Bioactive Tryptophan-Based Copper Complex with Auxiliary β-Carboline Spectacle Potential on Human Breast Cancer Cells: In Vitro and In Vivo Studies
Abstract
:1. Introduction
2. Results and Discussion
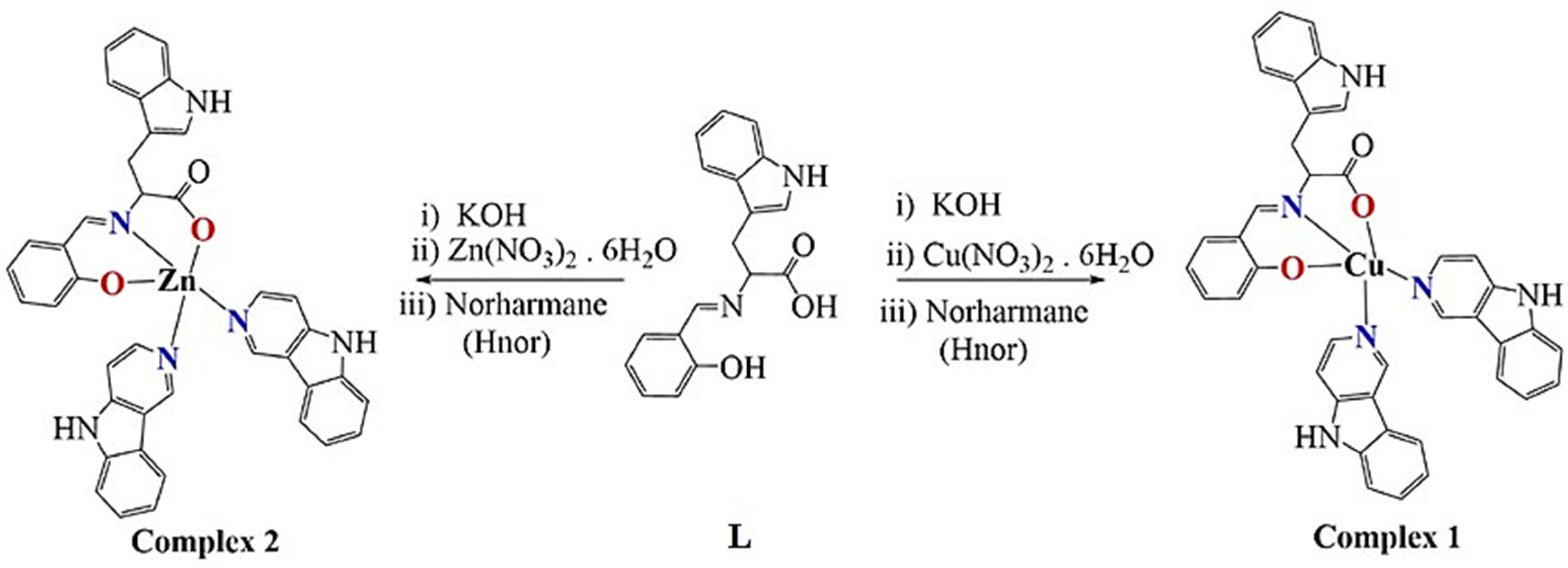
2.1. Synthesis and Characterization
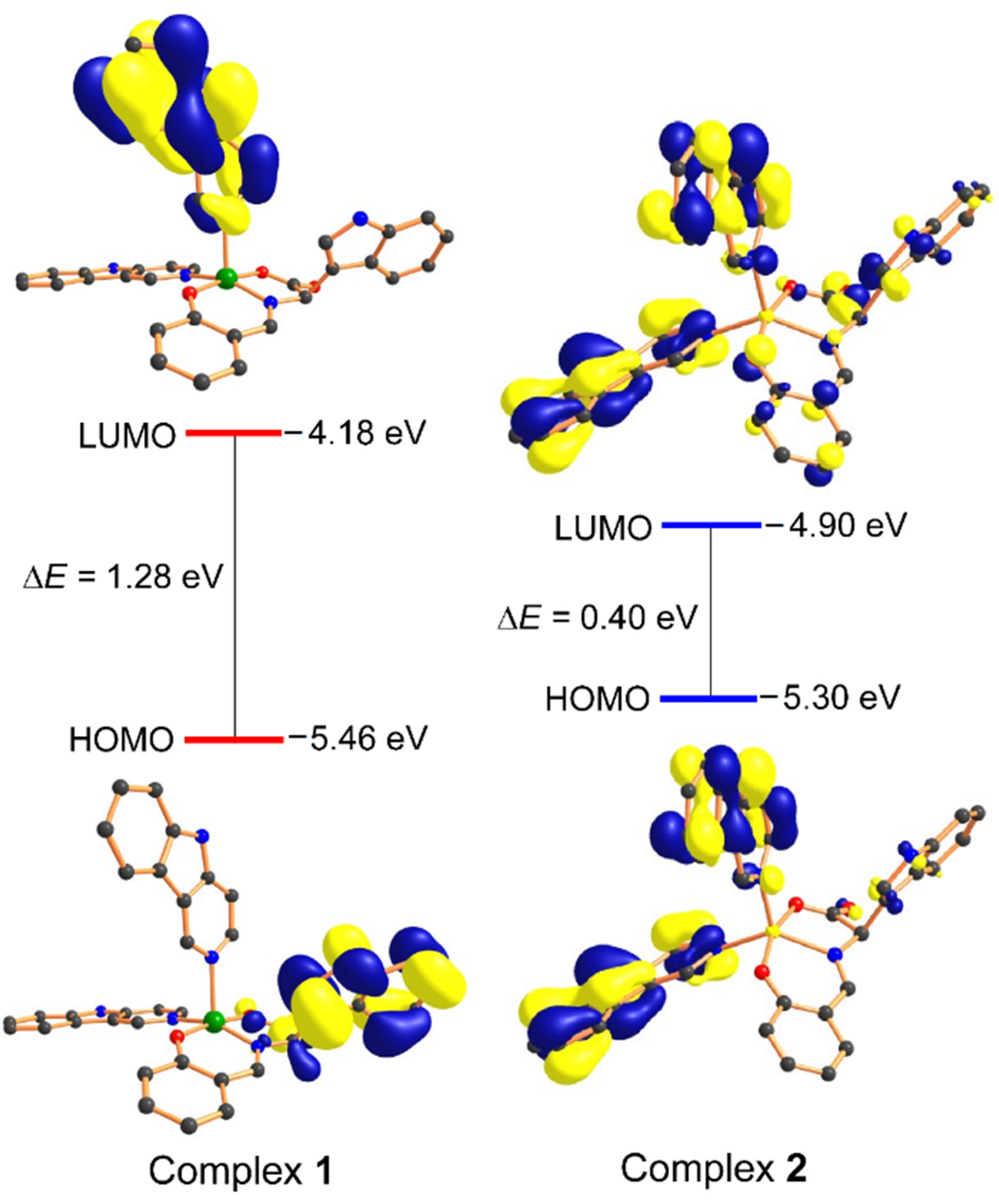
2.2. Computational Studies: Density Functional Theory
2.3. In-Vitro Anticancer Activity
2.3.1. MTT Assay
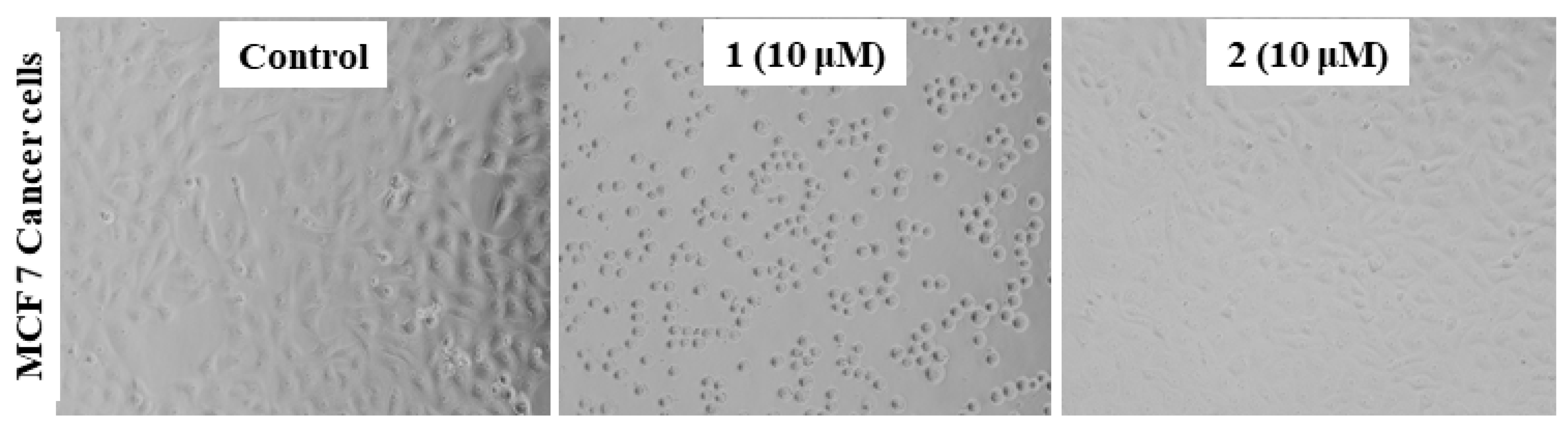
2.3.2. Morphology of the MCF7 Cancer Cells
2.3.3. Assessment of the Oxidative Stress against MCF7 Cells
2.4. In Vivo Studies
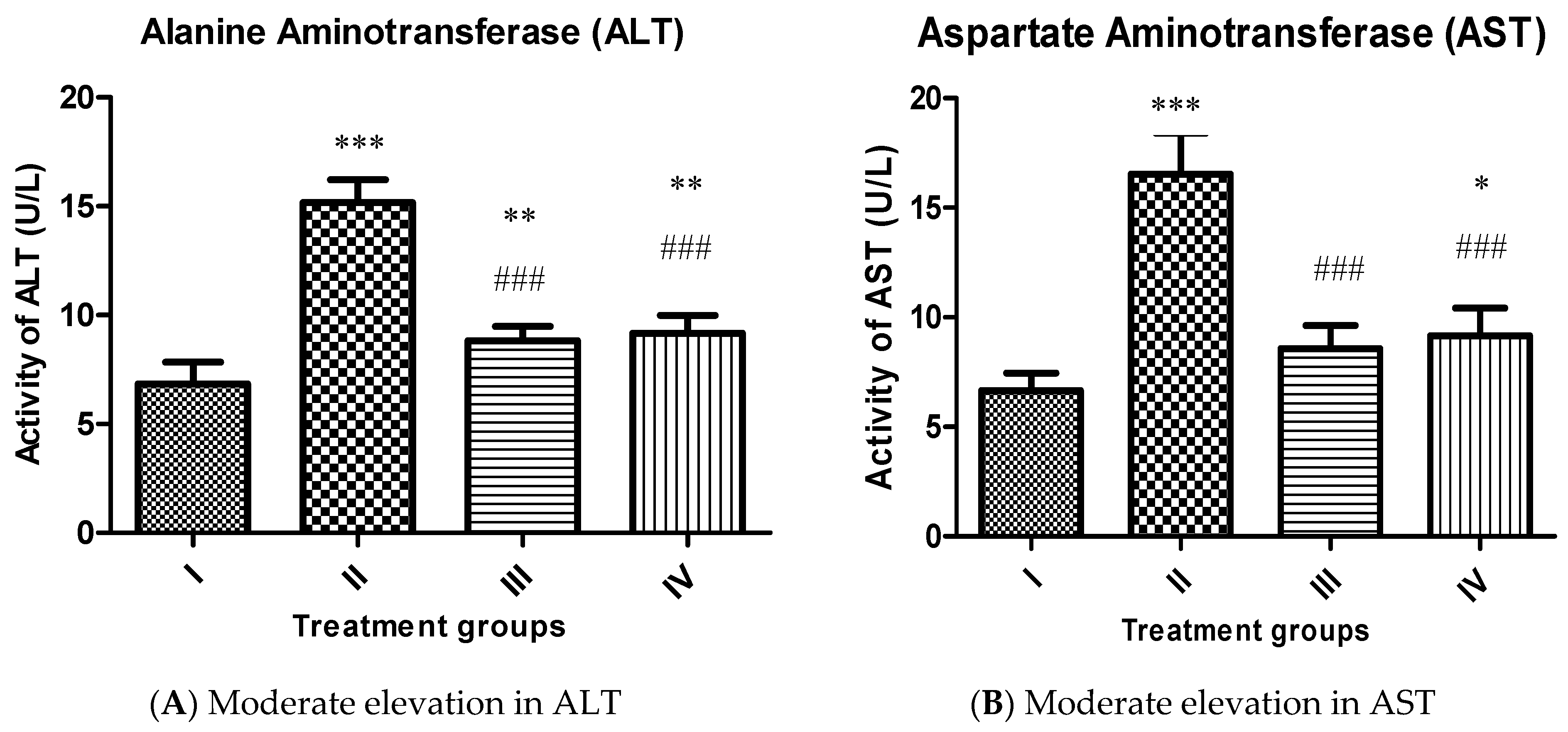
2.4.1. Effect on Liver Function Markers
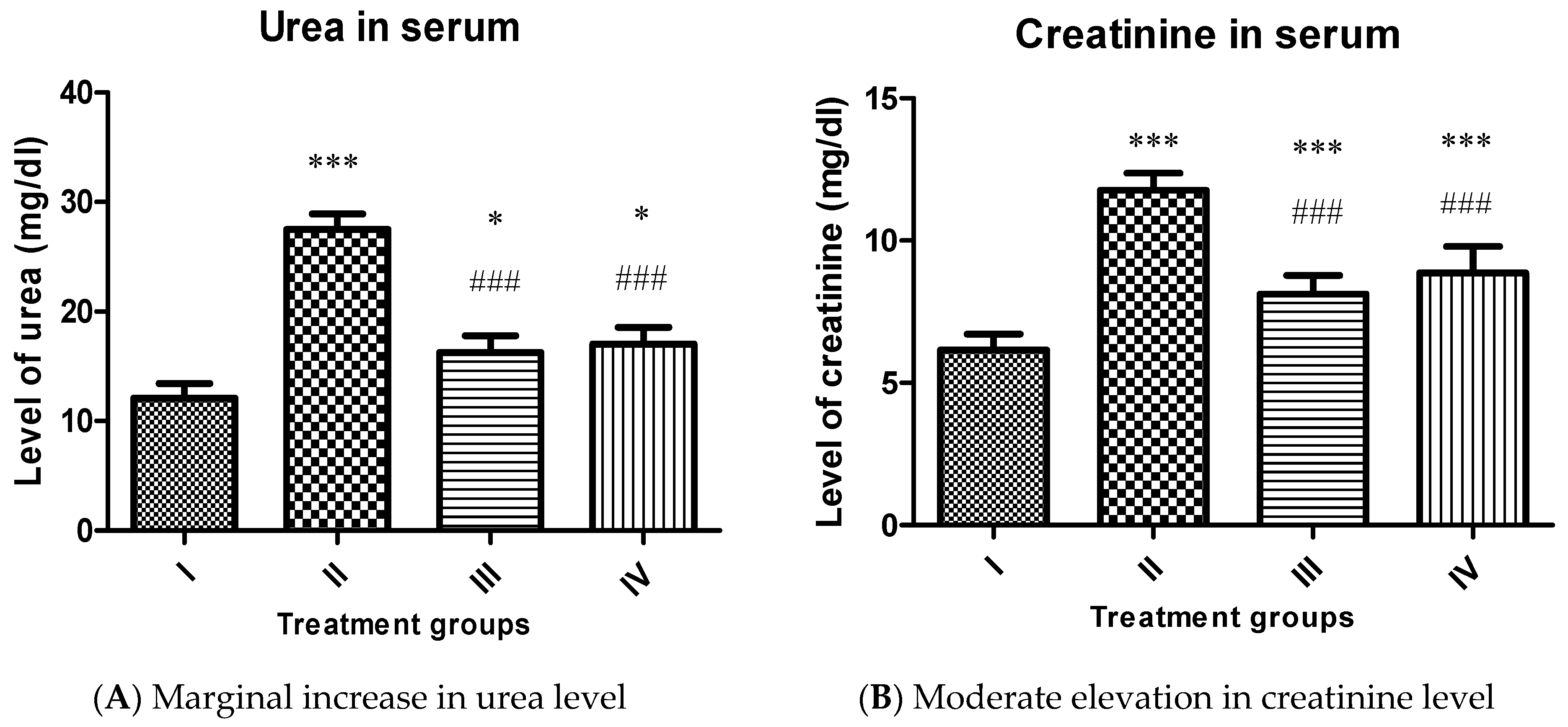
2.4.2. Effect on Renal Function Markers
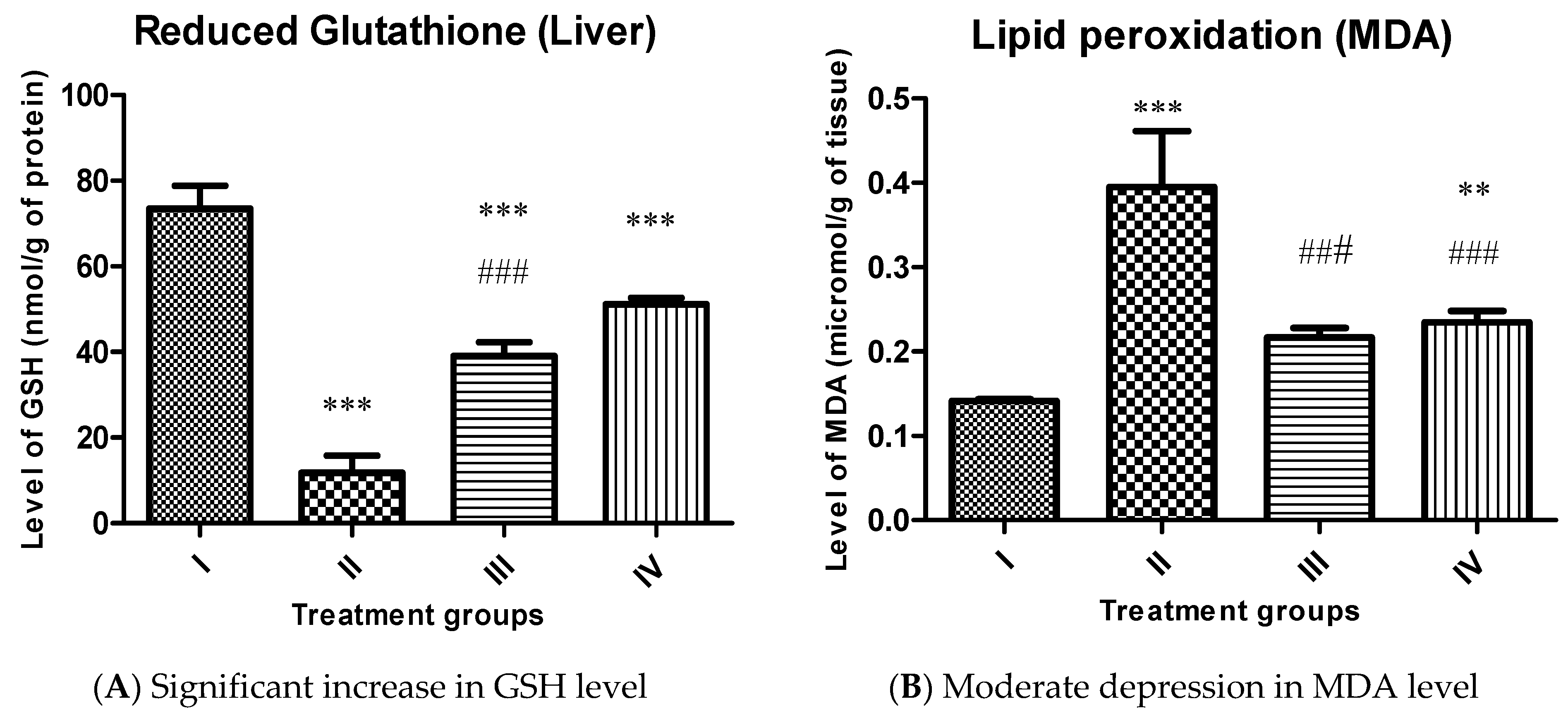
2.4.3. Effect on Key Antioxidant Parameters
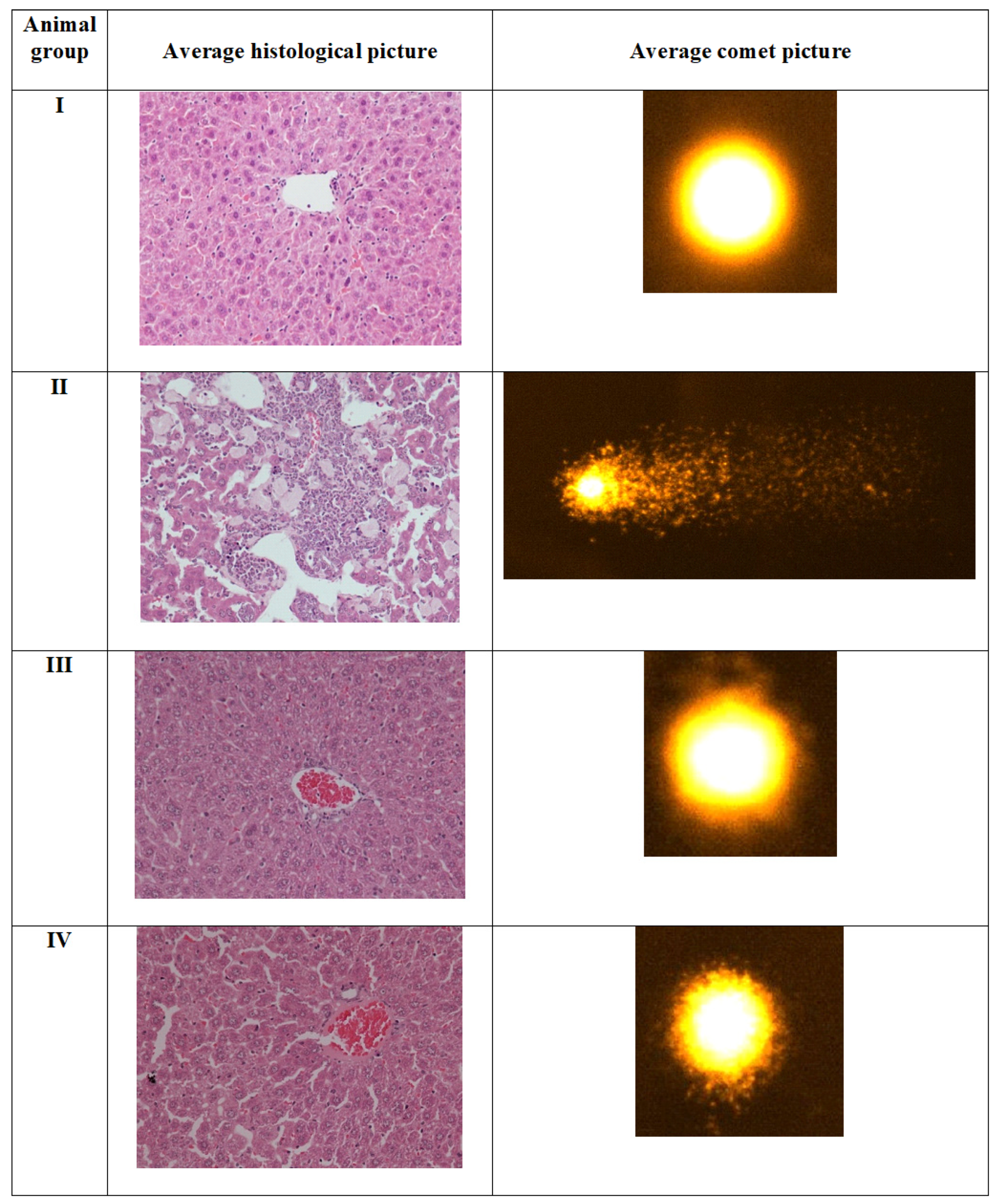
2.4.4. Comet Assay
2.5. Histopathological Evaluation in the Liver Samples
3. Materials and Methods
3.1. Chemicals and Instrumentation
3.2. Synthesis
3.2.1. Synthesis of [Cu(L)(Hnor)2] Complex (1)
3.2.2. Synthesis of [Zn(L)(Hnor)2] Complex (2)
3.3. Computational Methodology
3.4. In Vitro Cytotoxicity
3.5. In Vivo Toxicity Profiling
3.5.1. Animal Treatment and Sample Preparation
3.5.2. Estimation of Aspartate Aminotransferase (AST) and Alanine Aminotransferase (ALT) as Liver Function Markers
3.5.3. Measurement of Reduced Glutathione (GSH) and Malondialdehyde (MDA)
3.5.4. Comet Assay
3.5.5. Histopathological Assessment of Liver Tissues
3.5.6. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Parveen, S.; Arjmand, F.; Tabassum, S. Clinical developments of antitumor polymer therapeutics. RSC Adv. 2019, 9, 24699–24721. [Google Scholar] [CrossRef] [Green Version]
- Lippert, B. Cisplatin: Chemistry and Biochemistry of a Leading Anticancer Drug; John Wiley & Sons: Hoboken, NJ, USA, 2006; ISBN 9783906390420. [Google Scholar]
- Rosenberg, B.; Van Camp, L.; Krigas, T. Inhibition of cell division in Escherichia coli by electrolysis products from a platinum electrode [17]. Nature 1965, 205, 698–699. [Google Scholar] [CrossRef]
- Wang, X.; Wang, X.; Guo, Z. Functionalization of Platinum Complexes for Biomedical Applications. Acc. Chem. Res. 2015, 48, 2622–2631. [Google Scholar] [CrossRef]
- Johnstone, T.C.; Suntharalingam, K.; Lippard, S.J. The next generation of platinum drugs: Targeted Pt (II) agents, nanoparticle delivery, and Pt (IV) prodrugs. Chem. Rev. 2016, 116, 3436–3486. [Google Scholar] [CrossRef] [Green Version]
- Dasari, S.; Bernard Tchounwou, P. Cisplatin in cancer therapy: Molecular mechanisms of action. Eur. J. Pharmacol. 2014, 740, 364–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santini, C.; Pellei, M.; Gandin, V.; Porchia, M.; Tisato, F.; Marzano, C. Advances in copper complexes as anticancer agents. Chem. Rev. 2014, 114, 815–862. [Google Scholar] [CrossRef] [PubMed]
- Pearson, R.G. Hard and Soft Acids and Bases. J. Am. Chem. Soc. 1963, 85, 3533–3539. [Google Scholar] [CrossRef]
- Vella, F. Principles of Bioinorganic Chemistry; University Science Books: Mill Valley, CA, USA, 1995; Volume 23. [Google Scholar]
- Holm, R.H.; Kennepohl, P.; Solomon, E.I. Structural and functional aspects of metal sites in biology. Chem. Rev. 1996, 96, 2239–2314. [Google Scholar] [CrossRef]
- Metzler-Nolte, N.; Kraatz, H.B. Concepts and Models in Bioinorganic Chemistry; Wiley-Vch: New York, NY, USA, 2007; Volume 44. [Google Scholar]
- Solomon, E.I.; Heppner, D.E.; Johnston, E.M.; Ginsbach, J.W.; Cirera, J.; Qayyum, M.; Kieber-Emmons, M.T.; Kjaergaard, C.H.; Hadt, R.G.; Tian, L. Copper active sites in biology. Chem. Rev. 2014, 114, 3659–3853. [Google Scholar] [CrossRef] [Green Version]
- Denoyer, D.; Masaldan, S.; La Fontaine, S.; Cater, M.A. Targeting copper in cancer therapy: “Copper That Cancer”. Metallomics 2015, 7, 1459–1476. [Google Scholar] [CrossRef] [PubMed]
- Tardito, S.; Bassanetti, I.; Bignardi, C.; Elviri, L.; Tegoni, M.; Mucchino, C.; Bussolati, O.; Franchi-Gazzola, R.; Marchiò, L. Copper binding agents acting as copper ionophores lead to caspase inhibition and paraptotic cell death in human cancer cells. J. Am. Chem. Soc. 2011, 133, 6235–6242. [Google Scholar] [CrossRef]
- Zuo, J.; Bi, C.; Fan, Y.; Buac, D.; Nardon, C.; Daniel, K.G.; Dou, Q.P. Cellular and computational studies of proteasome inhibition and apoptosis induction in human cancer cells by amino acid Schiff base-copper complexes. J. Inorg. Biochem. 2013, 118, 83–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ng, C.H.; Kong, S.M.; Tiong, Y.L.; Maah, M.J.; Sukram, N.; Ahmad, M.; Khoo, A.S.B. Selective anticancer copper(ii)-mixed ligand complexes: Targeting of ROS and proteasomes. Metallomics 2014, 6, 892–906. [Google Scholar] [CrossRef]
- Siegel, R.; Naishadham, D.; Jemal, A. Cancer statistics, 2013. CA Cancer J. Clin. 2013, 63, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haskel, C.M. Breast cancer. In Cancer Treatment, 5th ed.; Haskell, C.M., Ed.; W.B. Saunders Company: Philadelphia, PA, USA, 2001. [Google Scholar]
- Belluti, F.; Fontana, G.; Bo, L.D.; Carenini, N.; Giommarelli, C.; Zunino, F. Design, synthesis and anticancer activities of stilbene-coumarin hybrid compounds: Identification of novel proapoptotic agents. Bioorganic Med. Chem. 2010, 18, 3543–3550. [Google Scholar] [CrossRef] [PubMed]
- Brummond, K.M.; Goodell, J.R.; La Porte, M.G.; Wang, L.; Xie, X.Q. Synthesis and in silico screening of a library of β-carboline-containing compounds. Beilstein J. Org. Chem. 2012, 8, 1048–1058. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, R.; Kimura, T.; Tsutsumi, T.; Tamura, S.; Sakai, N.; Konakahara, T. Structure-activity relationship in the antitumor activity of 6-, 8- or 6,8-substituted 3-benzylamino-β-carboline derivatives. Bioorg. Med. Chem. Lett. 2012, 22, 3506–3515. [Google Scholar] [CrossRef]
- Lunagariya, N.A.; Gohil, V.M.; Kushwah, V.; Neelagiri, S.; Jain, S.; Singh, S.; Bhutani, K.K. Design, synthesis and biological evaluation of 1,3,6-trisubstituted β-carboline derivatives for cytotoxic and anti-leishmanial potential. Bioorganic Med. Chem. Lett. 2016, 26, 789–794. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.M.B.L.; Garcia, F.P.; Da Silva Rodrigues, J.H.; Nakamura, C.V.; Ueda-Nakamura, T.; Meyer, E.; Tasca Gois Ruiz, A.L.; Foglio, M.A.; De Carvalho, J.E.; Da Costa, W.F.; et al. Synthesis, antitumor, antitrypanosomal and antileishmanial activities of benzo[4,5]canthin-6-ones bearing the N′-(substituted benzylidene)-carbohydrazide and N-alkylcarboxamide groups at C-2. Chem. Pharm. Bull. 2012, 60, 1372–1379. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.L.; Kuo, P.C.; Hwang, T.L.; Chiou, W.F.; Qian, K.; Lai, C.Y.; Lee, K.H.; Wu, T.S. Synthesis, in vitro anti-inflammatory and cytotoxic evaluation, and mechanism of action studies of 1-benzoyl-β-carboline and 1-benzoyl-3-carboxy-β-carboline derivatives. Bioorganic Med. Chem. 2011, 19, 1674–1682. [Google Scholar] [CrossRef]
- Guan, H.; Chen, H.; Peng, W.; Ma, Y.; Cao, R.; Liu, X.; Xu, A. Design of β-carboline derivatives as DNA-targeting antitumor agents. Eur. J. Med. Chem. 2006, 41, 1167–1179. [Google Scholar] [CrossRef]
- Funayama, Y.; Nishio, K.; Wakabayashi, K.; Nagao, M.; Shimoi, K.; Ohira, T.; Hasegawa, S.; Saijo, N. Effects of β- and γ-carboline derivatives on DNA topoisomerase activities. Mutat. Res. Fundam. Mol. Mech. Mutagen. 1996, 349, 183–191. [Google Scholar] [CrossRef]
- Song, Y.; Kesuma, D.; Wang, J.; Deng, Y.; Duan, J.; Wang, J.H.; Qi, R.Z. Specific inhibition of cyclin-dependent kinases and cell proliferation by harmine. Biochem. Biophys. Res. Commun. 2004, 317, 128–132. [Google Scholar] [CrossRef] [PubMed]
- Castro, A.C.; Dang, L.C.; Soucy, F.; Grenier, L.; Mazdiyasni, H.; Hottelet, M.; Parent, L.; Pien, C.; Palombella, V.; Adams, J. Novel IKK inhibitors: β-carbolines. Bioorg. Med. Chem. Lett. 2003, 13, 2419–2422. [Google Scholar] [CrossRef]
- Sears, P.G.; Lester, G.R.; Dawson, L.R. A study of the conductance behavior of some uni-univalent electrolytes in dimethyl sulfoxide at 25°. J. Phys. Chem. 1956, 60, 1433–1436. [Google Scholar] [CrossRef]
- Patra, A.K.; Bhowmick, T.; Ramakumar, S.; Nethaji, M.; Chakravarty, A.R. DNA cleavage in red light promoted by copper(II) complexes of α-amino acids and photoactive phenanthroline bases. Dalton Trans. 2008, 6966–6976. [Google Scholar] [CrossRef]
- Banaspati, A.; Das, D.; Choudhury, C.J.; Bhattacharyya, A.; Goswami, T.K. Photocytotoxic copper(II) complexes of N-salicylyl-L-tryptophan and phenanthroline bases. J. Inorg. Biochem. 2019, 191, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Ahmad Khan, R.; Al-Farhan, K.; De Almeida, A.; Alsalme, A.; Casini, A.; Ghazzali, M.; Reedijk, J. Light-stable bis(norharmane)silver(I) compounds: Synthesis, characterization and antiproliferative effects in cancer cells. J. Inorg. Biochem. 2014, 140, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Khan, R.A.; de Almeida, A.; Al-Farhan, K.; Alsalme, A.; Casini, A.; Ghazzali, M.; Reedijk, J. Transition-metal norharmane compounds as possible cytotoxic agents: New insights based on a coordination chemistry perspective. J. Inorg. Biochem. 2016, 165, 128–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahato, S.; Meheta, N.; Kotakonda, M.; Joshi, M.; Shit, M.; Choudhury, A.R.; Biswas, B. Synthesis, structure, polyphenol oxidase mimicking and bactericidal activity of a zinc-schiff base complex. Polyhedron 2020, 114933. [Google Scholar] [CrossRef]
- Amaral, T.C.; Miguel, F.B.; Couri, M.R.C.; Corbi, P.P.; Carvalho, M.A.; Campos, D.L.; Pavan, F.R.; Cuin, A. Silver(I) and zinc(II) complexes with symmetrical cinnamaldehyde Schiff base derivative: Spectroscopic, powder diffraction characterization, and antimycobacterial studies. Polyhedron 2018, 146, 166–171. [Google Scholar] [CrossRef] [Green Version]
- Maxim, C.; Pasatoiu, T.D.; Kravtsov, V.C.; Shova, S.; Muryn, C.A.; Winpenny, R.E.P.; Tuna, F.; Andruh, M. Copper(II) and zinc(II) complexes with Schiff-base ligands derived from salicylaldehyde and 3-methoxysalicylaldehyde: Synthesis, crystal structures, magnetic and luminescence properties. Inorg. Chim. Acta 2008, 361, 3903–3911. [Google Scholar] [CrossRef]
- Khan, R.A.; Khan, M.R.; Usman, M.; Sayeed, F.; Alghamdi, H.A.; Alrumman, S.; Alharbi, W.; Farshori, N.N.; Al-Oqail, M.M.; Siddiqui, M.R.; et al. β-Carboline copper complex as a potential mitochondrial-targeted anticancer chemotherapeutic agent: Favorable attenuation of human breast cancer MCF7 cells via apoptosis. Saudi J. Biol. Sci. 2020, 27, 2164–2173. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.M.; Peng, F.; Li, G.D.; Jie, X.M.; Cai, K.R.; Cai, C.; Zhong, Y.; Zeng, H.; Li, W.; Zhang, Z.; et al. The studies on the cytotoxicity in vitro, cellular uptake, cell cycle arrest and apoptosis-inducing properties of ruthenium methylimidazole complex [Ru(MeIm)4(p-cpip)]2+. J. Inorg. Biochem. 2016, 156, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Karmakar, S.; Purkait, K.; Chatterjee, S.; Mukherjee, A. Anticancer activity of a cis-dichloridoplatinum(ii) complex of a chelating nitrogen mustard: Insight into unusual guanine binding mode and low deactivation by glutathione. Dalton Trans. 2016, 45, 3599–3615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krȩzel, A.; Bal, W. Coordination chemistry of glutathione. Acta Biochim. Pol. 1999, 46, 567–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Florea, A.M.; Büsselberg, D. Cisplatin as an anti-tumor drug: Cellular mechanisms of activity, drug resistance and induced side effects. Cancers 2011, 3, 1351–1371. [Google Scholar] [CrossRef]
- Cadoni, E.; Valletta, E.; Caddeo, G.; Isaia, F.; Cabiddu, M.G.; Vascellari, S.; Pivetta, T. Competitive reactions among glutathione, cisplatin and copper-phenanthroline complexes. J. Inorg. Biochem. 2017, 173, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Cotgreave, I.A.; Moldeus, P.; Orrenius, S. Host biochemical defense mechanisms against prooxidants. Annu. Rev. Pharmacol. Toxicol. 1988, 28, 189–212. [Google Scholar] [CrossRef]
- Ortega, A.L.; Mena, S.; Estrela, J.M. Glutathione in cancer cell death. Cancers 2011, 3, 1285–1310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masella, R.; Di Benedetto, R.; Varì, R.; Filesi, C.; Giovannini, C. Novel mechanisms of natural antioxidant compounds in biological systems: Involvement of glutathione and glutathione-related enzymes. J. Nutr. Biochem. 2005, 16, 577–586. [Google Scholar] [CrossRef]
- Gomes, A.; Fernandes, E.; Lima, J.L.F.C. Fluorescence probes used for detection of reactive oxygen species. J. Biochem. Biophys. Methods 2005, 65, 45–80. [Google Scholar] [CrossRef] [PubMed]
- Tisato, F.; Marzano, C.; Porchia, M.; Pellei, M.; Santini, C. Copper in diseases and treatments, and copper-based anticancer strategies. Med. Res. Rev. 2010, 30, 708–749. [Google Scholar] [CrossRef]
- Zimmermann, T.; Burda, J.V. Cisplatin interaction with amino acids cysteine and methionine from gas phase to solutions with constant pH. Interdiscip. Sci. Comput. Life Sci. 2010, 2, 98–114. [Google Scholar] [CrossRef] [PubMed]
- Sajid, M.; Khan, M.R.; Shah, N.A.; Ullah, S.; Younis, T.; Majid, M.; Ahmad, B.; Nigussie, D. Proficiencies of Artemisia scoparia against CCl4 induced DNA damages and renal toxicity in rat. BMC Complement. Altern. Med. 2016, 16, 149. [Google Scholar] [CrossRef] [Green Version]
- Ebaid, H.; Habila, M.; Hassan, I.; Al-Tamimi, J.; Omar, M.S.; Rady, A.; Alhazza, I.M. Curcumin-containing Silver Nanoparticles prevent carbon tetrachlorideinduced hepatotoxicity in mice. Comb. Chem. High. Throughput Screen. 2020, 23. [Google Scholar] [CrossRef] [PubMed]
- Ebaid, H.; Al-Tamimi, J.; Hassan, I.; Alhazza, I.; Al-Khalifa, M. Antioxidant bioactivity of samsum Ant (Pachycondyla sennaarensis) Venom protects against CCL4-induced nephrotoxicity in mice. Oxid. Med. Cell. Longev. 2014, 2014, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassan, I.; Husain, F.M.; Khan, R.A.; Ebaid, H.; Al-Tamimi, J.; Alhazza, I.M.; Aman, S.; Ibrahim, K.E. Ameliorative effect of zinc oxide nanoparticles against potassium bromate-mediated toxicity in Swiss albino rats. Environ. Sci. Pollut. Res. 2019, 26, 9966–9980. [Google Scholar] [CrossRef]
- Das, R.K.; Hossain, S.U.; Bhattacharya, S. Protective effect of diphenylmethyl selenocyanate against CCl 4-induced hepatic injury. J. Appl. Toxicol. 2007, 27, 527–537. [Google Scholar] [CrossRef]
- Zhong-Feng, W.; Mao-Yu, W.; De-Hai, Y.; Zhao, Y.; Hong-Mei, X.; Zhong, S.; Wen-Yi, S.; Yu-Fang, H.; Jun-Qi, N.; Pu-Jun, G.; et al. Therapeutic effect of chitosan on CCl4-induced hepatic fibrosis in rats. Mol. Med. Rep. 2018, 18, 3211–3218. [Google Scholar] [CrossRef] [Green Version]
- Boll, M.; Weber, L.W.D.; Becker, E.; Stampfl, A. Mechanism of carbon tetrachloride-induced hepatotoxicity. Hepatocellular damage by reactive carbon tetrachloride metabolites. Zeitschrift Naturforsch. Sect. C J. Biosci. 2001, 56, 649–659. [Google Scholar] [CrossRef]
- Lee, C.; Yang, W.; Parr, R.G. Becke’s Three Parameter Hybrid Method Using the LYP. Phys. Rev. B 1988, 37, 785. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.; Yang, W.; Parr, R.G. Condens. Matter Mater. Phys. Phys. Rev. B 1988, 37, 785–789. [Google Scholar] [CrossRef] [Green Version]
- Stephens, P.J.; Devlin, F.J.; Chabalowski, C.F.; Frisch, M.J. Ab Initio calculation of vibrational absorption and circular dichroism spectra using density functional force fields. J. Phys. Chem. 1994, 98, 11623–11627. [Google Scholar] [CrossRef]
- Frisch, M.J. Gaussian 09; Gaussian, Inc.: Wallingford, CT, USA, 2009; p. 32. [Google Scholar]
- Hay, P.J.; Wadt, W.R. Ab initio effective core potentials for molecular calculations. Potentials for the transition metal atoms Sc to Hg. J. Chem. Phys. 1985, 82, 270–283. [Google Scholar] [CrossRef]
- Wadt, W.R.; Hay, P.J. Ab initio effective core potentials for molecular calculations. Potentials for main group elements Na to Bi. J. Chem. Phys. 1985, 82, 284–298. [Google Scholar] [CrossRef]
- Roy, L.E.; Hay, P.J.; Martin, R.L. Revised basis sets for the LANL effective core potentials. J. Chem. Theory Comput. 2008, 4, 1029–1031. [Google Scholar] [CrossRef] [PubMed]
- Barone, V.; Cossi, M. Quantum calculation of molecular energies and energy gradients in solution by a conductor solvent model. J. Phys. Chem. A 1998, 102, 1995–2001. [Google Scholar] [CrossRef]
- Cossi, M.; Barone, V. Time-dependent density functional theory for molecules in liquid solutions. J. Chem. Phys. 2001, 115, 4708–4717. [Google Scholar] [CrossRef]
- Cossi, M.; Rega, N.; Scalmani, G.; Barone, V. Energies, structures, and electronic properties of molecules in solution with the C-PCM solvation model. J. Comput. Chem. 2003, 24, 669–681. [Google Scholar] [CrossRef] [PubMed]
- Badawi, A.M.; Mohamed, M.A.S.; Mohamed, M.Z.; Khowdairy, M.M. Surface and antitumor activity of some novel metal-based cationic surfactants. J. Cancer Res. Ther. 2007, 3, 198–206. [Google Scholar] [CrossRef]
- Nagaraj, K.; Velmurugan, G.; Sakthinathan, S.; Venuvanalingam, P.; Arunachalam, S. Influence of self-assembly on intercalative DNA binding interaction of double-chain surfactant Co(iii) complexes containing imidazo[4,5-f][1,10]phenanthroline and dipyrido[3,2-d:2′-3′-f]quinoxaline ligands: Experimental and theoretical study. Dalton Trans. 2014, 43, 18074–18086. [Google Scholar] [CrossRef]
- Sharifi, N.; Aragon-Ching, J.B. Anti-Cancer Agents in Medicinal Chemistry: Editorial. Anticancer Agents Med. Chem. 2009, 9, 1039. [Google Scholar] [CrossRef] [PubMed]
- Shao, J.; Ma, Z.Y.; Li, A.; Liu, Y.H.; Xie, C.Z.; Qiang, Z.Y.; Xu, J.Y. Thiosemicarbazone Cu(II) and Zn(II) complexes as potential anticancer agents: Syntheses, crystal structure, DNA cleavage, cytotoxicity and apoptosis induction activity. J. Inorg. Biochem. 2014, 136, 13–23. [Google Scholar] [CrossRef]
- Kelkel, M.; Jacob, C.; Dicato, M.; Diederich, M. Potential of the dietary antioxidants resveratrol and curcumin in prevention and treatment of hematologic malignancies. Molecules 2010, 15, 7035–7074. [Google Scholar] [CrossRef] [Green Version]
- Linden, A.; Gülden, M.; Martin, H.J.; Maser, E.; Seibert, H. Peroxide-induced cell death and lipid peroxidation in C6 glioma cells. Toxicol. Vitr. 2008, 22, 1371–1376. [Google Scholar] [CrossRef]
- Draper, H.H.; Hadley, M. Malondialdehyde determination as index of lipid Peroxidation. Methods Enzymol. 1990, 186, 421–431. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, M.A.; Singh, G.; Kashyap, M.P.; Khanna, V.K.; Yadav, S.; Chandra, D.; Pant, A.B. Influence of cytotoxic doses of 4-hydroxynonenal on selected neurotransmitter receptors in PC-12 cells. Toxicol. Vitr. 2008, 22, 1681–1688. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, M.A.; Kashyap, M.P.; Kumar, V.; Al-Khedhairy, A.A.; Musarrat, J.; Pant, A.B. Protective potential of trans-resveratrol against 4-hydroxynonenal induced damage in PC12 cells. Toxicol. In Vitro 2010, 24, 1592–1598. [Google Scholar] [CrossRef]
- Siddiqui, M.A.; Ahmad, J.; Farshori, N.N.; Saquib, Q.; Jahan, S.; Kashyap, M.P.; Ahamed, M.; Musarrat, J.; Al-Khedhairy, A.A. Rotenone-induced oxidative stress and apoptosis in human liver HepG2 cells. Mol. Cell. Biochem. 2013, 384, 59–69. [Google Scholar] [CrossRef]
- Chandra, D.; Ramana, K.V.; Wang, L.; Christensen, B.N.; Bhatnagar, A.; Srivastava, S.K. Inhibition of fiber cell globulization and hyperglycemia-induced lens opacification by aminopeptidase inhibitor bestatin. Investig. Ophthalmol. Vis. Sci. 2002, 43, 2285–2292. [Google Scholar]
- Buege, J.A.; Aust, S.D. Microsomal lipid peroxidation. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1978; Volume 52, pp. 302–310. [Google Scholar]
- Tabassum, S.; Asim, A.; Khan, R.A.; Hussain, Z.; Srivastav, S.; Srikrishna, S.; Arjmand, F. Chiral heterobimetallic complexes targeting human DNA-topoisomerase Iα. Dalton Trans. 2013, 42, 16749–16761. [Google Scholar] [CrossRef]
- Khan, R.A.; Yadav, S.; Hussain, Z.; Arjmand, F.; Tabassum, S. Carbohydrate linked organotin(iv) complexes as human topoisomerase Iα inhibitor and their antiproliferative effects against the human carcinoma cell line. Dalton Trans. 2014, 43, 2534–2548. [Google Scholar] [CrossRef] [PubMed]
- Yousuf, I.; Arjmand, F.; Tabassum, S.; Toupet, L.; Khan, R.A.; Siddiqui, M.A. Mechanistic insights into a novel chromone-appended Cu(II) anticancer drug entity: In vitro binding profile with DNA/RNA substrates and cytotoxic activity against MCF-7 and HepG2 cancer cells. Dalton Trans. 2015, 44, 10330–10342. [Google Scholar] [CrossRef]
- Tabassum, S.; Asim, A.; Khan, R.A.; Arjmand, F.; Rajakumar, D.; Balaji, P.; Akbarsha, M.A. A multifunctional molecular entity CuII-SnIV heterobimetallic complex as a potential cancer chemotherapeutic agent: DNA binding/cleavage, SOD mimetic, topoisomerase Iα inhibitory and in vitro cytotoxic activities. RSC Adv. 2015, 5, 47439–47450. [Google Scholar] [CrossRef]
- Hassan, I.; Ebaid, H.; Alhazza, I.M.; Al-Tamimi, J.; Aman, S.; Abdel-Mageed, A.M. Copper mediates anti-inflammatory and antifibrotic activity of gleevec in hepatocellular carcinoma-induced male rats. Can. J. Gastroenterol. Hepatol. 2019, 2019, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Alhazza, I.M.; Hassan, I.; Ebaid, H.; Al-Tamimi, J.; Alwasel, S.H. Chemopreventive effect of riboflavin on the potassium bromate–induced renal toxicity in vivo. Naunyn. Schmiedebergs. Arch. Pharmacol. 2020, 393, 2355–2364. [Google Scholar] [CrossRef]
- Jollow, D.; Mitchell, J.R.; Zampaglione, N.; Gillette, J.R. Bromobenzene-induced liver necrosis. Protective role of glutathione and evidence for 3,4-bromobenzene oxide as the hepatotoxic metabolite. Pharmacology 1974, 11, 151–169. [Google Scholar] [CrossRef]
- Singh, N.P.; McCoy, M.T.; Tice, R.R.; Schneider, E.L. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp. Cell Res. 1988, 175, 184–191. [Google Scholar] [CrossRef] [Green Version]
- Hassan, I.; Chibber, S.; Khan, A.A.; Naseem, I. Riboflavin ameliorates cisplatin induced toxicities under photoillumination. PLoS ONE 2012, 7, e36273. [Google Scholar] [CrossRef] [PubMed]
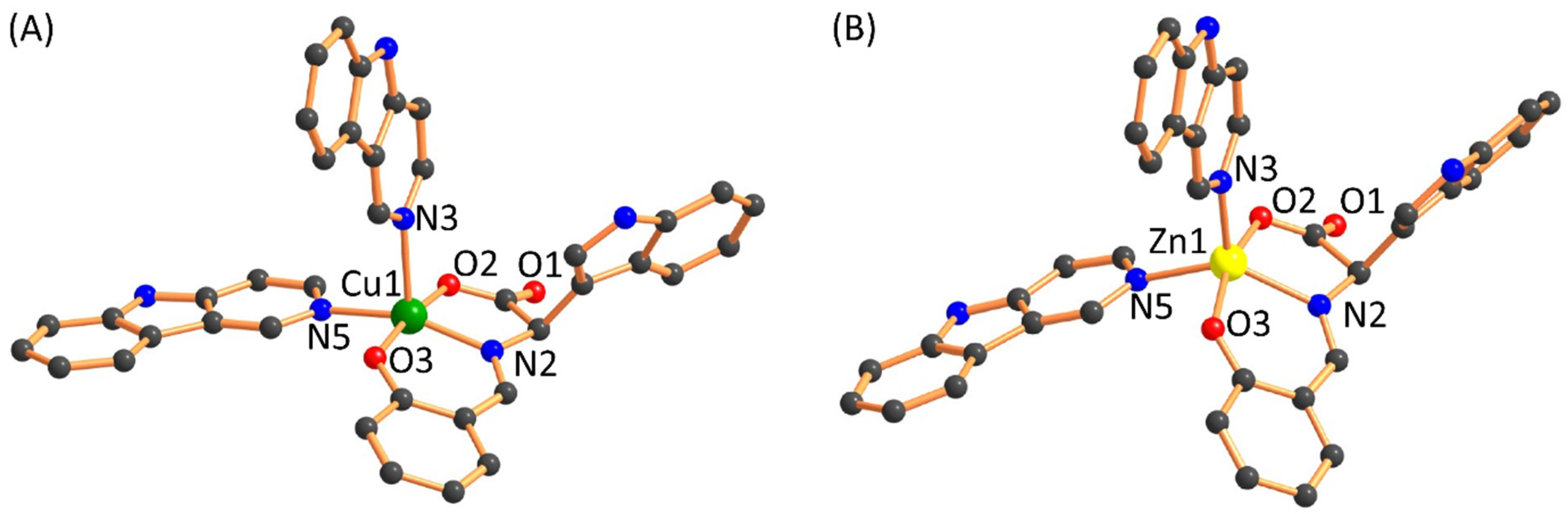

| Bonds | 1 (Calculated) | 2 (Calculated) | 1 (Experimental) | 2 (Experimental) | References |
|---|---|---|---|---|---|
| Cu(1)-N(2)/Zn(1)-N(2) | 1.99 | 2.136 | ~1.981–2.015 | ~2.000–2.280 | [31,33,34,35] |
| Cu(1)-N(3)/Zn(1)-N(3) | 2.346 | 2.134 | |||
| Cu(1)-N(5)/Zn(1)-N(5) | 2.116 | 2.161 | |||
| Cu(1)-O(2)/Zn(1)-O(2) | 1.984 | 2.061 | ~1.952–2.325 | ~1.990–2.089 | [31,33,34,36] |
| Cu(1)-O(3)/Zn(1)-O(3) | 1.958 | 2.069 |
| Vibrational Band | Complex 1 (Experimental) | Complex 1 (Calculated) | Complex 2 (Experimental) | Complex 2 (Calculated) |
|---|---|---|---|---|
| ν(N-H) stretching | 3418 | 3509 | 3412 | 3502 |
| ν(N=CH) stretching | 1601 | 1610 | 1597 | 1609 |
| ν(-O˗C=O) stretching | 1627 | 1656 | 1629 | 1653 |
| Complex | HepG2 (µM) | MCF7 (µM) | HEK293 (µM) | References |
|---|---|---|---|---|
| [Cu(L)(Hnor)2] (1) | 21.7 ± 0.9 | 7.8 ± 0.4 | >100 | This work |
| [Zn(L)(Hnor)2] (2) | 50 ± 1.5 | 55 ± 1.9 | >200 | This work |
| [Cu(tryp) (Hnor)2] | 27.0 ± 1.1 | 10 ± 1.3 | >100 | [37] |
| [Zn(tryp)(Hnor)2] | 88 ± 1.9 | 24 ± 1.7 | >150 | [37] |
| [Cu(Sal-Trp)(dppz)] | ND | 32.56 ± 3.05 | ND | [31] |
| [Cu(Sal-Trp)(nip)] | ND | 20.33 ± 5.50 | ND | [31] |
| Cisplatin | 7.63 ± 1.6 | 38 ± 1.23 | >50 | [38,39] |
| Cu(NO3)2 | >200 | >200 | >200 | [37] |
| Zn(NO3)2 | >200 | >200 | >200 | [37] |
| Hnor | >200 | >200 | >200 | This work |
| L | >200 | >200 | >200 | This work |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alharbi, W.; Hassan, I.; Khan, R.A.; Parveen, S.; Alharbi, K.H.; Bin Sharfan, I.I.; Alhazza, I.M.; Ebaid, H.; Alsalme, A. Bioactive Tryptophan-Based Copper Complex with Auxiliary β-Carboline Spectacle Potential on Human Breast Cancer Cells: In Vitro and In Vivo Studies. Molecules 2021, 26, 1606. https://doi.org/10.3390/molecules26061606
Alharbi W, Hassan I, Khan RA, Parveen S, Alharbi KH, Bin Sharfan II, Alhazza IM, Ebaid H, Alsalme A. Bioactive Tryptophan-Based Copper Complex with Auxiliary β-Carboline Spectacle Potential on Human Breast Cancer Cells: In Vitro and In Vivo Studies. Molecules. 2021; 26(6):1606. https://doi.org/10.3390/molecules26061606
Chicago/Turabian StyleAlharbi, Walaa, Iftekhar Hassan, Rais Ahmad Khan, Shazia Parveen, Khadijah H. Alharbi, Ibtisam I. Bin Sharfan, Ibrahim M. Alhazza, Hossam Ebaid, and Ali Alsalme. 2021. "Bioactive Tryptophan-Based Copper Complex with Auxiliary β-Carboline Spectacle Potential on Human Breast Cancer Cells: In Vitro and In Vivo Studies" Molecules 26, no. 6: 1606. https://doi.org/10.3390/molecules26061606






