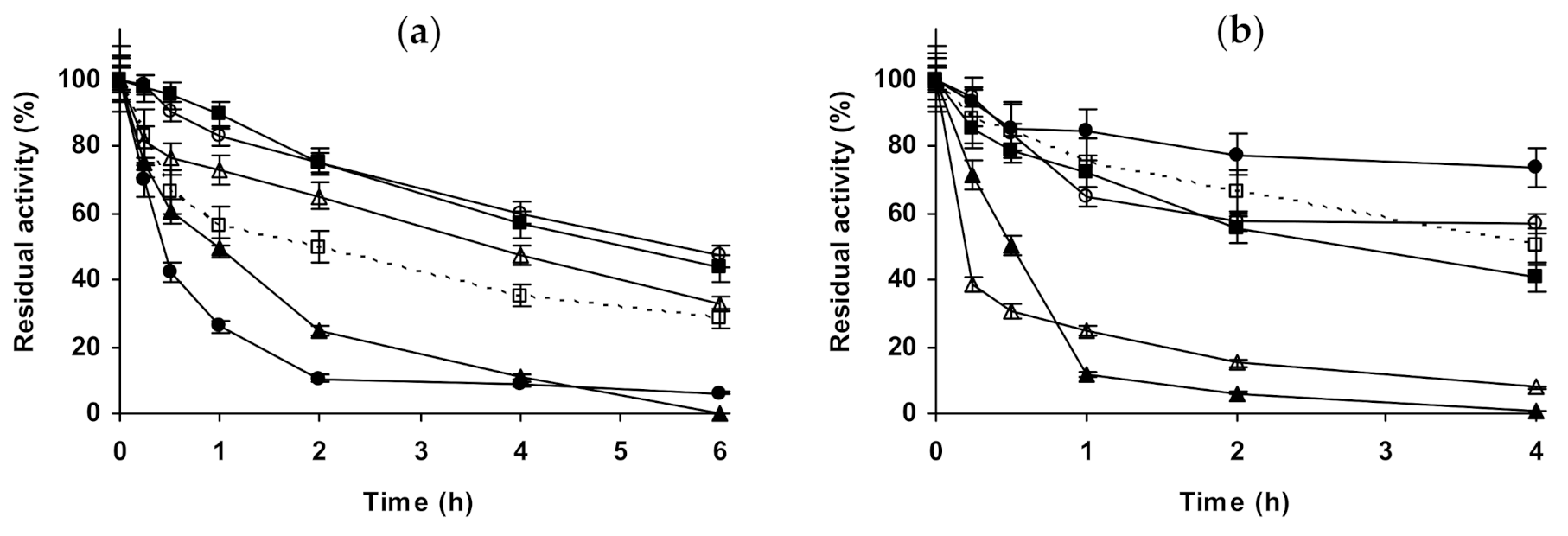
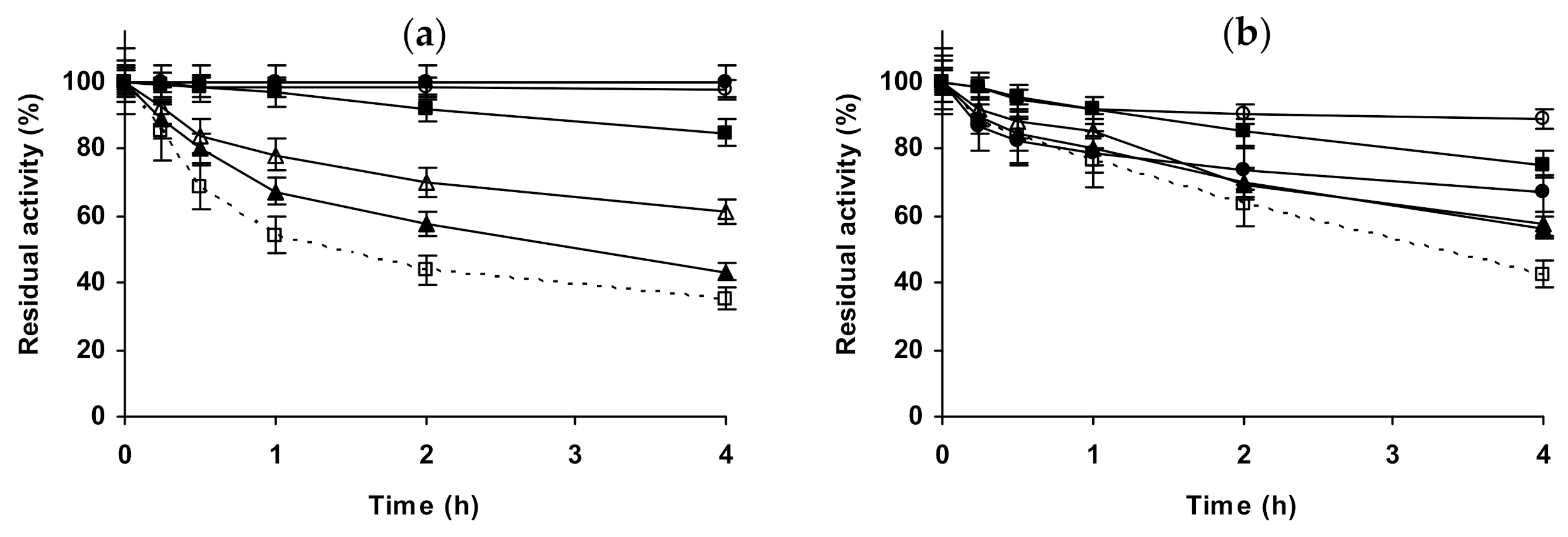
2.3. Effect of Different Salts on the Stability of Some Additional Immobilized Monomeric Enzymes
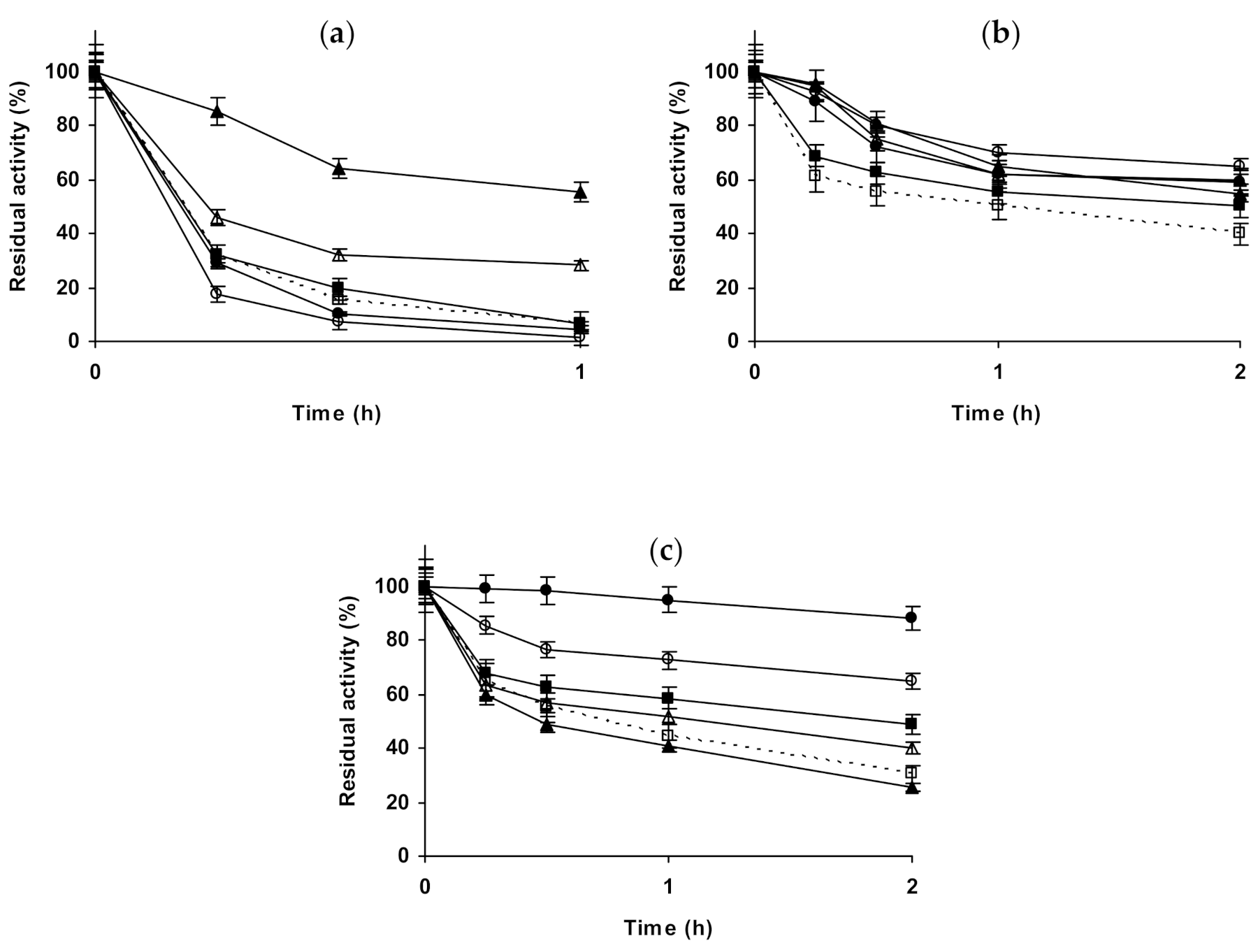
2.4. Effect of Different Salts on the Stability of Some Glutaraldehyde-Amino Agarose Immobilized Multimeric Enzymes
3. Materials and Methods
3.1. Materials
3.2. Methods
3.2.1. Enzyme Activity Assay
3.2.2. Enzymes Immobilizations
Immobilization of the Lipases on Octyl Agarose Beads
Immobilization of Enzymes on Glutaraldehyde-Amino Agarose Beads
Immobilization of the Enzyme on Glyoxyl Agarose Beads
3.2.3. Stress Inactivation of Different Enzyme Preparations in the Presence of Different Salts on Biocatalyst Stability
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Huang, X.; Cao, M.; Zhao, H. Integrating biocatalysis with chemocatalysis for selective transformations. Curr. Opin. Chem. Biol. 2020, 55, 161–170. [Google Scholar]
- Fryszkowska, A.; Devine, P.N. Biocatalysis in drug discovery and development. Curr. Opin. Chem. Biol. 2020, 55, 151–160. [Google Scholar]
- Woodley, J.M. New frontiers in biocatalysis for sustainable synthesis. Curr. Opin. Green Sustain. Chem. 2020, 21, 22–26. [Google Scholar]
- Domínguez de María, P.; de Gonzalo, G.; Alcántara, A.R. Biocatalysis as useful tool in asymmetric synthesis: An assessment of recently granted patents (2014–2019). Catalysts 2019, 9, 802. [Google Scholar]
- Woodley, J.M. Accelerating the implementation of biocatalysis in industry. Appl. Microbiol. Biotechnol. 2019, 103, 4733–4739. [Google Scholar]
- Sheldon, R.A.; Woodley, J.M. Role of biocatalysis in sustainable chemistry. Chem. Rev. 2018, 118, 801–838. [Google Scholar]
- Almeida, J.M.; Alnoch, R.C.; Souza, E.M.; Mitchell, D.A.; Krieger, N. Metagenomics: Is it a powerful tool to obtain lipases for application in biocatalysis? Biochim. Biophys. Acta Proteins Proteom. 2020, 1868, 140320. [Google Scholar]
- Ferrer, M.; Beloqui, A.; Timmis, K.; Golyshin, P. Metagenomics for mining new genetic resources of microbial communities. J. Mol. Microbiol. Biotechnol. 2009, 16, 109–123. [Google Scholar]
- Fernández-Arrojo, L.; Guazzaroni, M.-E.; López-Cortés, N.; Beloqui, A.; Ferrer, M. Metagenomic era for biocatalyst identification. Curr. Opin. Biotechnol. 2010, 21, 725–733. [Google Scholar]
- Ferrer, M.; Martínez-Martínez, M.; Bargiela, R.; Streit, W.R.; Golyshina, O.V.; Golyshin, P.N. Estimating the success of enzyme bioprospecting through metagenomics: Current status and future trends. Microb. Biotechnol. 2016, 9, 22–34. [Google Scholar]
- Manning, M.C.; Chou, D.K.; Murphy, B.M.; Payne, R.W.; Katayama, D.S. Stability of protein pharmaceuticals: An update. Pharm. Res. 2010, 27, 544–575. [Google Scholar]
- Shortle, D.; Stites, W.E.; Meeker, A.K. Contributions of the large hydrophobic amino acids to the stability of staphylococcal nuclease. Biochemistry 1990, 29, 8033–8041. [Google Scholar]
- Cherry, J.R.; Fidantsef, A.L. Directed evolution of industrial enzymes: An update. Curr. Opin. Biotechnol. 2003, 14, 438–443. [Google Scholar]
- Eijsink, V.G.H.; GÅseidnes, S.; Borchert, T.V.; Van Den Burg, B. Directed evolution of enzyme stability. Biomol. Eng. 2005, 22, 21–30. [Google Scholar]
- Packer, M.S.; Liu, D.R. Methods for the directed evolution of proteins. Nat. Rev. Genet. 2015, 16, 379–394. [Google Scholar]
- Renata, H.; Wang, Z.J.; Arnold, F.H. Expanding the enzyme universe: Accessing non-natural reactions by mechanism-guided directed evolution. Angew. Chemie Int. Ed. 2015, 54, 3351–3367. [Google Scholar]
- Spicer, C.D.; Davis, B.G. Selective chemical protein modification. Nat. Commun. 2014, 5, 4740. [Google Scholar]
- Boutureira, O.; Bernardes, G.J.L. Advances in chemical protein modification. Chem. Rev. 2015, 115, 2174–2195. [Google Scholar]
- Chalker, J.M.; Bernardes, G.J.L.; Lin, Y.A.; Davis, B.G. Chemical modification of proteins at cysteine: Opportunities in chemistry and biology. Chem. Asian J. 2009, 4, 630–640. [Google Scholar]
- Mateo, C.; Palomo, J.M.; Fernandez-Lorente, G.; Guisan, J.M.; Fernandez-Lafuente, R. Improvement of enzyme activity, stability and selectivity via immobilization techniques. Enzyme Microb. Technol. 2007, 40, 1451–1463. [Google Scholar]
- Garcia-Galan, C.; Berenguer-Murcia, Á.; Fernandez-Lafuente, R.; Rodrigues, R.C. Potential of different enzyme immobilization strategies to improve enzyme performance. Adv. Synth. Catal. 2011, 353, 2885–2904. [Google Scholar]
- Barbosa, O.; Ortiz, C.; Berenguer-Murcia, Á.; Torres, R.; Rodrigues, R.C.; Fernandez-Lafuente, R. Strategies for the one-step immobilization-purification of enzymes as industrial biocatalysts. Biotechnol. Adv. 2015, 33, 435–456. [Google Scholar]
- Tamborini, L.; Fernandes, P.; Paradisi, F.; Molinari, F. Flow bioreactors as complementary tools for biocatalytic process intensification. Trends Biotechnol. 2018, 36, 73–88. [Google Scholar]
- Hu, Y.; Wang, X.C.; Ngo, H.H.; Sun, Q.; Yang, Y. Anaerobic dynamic membrane bioreactor (AnDMBR) for wastewater treatment: A review. Bioresour. Technol. 2018, 247, 1107–1118. [Google Scholar]
- Asif, M.B.; Hai, F.I.; Kang, J.; van de Merwe, J.P.; Leusch, F.D.L.; Price, W.E.; Nghiem, L.D. Biocatalytic degradation of pharmaceuticals, personal care products, industrial chemicals, steroid hormones and pesticides in a membrane distillation-enzymatic bioreactor. Bioresour. Technol. 2018, 247, 528–536. [Google Scholar]
- García-Pérez, T.; López, J.C.; Passos, F.; Lebrero, R.; Revah, S.; Muñoz, R. Simultaneous methane abatement and PHB production by Methylocystis hirsuta in a novel gas-recycling bubble column bioreactor. Chem. Eng. J. 2018, 334, 691–697. [Google Scholar]
- Hirata, D.B.; Albuquerque, T.L.; Rueda, N.; Virgen-Ortíz, J.J.; Tacias-Pascacio, V.G.; Fernandez-Lafuente, R. Evaluation of different immobilized lipases in transesterification reactions using tributyrin: Advantages of the heterofunctional octyl agarose beads. J. Mol. Catal. B Enzym. 2016, 133, 117–123. [Google Scholar]
- Virgen-Ortíz, J.J.; dos Santos, J.C.S.; Berenguer-Murcia, Á.; Barbosa, O.; Rodrigues, R.C.; Fernandez-Lafuente, R. Polyethylenimine: A very useful ionic polymer in the design of immobilized enzyme biocatalysts. J. Mater. Chem. B 2017, 5, 7461–7490. [Google Scholar]
- Rodrigues, R.C.; Barbosa, O.; Ortiz, C.; Berenguer-Murcia, Á.; Torres, R.; Fernandez-Lafuente, R. Amination of enzymes to improve biocatalyst performance: Coupling genetic modification and physicochemical tools. RSC Adv. 2014, 4, 38350–38374. [Google Scholar]
- Santiago, G.; Martínez-Martínez, M.; Alonso, S.; Bargiela, R.; Coscolín, C.; Golyshin, P.N.; Guallar, V.; Ferrer, M. Rational engineering of multiple active sites in an ester hydrolase. Biochemistry 2018, 57, 2245–2255. [Google Scholar]
- Alonso, S.; Santiago, G.; Cea-Rama, I.; Fernandez-Lopez, L.; Coscolín, C.; Modregger, J.; Ressmann, A.K.; Martínez-Martínez, M.; Marrero, H.; Bargiela, R.; et al. Genetically engineered proteins with two active sites for enhanced biocatalysis and synergistic chemo- and biocatalysis. Nat. Catal. 2020, 3, 319–328. [Google Scholar]
- Carpenter, J.F.; Arakawa, T.; Crowe, J.H. Interaction of stabilizing additives with proteins during freeze-thawing and freeze-drying. Dev. Biol. Stand. 1992, 74, 225–238. [Google Scholar]
- Han, Y.; Jin, B.-S.; Lee, S.-B.; Sohn, Y.; Joung, J.-W.; Lee, J.-H. Effects of sugar additives on protein stability of recombinant human serum albumin during lyophilization and storage. Arch. Pharm. Res. 2007, 30, 1124. [Google Scholar]
- Leibly, D.J.; Nguyen, T.N.; Kao, L.T.; Hewitt, S.N.; Barrett, L.K.; van Voorhis, W.C. Stabilizing additives added during cell lysis aid in the solubilization of recombinant proteins. PLoS ONE 2012, 7, e52482. [Google Scholar]
- Brennan, J.D.; Benjamin, D.; DiBattista, E.; Gulcev, M.D. Using sugar and amino acid additives to stabilize enzymes within sol−gel derived silica. Chem. Mater. 2003, 15, 737–745. [Google Scholar]
- Gray, C.J. Additives and enzyme stability. Biocatalysis 1988, 1, 187–196. [Google Scholar]
- Chang, Y.K.; Chase, H.A. Ion exchange purification of G6PDH from unclarified yeast cell homogenates using expanded bed adsorption. Biotechnol. Bioeng. 1996, 49, 204–216. [Google Scholar]
- Rao, C.S. Purification of large proteins using ion-exchange membranes. Process Biochem. 2001, 37, 247–256. [Google Scholar]
- Johansson, H.; Jägersten, C.; Shiloach, J. Large scale recovery and purification of periplasmic recombinant protein from E. coli using expanded bed adsorption chromatography followed by new ion exchange media. J. Biotechnol. 1996, 48, 9–14. [Google Scholar]
- Perosa, F.; Carbone, R.; Ferrone, S.; Dammacco, F. Purification of human immunoglobulins by sequential precipitation with caprylic acid and ammonium sulphate. J. Immunol. Methods 1990, 128, 9–16. [Google Scholar]
- Rothenberg, M.A.; Nachmansohn, D. Studies on cholinesterase. III. Purification of the enzyme by fractional ammonium sulfate precipitation. J. Biol. Chem. 1947, 168, 223–231. [Google Scholar]
- Coelho, D.F.; Silveira, E.; Pessoa Junior, A.; Tambourgi, E.B. Bromelain purification through unconventional aqueous two-phase system (PEG/ammonium sulphate). Bioprocess Biosyst. Eng. 2013, 36, 185–192. [Google Scholar]
- Pereira, M.M.; Cruz, R.A.P.; Almeida, M.R.; Lima, Á.S.; Coutinho, J.A.P.; Freire, M.G. Single-step purification of ovalbumin from egg white using aqueous biphasic systems. Process Biochem. 2016, 51, 781–791. [Google Scholar]
- Wu, W.C.; Ng, H.S.; Sun, I.M.; Lan, J.C.W. Single step purification of bromelain from Ananas comosus pulp using a polymer/salt aqueous biphasic system. J. Taiwan Inst. Chem. Eng. 2017, 79, 158–162. [Google Scholar]
- Ho, S.L.; Lan, J.C.W.; Tan, J.S.; Yim, H.S.; Ng, H.S. Aqueous biphasic system for the partial purification of Bacillus subtilis carboxymethyl cellulase. Process Biochem. 2017, 58, 276–281. [Google Scholar]
- He, J.Y.; Sun, Z.H.; Ruan, W.Q.; Xu, Y. Biocatalytic synthesis of ethyl (S)-4-chloro-3-hydroxy-butanoate in an aqueous-organic solvent biphasic system using Aureobasidium pullulans CGMCC 1244. Process Biochem. 2006, 41, 244–249. [Google Scholar]
- Del-Val, M.I.; Otero, C. Biphasic aqueous media containing polyethylene glycol for the enzymatic synthesis of oligosaccharides from lactose. Enzyme Microb. Technol. 2003, 33, 118–126. [Google Scholar]
- Hernandez-Justiz, O.; Fernandez-Lafuente, R.; Terreni, M.; Guisan, J.M. Use of aqueous two-phase systems for in situ extraction of water soluble antibiotics during their synthesis by enzymes immobilized on porous supports. Biotechnol. Bioeng. 1998, 59, 73–79. [Google Scholar]
- Andersson, E.; Hahn-Hägerdal, B. Enzyme action in polymer and salt solutions. I. Stability of penicillin acylase in poly(ethylene glycol) and potassium phosphate solutions in relation to water activity. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 1987, 912, 317–324. [Google Scholar]
- Andersson, E.; Hahn-Hägerdal, B. Enzyme action in polymer and salt solutions. II. Activity of penicillin acylase in poly(ethylene glycol) and potassium phosphate solutions in relation to water activity. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 1987, 912, 325–328. [Google Scholar]
- Andersson, E.; Hahn-Hägerdal, B. Bioconversions in aqueous two-phase systems. Enzyme Microb. Technol. 1990, 12, 242–254. [Google Scholar]
- Andersson, E.; Mattiasson, B.; Hahn-Hägerdal, B. Enzymatic conversion in aqueous two-phase systems: Deacylation of benzylpenicillin to 6-aminopenicillanic acid with penicillin acylase. Enzyme Microb. Technol. 1984, 6, 301–306. [Google Scholar]
- Pereira, J.F.B.; Freire, M.G.; Coutinho, J.A.P. Aqueous two-phase systems: Towards novel and more disruptive applications. Fluid Phase Equilib. 2020, 505, 112341. [Google Scholar]
- Vobecká, L.; Romanov, A.; Slouka, Z.; Hasal, P.; Přibyl, M. Optimization of aqueous two-phase systems for the production of 6-aminopenicillanic acid in integrated microfluidic reactors-separators. New Biotechnol. 2018, 47, 73–79. [Google Scholar]
- Wang, K.; Lu, Y.; Liang, W.Q.; Wang, S.D.; Jiang, Y.; Huang, R.; Liu, Y.H. Enzymatic synthesis of galacto-oligosaccharides in an organic–aqueous biphasic system by a novel β-galactosidase from a metagenomic library. J. Agric. Food Chem. 2012, 60, 3940–3946. [Google Scholar]
- Shahid, S.; Ahmad, F.; Hassan, M.I.; Islam, A. Mixture of macromolecular crowding agents has a non-additive effect on the stability of proteins. Appl. Biochem. Biotechnol. 2019, 188, 927–941. [Google Scholar]
- Dufrechou, M.; Poncet-Legrand, C.; Sauvage, F.-X.; Vernhet, A. Stability of white wine proteins: Combined effect of pH, ionic strength, and temperature on their aggregation. J. Agric. Food Chem. 2012, 60, 1308–1319. [Google Scholar]
- Porter, W.R.; Staack, H.; Brandt, K.; Manning, M.C. Thermal stability of low molecular weight urokinase during heat treatment. I. Effects of protein concentration, pH and ionic strength. Thromb. Res. 1993, 71, 265–279. [Google Scholar]
- Stigter, D.; Alonso, D.O.; Dill, K.A. Protein stability: Electrostatics and compact denatured states. Proc. Natl. Acad. Sci. USA 1991, 88, 4176–4180. [Google Scholar]
- Ibragimova, G.T.; Wade, R.C. Importance of explicit salt ions for protein stability in molecular dynamics simulation. Biophys. J. 1998, 74, 2906–2911. [Google Scholar]
- Nick Pace, C.; Alston, R.W.; Shaw, K.L. Charge–charge interactions influence the denatured state ensemble and contribute to protein stability. Protein Sci. 2000, 9, 1395–1398. [Google Scholar]
- Yang, A.-S.; Honig, B. Structural origins of pH and ionic strength effects on protein stability: Acid denaturation of sperm whale apomyoglobin. J. Mol. Biol. 1994, 237, 602–614. [Google Scholar]
- Dominy, B.N.; Perl, D.; Schmid, F.X.; Brooks, C.L. The effects of ionic strength on protein stability: The cold shock protein family. J. Mol. Biol. 2002, 319, 541–554. [Google Scholar]
- Zhou, R.; Nashine, V.; Palm, T.; Gandhi, R.; Adams, M. Utilization of zwitterion-based solutions to dissect the relative effects of solution pH and ionic strength on the aggregation behavior and conformational stability of a fusion protein. J. Pharm. Sci. 2014, 103, 3065–3074. [Google Scholar]
- Baldwin, R.L. How Hofmeister ion interactions affect protein stability. Biophys. J. 1996, 71, 2056–2063. [Google Scholar]
- Hjörleifsson, J.G.; Ásgeirsson, B. pH-dependent binding of chloride to a marine alkaline phosphatase affects the catalysis, active site stability, and dimer equilibrium. Biochemistry 2017, 56, 5075–5089. [Google Scholar]
- Flores, M.V.; Ertola, R.J.; Voget, C.E. Effect of monovalent cations on the stability and activity of Kluyveromyces lactis β-galactosidase. LWT Food Sci. Technol. 1996, 29, 503–506. [Google Scholar]
- Jurado, E.; Camacho, F.; Luzón, G.; Vicaria, J.M. Kinetic models of activity for β-galactosidases: Influence of pH, ionic concentration and temperature. Enzyme Microb. Technol. 2004, 34, 33–40. [Google Scholar]
- Ullmann, A.; Monod, J. On the effect of divalent cations and protein concentration upon renaturation of β-galactosidase from E. coli. Biochem. Biophys. Res. Commun. 1969, 35, 35–42. [Google Scholar]
- Fernández-Lafuente, R.; Hernández-Jústiz, O.; Mateo, C.; Terreni, M.; Fernández-Lorente, G.; Moreno, M.A.; Alonso, J.; García-López, J.L.; Guisan, J.M. Biotransformations catalyzed by multimeric enzymes: Stabilization of tetrameric ampicillin acylase permits the optimization of ampicillin synthesis under dissociation conditions. Biomacromolecules 2001, 2, 95–104. [Google Scholar]
- Kaddour, S.; López-Gallego, F.; Sadoun, T.; Fernandez-Lafuente, R.; Guisan, J.M. Preparation of an immobilized-stabilized catalase derivative from Aspergillus niger having its multimeric structure stabilized: The effect of Zn2+ on enzyme stability. J. Mol. Catal. B Enzym. 2008, 55, 142–145. [Google Scholar]
- Zaak, H.; Siar, E.-H.; Kornecki, J.F.; Fernandez-Lopez, L.; Pedrero, S.G.; Virgen-Ortíz, J.J.; Fernandez-Lafuente, R. Effect of immobilization rate and enzyme crowding on enzyme stability under different conditions. The case of lipase from Thermomyces lanuginosus immobilized on octyl agarose beads. Process Biochem. 2017, 56, 117–123. [Google Scholar]
- Zaak, H.; Fernandez-Lopez, L.; Velasco-Lozano, S.; Alcaraz-Fructuoso, M.T.; Sassi, M.; Lopez-Gallego, F.; Fernandez-Lafuente, R. Effect of high salt concentrations on the stability of immobilized lipases: Dramatic deleterious effects of phosphate anions. Process Biochem. 2017, 62, 128–134. [Google Scholar]
- Fernandez-Lopez, L.; Bartolome-Cabrero, R.; Rodriguez, M.D.; Dos Santos, C.S.; Rueda, N.; Fernandez-Lafuente, R. Stabilizing effects of cations on lipases depend on the immobilization protocol. RSC Adv. 2015, 5, 83868–83875. [Google Scholar]
- Cipolatti, E.P.; Pinto, M.C.C.; Robert, J.D.M.; da Silva, T.P.; Beralto, T.D.C.; Santos, J.G.F.; de Castro, R.P.V.; Fernandez-Lafuente, R.; Manoel, E.A.; Pinto, J.C.; et al. Pilot-scale development of core–shell polymer supports for the immobilization of recombinant lipase B from Candida antarctica and their application in the production of ethyl esters from residual fatty acids. J. Appl. Polym. Sci. 2018, 135, 1–13. [Google Scholar]
- Kornecki, J.F.; Carballares, D.; Morellon-Sterling, R.; Siar, E.H.; Kashefi, S.; Chafiaa, M.; Arana-Peña, S.; Rios, N.S.; Gonçalves, L.R.B.; Fernandez-Lafuente, R. Influence of phosphate anions on the stability of immobilized enzymes. Effect of enzyme nature, immobilization protocol and inactivation conditions. Process Biochem. 2020, 95, 288–296. [Google Scholar]
- Manoel, E.A.; dos Santos, J.C.S.; Freire, D.M.G.; Rueda, N.; Fernandez-Lafuente, R. Immobilization of lipases on hydrophobic supports involves the open form of the enzyme. Enzyme Microb. Technol. 2015, 71, 53–57. [Google Scholar]
- Rodrigues, R.C.; Virgen-Ortíz, J.J.; dos Santos, J.C.S.; Berenguer-Murcia, Á.; Alcantara, A.R.; Barbosa, O.; Ortiz, C.; Fernandez-Lafuente, R. Immobilization of lipases on hydrophobic supports: Immobilization mechanism, advantages, problems, and solutions. Biotechnol. Adv. 2019, 37, 746–770. [Google Scholar]
- Brzozowski, A.M.; Derewenda, U.; Derewenda, Z.S.; Dodson, G.G.; Lawson, D.M.; Turkenburg, J.P.; Bjorkling, F.; Huge-Jensen, B.; Patkar, S.A.; Thim, L. A model for interfacial activation in lipases from the structure of a fungal lipase-inhibitor complex. Nature 1991, 351, 491–494. [Google Scholar]
- Verger, R. ‘Interfacial activation’ of lipases: Facts and artifacts. Trends Biotechnol. 1997, 15, 32–38. [Google Scholar]
- van Tilbeurgh, H.; Egloff, M.-P.; Martinez, C.; Rugani, N.; Verger, R.; Cambillau, C. Interfacial activation of the lipase–procolipase complex by mixed micelles revealed by X-ray crystallography. Nature 1993, 362, 814–820. [Google Scholar]
- Uppenberg, J.; Hansen, M.T.; Patkar, S.; Jones, T.A. The sequence, crystal structure determination and refinement of two crystal forms of lipase B from Candida antarctica. Structure 1994, 2, 293–308. [Google Scholar]
- Anderson, E.M.; Larsson, K.M.; Kirk, O. One biocatalyst—Many applications: The use of Candida antarctica B-lipase in organic synthesis. Biocatal. Biotransform. 1998, 16, 181–204. [Google Scholar]
- Gotor-Fernández, V.; Busto, E.; Gotor, V. Candida antarctica lipase B: An ideal biocatalyst for the preparation of nitrogenated organic compounds. Adv. Synth. Catal. 2006, 348, 797–812. [Google Scholar]
- Kirk, O.; Christensen, M.W. Lipases from Candida antarctica: Unique biocatalysts from a unique origin. Org. Process Res. Dev. 2002, 6, 446–451. [Google Scholar]
- Domínguez De María, P.; Carboni-Oerlemans, C.; Tuin, B.; Bargeman, G.; Van Der Meer, A.; Van Gemert, R. Biotechnological applications of Candida antarctica lipase A: State-of-the-art. J. Mol. Catal. B Enzym. 2005, 37, 36–46. [Google Scholar]
- Ericsson, D.J.; Kasrayan, A.; Johansson, P.; Bergfors, T.; Sandström, A.G.; Bäckvall, J.E.; Mowbray, S.L. X-ray structure of Candida antarctica lipase A shows a novel lid structure and a likely mode of interfacial activation. J. Mol. Biol. 2008, 376, 109–119. [Google Scholar]
- Monteiro, R.R.C.; Virgen-Ortiz, J.J.; Berenguer-Murcia, Á.; da Rocha, T.N.; dos Santos, J.C.S.; Alcántara, A.R.; Fernandez-Lafuente, R. Biotechnological relevance of the lipase A from Candida antarctica. Catal. Today 2021, 362, 141–154. [Google Scholar]
- Grochulski, P.; Li, Y.; Schrag, J.D.; Bouthillier, F.; Smith, P.; Harrison, D.; Rubin, B.; Cygler, M. Insights into interfacial activation from an open structure of Candida rugosa lipase. J. Biol. Chem. 1993, 268, 12843–12847. [Google Scholar]
- Rodrigues, R.C.; Fernandez-Lafuente, R. Lipase from Rhizomucor miehei as an industrial biocatalyst in chemical process. J. Mol. Catal. B Enzym. 2010, 64, 1–22. [Google Scholar]
- Rodrigues, R.C.; Fernandez-Lafuente, R. Lipase from Rhizomucor miehei as a biocatalyst in fats and oils modification. J. Mol. Catal. B Enzym. 2010, 66, 15–32. [Google Scholar]
- Monteiro, R.R.C.; Arana-Peña, S.; da Rocha, T.N.; Miranda, L.P.; Berenguer-Murcia, Á.; Tardioli, P.W.; dos Santos, J.C.S.; Fernandez-Lafuente, R. Liquid lipase preparations designed for industrial production of biodiesel. Is it really an optimal solution? Renew. Energy 2021, 164, 1566–1587. [Google Scholar]
- Mibielli, G.M.; Fagundes, A.P.; Bender, J.P.; Vladimir Oliveira, J. Lab and pilot plant FAME production through enzyme-catalyzed reaction of low-cost feedstocks. Bioresour. Technol. Rep. 2019, 5, 150–156. [Google Scholar]
- Englund, P.T.; King, T.P.; Craig, L.C.; Walti, A. Studies on ficin. I. Its isolation and characterization. Biochemistry 1968, 7, 163–175. [Google Scholar]
- Morellon-Sterling, R.; El-Siar, H.; Tavano, O.L.; Berenguer-Murcia, Á.; Fernández-Lafuente, R. Ficin: A protease extract with relevance in biotechnology and biocatalysis. Int. J. Biol. Macromol. 2020, 162, 394–404. [Google Scholar]
- Birktoft, J.J.; Blow, D.M. Structure of crystalline α-chymotrypsin: V. The atomic structure of tosyl-α-chymotrypsin at 2 Å resolution. J. Mol. Biol. 1972, 68, 187–240. [Google Scholar]
- Blow, D.M. Structure and mechanism of chymotrypsin. Acc. Chem. Res. 1976, 9, 145–152. [Google Scholar]
- Huber, R.; Kukla, D.; Bode, W.; Schwager, P.; Bartels, K.; Deisenhofer, J.; Steigemann, W. Structure of the complex formed by bovine trypsin and bovine pancreatic trypsin inhibitor: II. Crystallographic refinement at 1.9 Å resolution. J. Mol. Biol. 1974, 89, 73–101. [Google Scholar]
- Bode, W.; Schwager, P. The refined crystal structure of bovine β-trypsin at 1·8 Å resolution: II. Crystallographic refinement, calcium binding site, benzamidine binding site and active site at pH 7·0. J. Mol. Biol. 1975, 98, 693–717. [Google Scholar]
- Mateo, C.; Abian, O.; Bernedo, M.; Cuenca, E.; Fuentes, M.; Fernandez-Lorente, G.; Palomo, J.M.; Grazu, V.; Pessela, B.C.C.; Giacomini, C.; et al. Some special features of glyoxyl supports to immobilize proteins. Enzyme Microb. Technol. 2005, 37, 456–462. [Google Scholar]
- Mateo, C.; Palomo, J.M.; Fuentes, M.; Betancor, L.; Grazu, V.; López-Gallego, F.; Pessela, B.C.C.; Hidalgo, A.; Fernández-Lorente, G.; Fernández-Lafuente, R.; et al. Glyoxyl agarose: A fully inert and hydrophilic support for immobilization and high stabilization of proteins. Enzyme Microb. Technol. 2006, 39, 274–280. [Google Scholar]
- Barbosa, O.; Ortiz, C.; Berenguer-Murcia, Á.; Torres, R.; Rodrigues, R.C.; Fernandez-Lafuente, R. Glutaraldehyde in bio-catalysts design: A useful crosslinker and a versatile tool in enzyme immobilization. RSC Adv. 2014, 4, 1583–1600. [Google Scholar]
- Tanaka, Y.; Kagamiishi, A.; Kiuchi, A.; Horiuchi, T. Purification and properties of β-galactosidase from Aspergillus oryzae. J. Biochem. 1975, 77, 241–247. [Google Scholar]
- Berka, R.M.; Schneider, P.; Golightly, E.J.; Brown, S.H.; Madden, M.; Brown, K.M.; Halkier, T.; Mondorf, K.; Xu, F. Characterization of the gene encoding an extracellular laccase of Myceliophthora thermophila and analysis of the recombinant enzyme expressed in Aspergillus oryzae. Appl. Environ. Microbiol. 1997, 63, 3151–3157. [Google Scholar]
- Lima, M.A.; Oliveira-Neto, M.; Kadowaki, M.A.S.; Rosseto, F.R.; Prates, E.T.; Squina, F.M.; Leme, A.F.P.; Skaf, M.S.; Polikarpov, I. Aspergillus niger β-glucosidase has a cellulase-like tadpole molecular shape: Insights into glycoside hydrolase family 3 (gh3) β-glucosidase structure and function. J. Biol. Chem. 2013, 288, 32991–33005. [Google Scholar]
- Yazaki, T.; Ohnishi, M.; Rokushika, S.; Okada, G. Subsite structure of the β-glucosidase from Aspergillus niger, evaluated by steady-state kinetics with cello-oligosaccharides as substrates. Carbohydr. Res. 1997, 298, 51–57. [Google Scholar]
- Longley, W. The crystal structure of bovine liver catalase: A combined study by X-ray diffraction and electron microscopy. J. Mol. Biol. 1967, 30, 323-IN4. [Google Scholar]
- Kutzbach, C.; Rauenbusch, E. Preparation and general properties of crystalline penicillin acylase from Escherichia coli ATCC 11 105. Hoppe-Seyler’s Zeitschrift für Physiol. Chemie 1974, 355, 45–53. [Google Scholar]
- Schumacher, G.; Sizmann, D.; Haug, H.; Buckel, P.; Böck, A. Penicillin acylase from E. coli: Unique gene-protein relation. Nucleic Acids Res. 1986, 14, 5713–5727. [Google Scholar]
- Hewitt, L.; Kasche, V.; Lummer, K.; Lewis, R.J.; Murshudov, G.N.; Verma, C.S.; Dodson, G.G.; Wilson, K.S. Structure of a slow processing precursor penicillin acylase from Escherichia coli reveals the linker peptide blocking the active-site cleft. J. Mol. Biol. 2000, 302, 887–898. [Google Scholar]
- Barbosa, O.; Torres, R.; Ortiz, C.; Fernandez-Lafuente, R. Versatility of glutaraldehyde to immobilize lipases: Effect of the immobilization protocol on the properties of lipase B from Candida antarctica. Process Biochem. 2012, 47, 1220–1227. [Google Scholar]
- Böck, A.; Wirth, R.; Schmid, G.; Schumacher, G.; Lang, G.; Buckel, P. The two subunits of penicillin acylase are processed from a common precursor. FEMS Microbiol. Lett. 1983, 20, 141–144. [Google Scholar]
- Kasche, V.; Lummer, K.; Nurk, A.; Piotraschke, E.; Rieks, A.; Stoeva, S.; Voelter, W. Intramolecular autoproteolysis initiates the maturation of penicillin amidase from Escherichia coli. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 1999, 1433, 76–86. [Google Scholar]
- Lee, H.; Ku Park, O.; Sam Kang, H. Identification of a new active site for autocatalytic processing of penicillin acylase precursor in Escherichia coli ATCC11105. Biochem. Biophys. Res. Commun. 2000, 272, 199–204. [Google Scholar]
- Siar, E.-H.; Zaak, H.; Kornecki, J.F.; Zidoune, M.N.; Barbosa, O.; Fernandez-Lafuente, R. Stabilization of ficin extract by immobilization on glyoxyl agarose. Preliminary characterization of the biocatalyst performance in hydrolysis of proteins. Process Biochem. 2017, 58, 98–104. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar]
- Fernandez-Lafuente, R.; Rosell, C.M.; Rodriguez, V.; Santana, C.; Soler, G.; Bastida, A.; Guisán, J.M. Preparation of activated supports containing low pK amino groups. A new tool for protein immobilization via the carboxyl coupling method. Enzyme Microb. Technol. 1993, 15, 546–550. [Google Scholar]
- Guisán, J. Aldehyde-agarose gels as activated supports for immobilization-stabilization of enzymes. Enzyme Microb. Technol. 1988, 10, 375–382. [Google Scholar]
- Betancor, L.; López-Gallego, F.; Hidalgo, A.; Alonso-Morales, N.; Mateo, G.D.-O.C.; Fernández-Lafuente, R.; Guisán, J.M. Different mechanisms of protein immobilization on glutaraldehyde activated supports: Effect of support activation and immobilization conditions. Enzyme Microb. Technol. 2006, 39, 877–882. [Google Scholar]
- Vazquez-Ortega, P.G.; Alcaraz-Fructuoso, M.T.; Rojas-Contreras, J.A.; López-Miranda, J.; Fernandez-Lafuente, R. Stabilization of dimeric β-glucosidase from Aspergillus niger via glutaraldehyde immobilization under different conditions. Enzyme Microb. Technol. 2018, 110, 38–45. [Google Scholar]
- Zaak, H.; Peirce, S.; de Albuquerque, T.; Sassi, M.; Fernandez-Lafuente, R. Exploiting the versatility of aminated supports activated with glutaraldehyde to immobilize β-galactosidase from Aspergillus oryzae. Catalysts 2017, 7, 250. [Google Scholar]
- Wood, A.N.P.; Fernandez-Lafuente, R.; Cowan, D.A. Purification and partial characterization of a novel thermophilic carboxylesterase with high mesophilic specific activity. Enzyme Microb. Technol. 1995, 17, 816–825. [Google Scholar]
- Rocha-Martin, J.; Fernández-Lorente, G.; Guisan, J.M. Sequential hydrolysis of commercial casein hydrolysate by immobilized trypsin and thermolysin to produce bioactive phosphopeptides. Biocatal. Biotransform. 2018, 36, 159–171. [Google Scholar]
- Pedroche, J.; del Mar Yust, M.; Mateo, C.; Fernández-Lafuente, R.; Girón-Calle, J.; Alaiz, M.; Vioque, J.; Guisán, J.M.; Millán, F. Effect of the support and experimental conditions in the intensity of the multipoint covalent attachment of proteins on glyoxyl-agarose supports: Correlation between enzyme-support linkages and thermal stability. Enzyme Microb. Technol. 2007, 40, 1160–1166. [Google Scholar]
- Moyano, F.; Setien, E.; Silber, J.J.; Correa, N.M. Enzymatic Hydrolysis of N-Benzoyl-l-Tyrosine p-Nitroanilide by α-Chymotrypsin in DMSO-Water/AOT/n-Heptane Reverse Micelles. A Unique Interfacial Effect on the Enzymatic Activity. Langmuir 2013, 29, 8245–8254. [Google Scholar]
- De Albuquerque, T.L.; Peirce, S.; Rueda, N.; Marzocchella, A.; Gonçalves, L.R.B.; Rocha, M.V.P.; Fernandez-Lafuente, R. Ion exchange of β-galactosidase: The effect of the immobilization pH on enzyme stability. Process Biochem. 2016, 51, 875–880. [Google Scholar]
- Boudrant, J.; Woodley, J.M.; Fernandez-Lafuente, R. Parameters necessary to define an immobilized enzyme preparation. Process Biochem. 2020, 90, 66–80. [Google Scholar]
- Bastida, A.; Sabuquillo, P.; Armisen, P.; Fernández-Lafuente, R.; Huguet, J.; Guisán, J.M. A single step purification, immobilization, and hyperactivation of lipases via interfacial adsorption on strongly hydrophobic supports. Biotechnol. Bioeng. 1998, 58, 486–493. [Google Scholar]
- Siar, E.-H.; Arana-Peña, S.; Barbosa, O.; Zidoune, M.; Fernandez-Lafuente, R. Immobilization/stabilization of ficin extract on glutaraldehyde-activated agarose beads. Variables that control the final stability and activity in protein hydrolyses. Catalysts 2018, 8, 149. [Google Scholar]
- Blanco, R.M.; Guisán, J. Protecting effect of competitive inhibitors during very intense insolubilized enzyme-activated support multipoint attachments: Trypsin (amine)-agarose (aldehyde) system. Enzyme Microb. Technol. 1988, 10, 227–232. [Google Scholar]
- Fernández-Lafuente, R.; Rodriguez, V.; Guisán, J.M. The coimmobilization of d-amino acid oxidase and catalase enables the quantitative transformation of d-amino acids (d-phenylalanine) into α-keto acids (phenylpyruvic acid). Enzyme Microb. Technol. 1998, 23, 28–33. [Google Scholar]
- Guisán, J.M.; Bastida, A.; Cuesta, C.; Fernandez-Lufuente, R.; Rosell, C.M. Immobilization-stabilization of α-chymotrypsin by covalent attachment to aldehyde-agarose gels. Biotechnol. Bioeng. 1991, 38, 1144–1152. [Google Scholar]
- Blanco, R.M.; Calvete, J.J.; Guisán, J. Immobilization-stabilization of enzymes; variables that control the intensity of the trypsin (amine)-agarose (aldehyde) multipoint attachment. Enzyme Microb. Technol. 1989, 11, 353–359. [Google Scholar]
- Abian, O.; Grazú, V.; Hermoso, J.; González, R.; García, J.L.; Fernández-Lafuente, R.; Guisán, J.M. Stabilization of penicillin G acylase from Escherichia coli: Site-directed mutagenesis of the protein surface to increase multipoint covalent attachment. Appl. Environ. Microbiol. 2004, 70, 1249–1251. [Google Scholar]
- Rosell, C.M.; Fernandez-Lafuente, R.; Guisan, J.M. Modification of enzyme properties by the use of inhibitors during their stabilisation by multipoint covalent attachment. Biocatal. Biotransform. 1995, 12, 67–76. [Google Scholar]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Braham, S.A.; Siar, E.-H.; Arana-Peña, S.; Carballares, D.; Morellon-Sterling, R.; Bavandi, H.; de Andrades, D.; Kornecki, J.F.; Fernandez-Lafuente, R. Effect of Concentrated Salts Solutions on the Stability of Immobilized Enzymes: Influence of Inactivation Conditions and Immobilization Protocol. Molecules 2021, 26, 968. https://doi.org/10.3390/molecules26040968
Braham SA, Siar E-H, Arana-Peña S, Carballares D, Morellon-Sterling R, Bavandi H, de Andrades D, Kornecki JF, Fernandez-Lafuente R. Effect of Concentrated Salts Solutions on the Stability of Immobilized Enzymes: Influence of Inactivation Conditions and Immobilization Protocol. Molecules. 2021; 26(4):968. https://doi.org/10.3390/molecules26040968
Chicago/Turabian StyleBraham, Sabrina Ait, El-Hocine Siar, Sara Arana-Peña, Diego Carballares, Roberto Morellon-Sterling, Hossein Bavandi, Diandra de Andrades, Jakub F. Kornecki, and Roberto Fernandez-Lafuente. 2021. "Effect of Concentrated Salts Solutions on the Stability of Immobilized Enzymes: Influence of Inactivation Conditions and Immobilization Protocol" Molecules 26, no. 4: 968. https://doi.org/10.3390/molecules26040968
APA StyleBraham, S. A., Siar, E. -H., Arana-Peña, S., Carballares, D., Morellon-Sterling, R., Bavandi, H., de Andrades, D., Kornecki, J. F., & Fernandez-Lafuente, R. (2021). Effect of Concentrated Salts Solutions on the Stability of Immobilized Enzymes: Influence of Inactivation Conditions and Immobilization Protocol. Molecules, 26(4), 968. https://doi.org/10.3390/molecules26040968






