Anti-Allergic Diarrhea Effect of Diosgenin Occurs via Improving Gut Dysbiosis in a Murine Model of Food Allergy
Abstract
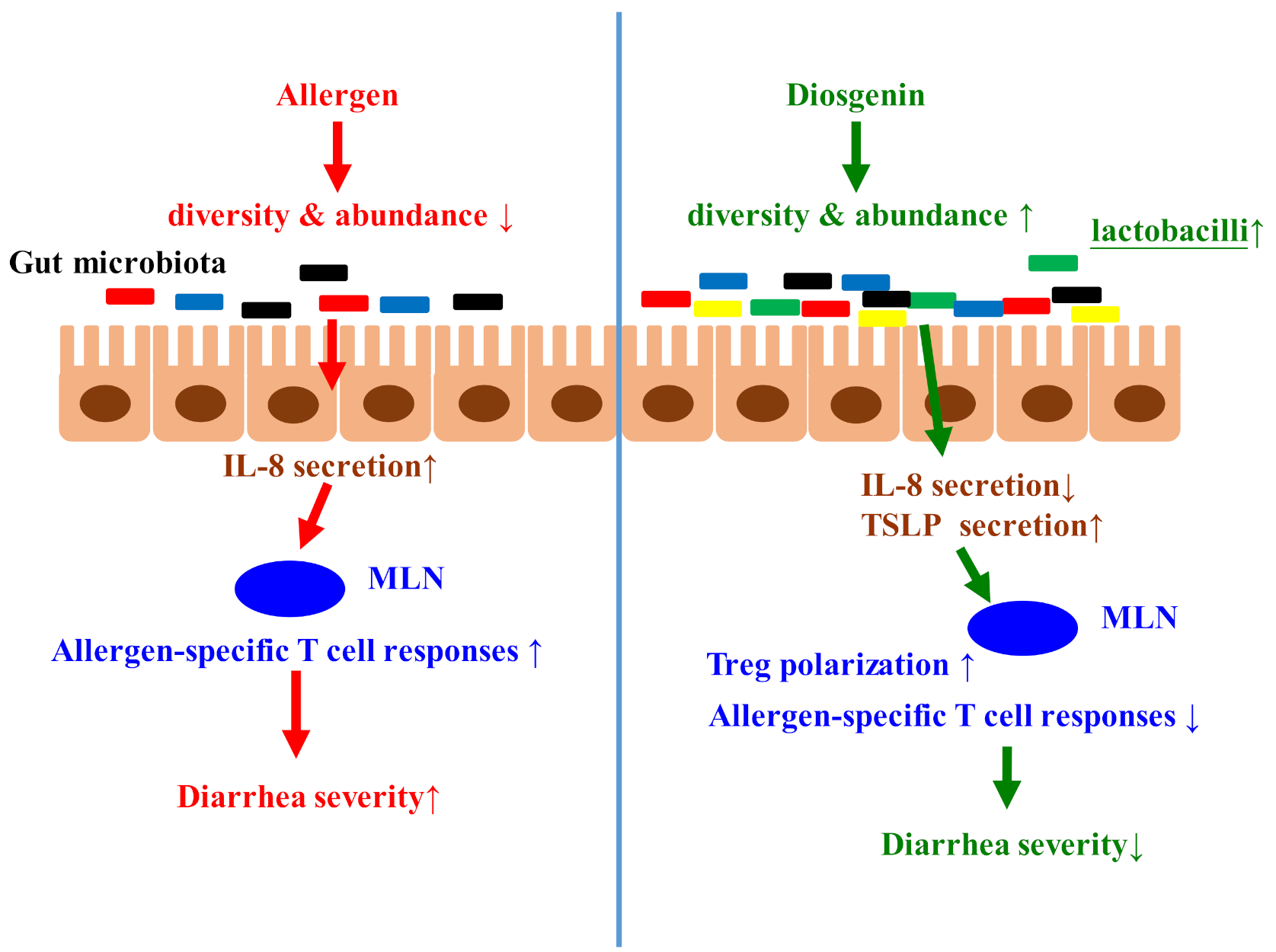
1. Introduction
2. Materials and Methods
2.1. Chemicals, Reagents and Cell Lines
2.2. Mice and Ethics Statement
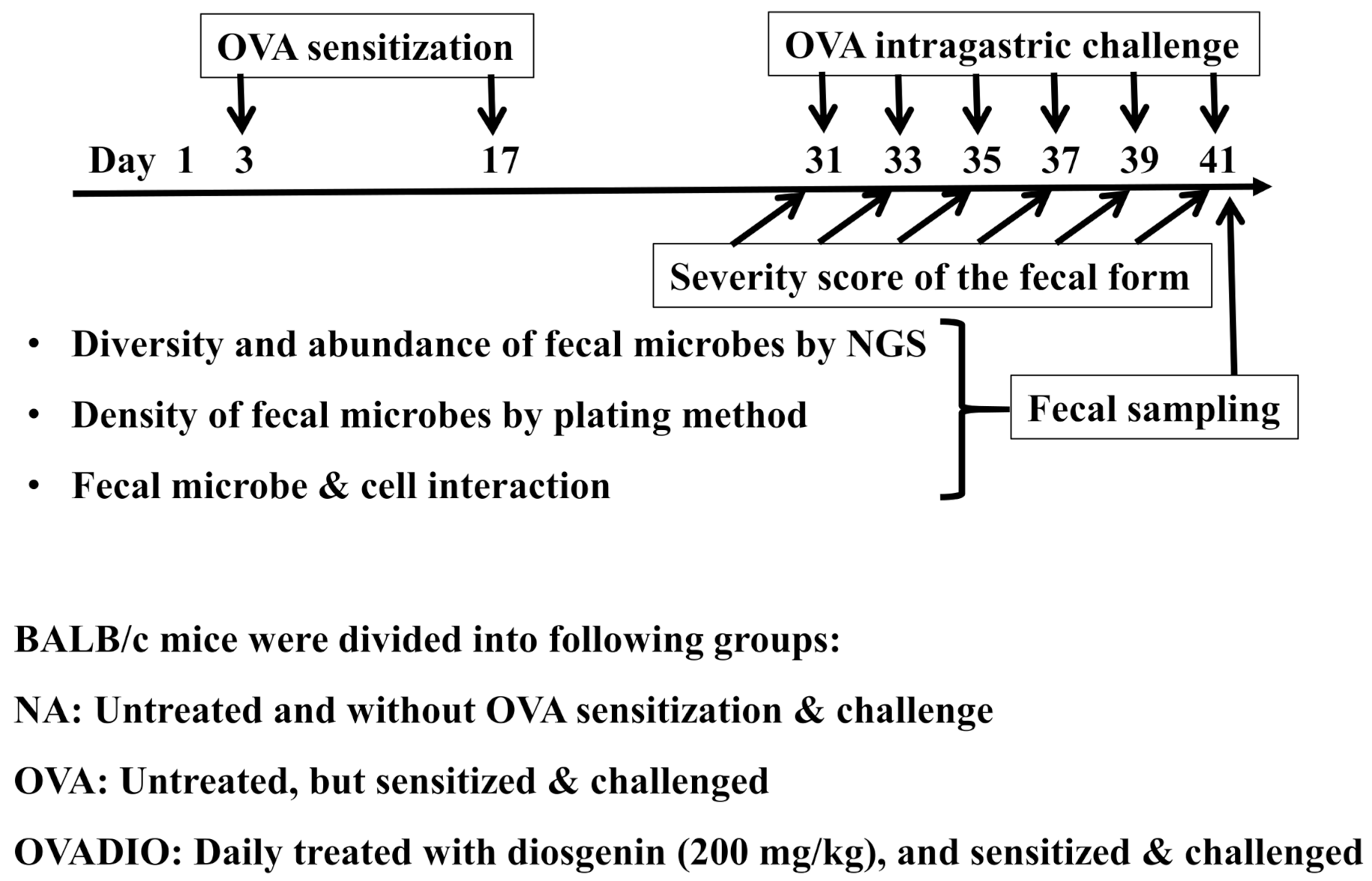
2.3. Protocol of Animal Experiments
2.4. Next-Generation Sequencing (NGS) Analysis
2.5. Culture of Fecal Bacteria
2.6. Identification of Relevant Fecal Microbes
2.7. Cell Culture Experiments
2.8. Statistical Analysis
3. Results
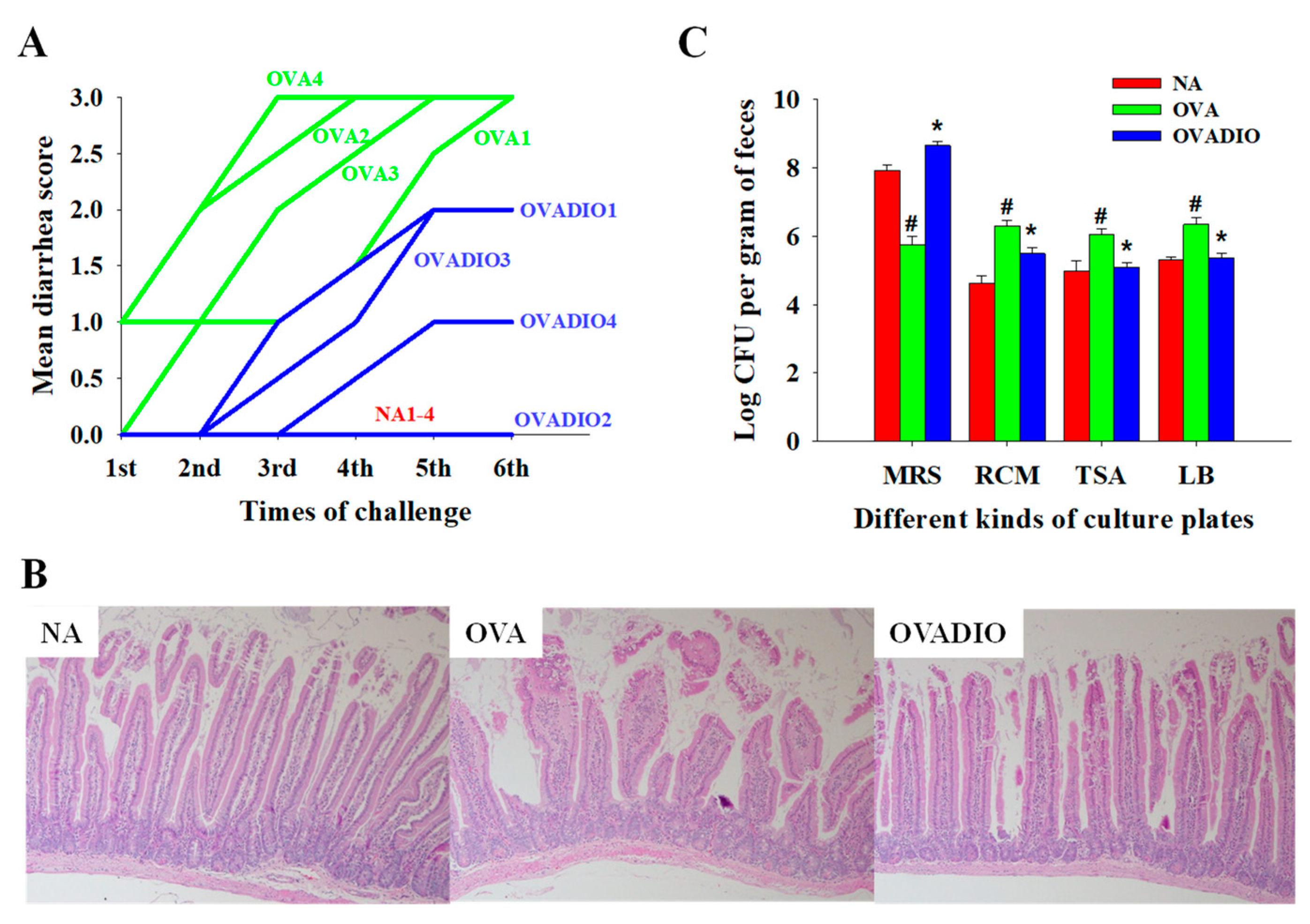
3.1. Severity of Allergic Diarrhea and Density of Fecal Bacteria
3.2. NGS Analysis of Fecal Microbes
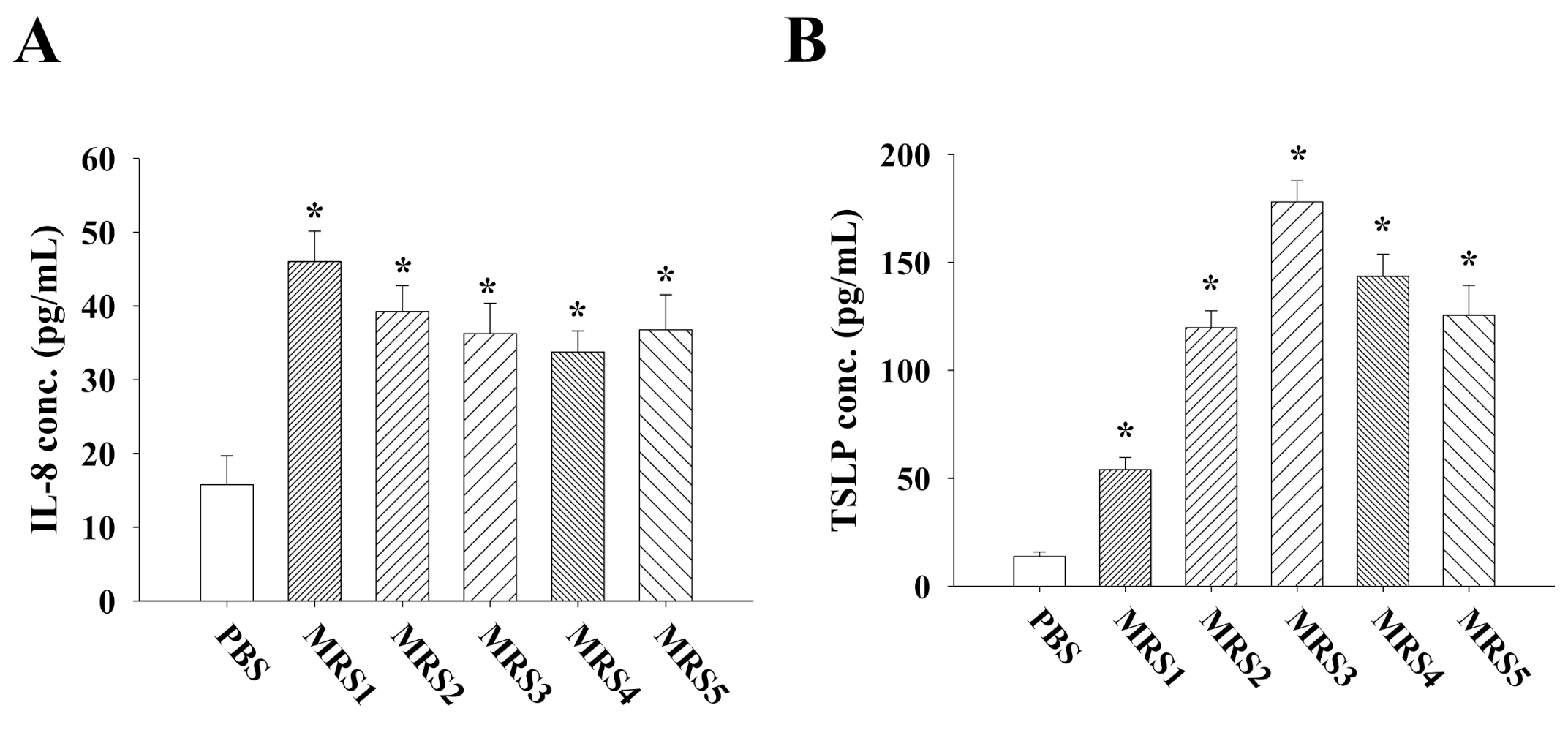
3.3. Impact of Fecal MRS Strains from Diosgenin-Treated Mice on Cytokine/Chemokine Production by Caco-2 Cells
3.4. Impact of Fecal MRS Strains from Diosgenin-Treated Mice on Cytokine Production by Allergen-Primed MLN Cells
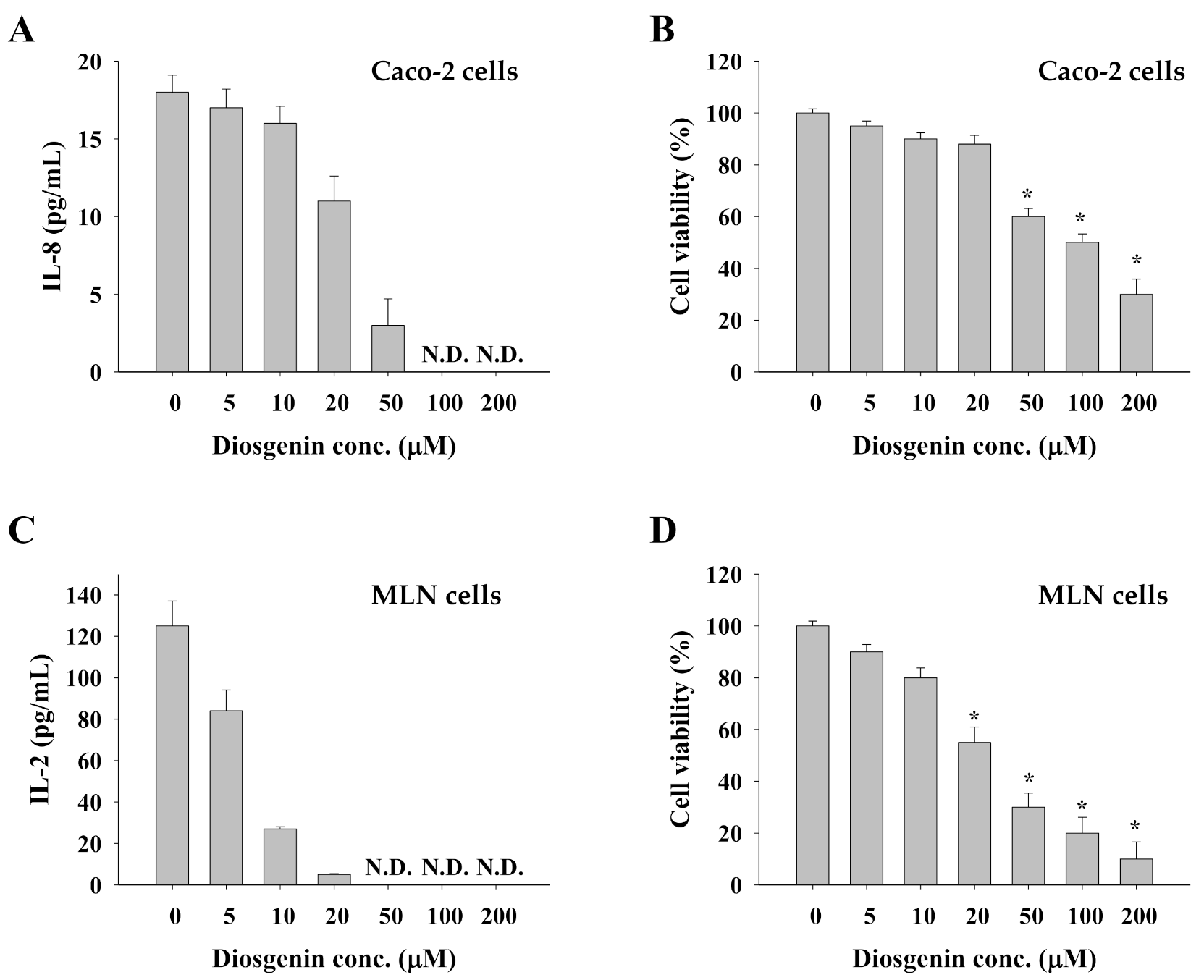
3.5. Impact of Diosgenin on Caco-2 Cells and Allergen-Primed MLN Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- De Martinis, M.; Sirufo, M.M.; Suppa, M.; Ginaldi, L. New perspectives in food allergy. Int. J. Mol. Sci. 2020, 21, 1474. [Google Scholar] [CrossRef]
- Björkstén, B.; Sepp, E.; Julge, K.; Voor, T.; Mikelsaar, M. Allergy development and the intestinal microflora during the first year of life. J. Allergy Clin. Immunol. 2001, 108, 516–520. [Google Scholar] [CrossRef]
- Rivas, M.N.; Burton, O.T.; Wise, P.; Zhang, Y.-q.; Hobson, S.A.; Lloret, M.G.; Chehoud, C.; Kuczynski, J.; DeSantis, T.; Warrington, J. A microbiota signature associated with experimental food allergy promotes allergic sensitization and anaphylaxis. J. Allergy Clin. Immunol. 2013, 131, 201–212. [Google Scholar] [CrossRef]
- Huang, C.-H.; Lu, S.-Y.; Tsai, W.-C. Relevant fecal microbes isolated from mice with food allergy elicited intestinal cytokine/chemokine network and T-cell immune responses. Biosci. Microbiota Food Health 2020, 39, 234–242. [Google Scholar] [CrossRef]
- Potaczek, D.P.; Harb, H.; Michel, S.; Alhamwe, B.A.; Renz, H.; Tost, J. Epigenetics and allergy: From basic mechanisms to clinical applications. Epigenomics 2017, 9, 539–571. [Google Scholar] [CrossRef]
- Alhamwe, B.A.; Khalaila, R.; Wolf, J.; von Bulow, V.; Harb, H.; Alhamdan, F.; Hii, C.S.; Prescott, S.L.; Ferrante, A.; Renz, H.; et al. Histone modifications and their role in epigenetics of atopy and allergic diseases. Allergy Asthma Clin. Immunol. 2018, 14, 39. [Google Scholar] [CrossRef]
- Tan, T.H.; Ellis, J.A.; Saery, R.; Allen, K.J. The role of genetics and environment in the rise of childhood food allergy. Clin. Exp. Allergy 2012, 42, 20–29. [Google Scholar] [CrossRef]
- Du Toit, G.; Tsakok, T.; Lack, S.; Lack, G. Prevention of food allergy. J. Allergy Clin. Immunol. 2016, 137, 998–1010. [Google Scholar] [CrossRef]
- Tost, J. A translational perspective on epigenetics in allergic diseases. J. Allergy Clin. Immunol. 2018, 142, 715–726. [Google Scholar] [CrossRef] [PubMed]
- Berni Canani, R.; Paparo, L.; Nocerino, R.; Cosenza, L.; Pezzella, V.; Di Costanzo, M.; Capasso, M.; Del Monaco, V.; D’Argenio, V.; Greco, L.; et al. Differences in DNA methylation profile of Th1 and Th2 cytokine genes are associated with tolerance acquisition in children with IgE-mediated cow’s milk allergy. Clin. Epigenet. 2015, 7, 38. [Google Scholar] [CrossRef]
- Alhamwe, B.A.; Meulenbroek, L.A.P.M.; Veening-Griffioen, D.H.; Wehkamp, T.M.D.; Alhamdan, F.; Miethe, S.; Harb, H.; Hogenkamp, A.; Knippels, L.M.J.; von Strandmann, E.P.; et al. Decreased histone acetylation levels at th1 and regulatory loci after induction of food allergy. Nutrients 2020, 12, 3193. [Google Scholar] [CrossRef]
- Abbring, S.; Wolf, J.; Ayechu-Muruzabal, V.; Diks, M.A.P.; Alhamwe, B.A.; Alhamdan, F.; Harb, H.; Renz, H.; Garn, H.; Garssen, J.; et al. Raw cow’s milk reduces allergic symptoms in a murine model for food allergy—A potential role for epigenetic modifications. Nutrients 2019, 11, 1721. [Google Scholar] [CrossRef]
- Huang, C.-H.; Ku, C.-Y.; Jan, T.-R. Diosgenin attenuates allergen-induced intestinal inflammation and IgE production in a murine model of food allergy. Planta Med. 2009, 75, 1300–1305. [Google Scholar] [CrossRef] [PubMed]
- Yamada, T.; Hoshino, M.; Hayakawa, T.; Ohhara, H.; Yamada, H.; Nakazawa, T.; Inagaki, T.; Iida, M.; Ogasawara, T.; Uchida, A. Dietary diosgenin attenuates subacute intestinal inflammation associated with indomethacin in rats. Am. J. Physiol. Gastrointest. Liver Physiol. 1997, 273, G355–G364. [Google Scholar] [CrossRef]
- Huang, C.-H.; Cheng, J.-Y.; Deng, M.-C.; Chou, C.-H.; Jan, T.-R. Prebiotic effect of diosgenin, an immunoactive steroidal sapogenin of the Chinese yam. Food Chem. 2012, 132, 428–432. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-H.; Shen, C.-C.; Liang, Y.-C.; Jan, T.-R. The probiotic activity of Lactobacillus murinus against food allergy. J. Funct. Foods 2016, 25, 231–241. [Google Scholar] [CrossRef]
- Huang, C.-H.; Lin, Y.-C.; Jan, T.-R. Lactobacillus reuteri induces intestinal immune tolerance against food allergy in mice. J. Funct. Foods 2017, 31, 44–51. [Google Scholar] [CrossRef]
- Dong, M.; Meng, Z.; Kuerban, K.; Qi, F.; Liu, J.; Wei, Y.; Wang, Q.; Jiang, S.; Feng, M.; Ye, L. Diosgenin promotes antitumor immunity and PD-1 antibody efficacy against melanoma by regulating intestinal microbiota. Cell Death Dis. 2018, 9, 1039. [Google Scholar] [CrossRef] [PubMed]
- Okawara, M.; Tokudome, Y.; Todo, H.; Sugibayashi, K.; Hashimoto, F. Enhancement of diosgenin distribution in the skin by cyclodextrin complexation following oral administration. Biol. Pharm. Bull. 2013, 36, 36–40. [Google Scholar] [CrossRef]
- Hasain, Z.; Mokhtar, N.M.; Kamaruddin, N.A.; Ismail, N.A.M.; Razalli, N.H.; Gnanou, J.V.; Ali, R.A.R. Gut microbiota and gestational diabetes mellitus: A review of host-gut microbiota interactions and their therapeutic potential. Front. Cell. Infect. Microbiol. 2020, 10, 188. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Liu, Y.; Guo, X.; Bai, Q.; Zhu, X.; Ren, H.; Chen, Q.; Yue, T.; Long, F. Antiallergic activity of Lactobacillus plantarum against peanut allergy in a Balb/c mouse model. Food Agric. Immunol. 2019, 30, 762–773. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Wang, X.; Gibson, G. Effects of the in vitro fermentation of oligofructose and inulin by bacteria growing in the human large intestine. J. Appl. Microbiol. 1993, 75, 373–380. [Google Scholar]
- Edwards, U.; Rogall, T.; Blöcker, H.; Emde, M.; Böttger, E.C. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989, 17, 7843–7853. [Google Scholar] [CrossRef]
- Chiu, Y.-H.; Lu, Y.-C.; Ou, C.-C.; Lin, S.-L.; Tsai, C.-C.; Huang, C.-T.; Lin, M.-Y. Lactobacillus plantarum MYL26 induces endotoxin tolerance phenotype in Caco-2 cells. BMC Microbiol. 2013, 13, 1–9. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, Z.; Yan, J.; Wu, X.; Xu, G. Diosgenin exerts antitumor activity via downregulation of Skp2 in breast cancer cells. BioMed Res. Int. 2020, 2020, 1–10. [Google Scholar] [CrossRef]
- Plunkett, C.H.; Nagler, C.R. The influence of the microbiome on allergic sensitization to food. J. Immunol. 2017, 198, 581–589. [Google Scholar] [CrossRef]
- Eckmann, L.; Jung, H.C.; Schürer-Maly, C.; Panja, A.; Morzycka-Wroblewska, E.; Kagnoff, M.F. Differential cytokine expression by human intestinal epithelial cell lines: Regulated expression of interleukin 8. Gastroenterology 1993, 105, 1689–1697. [Google Scholar] [CrossRef]
- Rimoldi, M.; Chieppa, M.; Salucci, V.; Avogadri, F.; Sonzogni, A.; Sampietro, G.M.; Nespoli, A.; Viale, G.; Allavena, P.; Rescigno, M. Intestinal immune homeostasis is regulated by the crosstalk between epithelial cells and dendritic cells. Nat. Immunol. 2005, 6, 507–514. [Google Scholar] [CrossRef]
- Iliev, I.D.; Spadoni, I.; Mileti, E.; Matteoli, G.; Sonzogni, A.; Sampietro, G.M.; Foschi, D.; Caprioli, F.; Viale, G.; Rescigno, M. Human intestinal epithelial cells promote the differentiation of tolerogenic dendritic cells. Gut 2009, 58, 1481–1489. [Google Scholar] [CrossRef]
- Ziegler, S.F.; Roan, F.; Bell, B.D.; Stoklasek, T.A.; Kitajima, M.; Han, H. The biology of thymic stromal lymphopoietin (TSLP). Adv. Pharmacol. 2013, 66, 129–155. [Google Scholar]
- Zimmerman, N.P.; Vongsa, R.A.; Wendt, M.K.; Dwinell, M.B. Chemokines and chemokine receptors in mucosal homeostasis at the intestinal epithelial barrier in inflammatory bowel disease. Inflamm. Bowel Dis. 2008, 14, 1000–1011. [Google Scholar] [CrossRef]
- Nelson, B.H. IL-2, regulatory T cells, and tolerance. J. Immunol. 2004, 172, 3983–3988. [Google Scholar] [CrossRef] [PubMed]
- Talaat, R.M.; Mohamed, S.F.; Bassyouni, I.H.; Raouf, A.A. Th1/Th2/Th17/Treg cytokine imbalance in systemic lupus erythematosus (SLE) patients: Correlation with disease activity. Cytokine 2015, 72, 146–153. [Google Scholar] [CrossRef]
- Ikarashi, N.; Fujitate, N.; Togashi, T.; Takayama, N.; Fukuda, N.; Kon, R.; Sakai, H.; Kamei, J.; Sugiyama, K. Acacia polyphenol ameliorates atopic dermatitis in trimellitic anhydride-induced model mice via changes in the gut microbiota. Foods 2020, 9, 773. [Google Scholar] [CrossRef]
- Li, B.-Y.; Xu, X.-Y.; Gan, R.-Y.; Sun, Q.-C.; Meng, J.-M.; Shang, A.; Mao, Q.-Q.; Li, H.-B. Targeting gut microbiota for the prevention and management of diabetes mellitus by dietary natural products. Foods 2019, 8, 440. [Google Scholar] [CrossRef]
- Bloomfield, S.F.; Rook, G.A.; Scott, E.A.; Shanahan, F.; Stanwell-Smith, R.; Turner, P. Time to abandon the hygiene hypothesis: New perspectives on allergic disease, the human microbiome, infectious disease prevention and the role of targeted hygiene. Perspect. Public Health 2016, 136, 213–224. [Google Scholar] [CrossRef]
- Stefka, A.T.; Feehley, T.; Tripathi, P.; Qiu, J.; McCoy, K.; Mazmanian, S.K.; Tjota, M.Y.; Seo, G.-Y.; Cao, S.; Theriault, B.R. Commensal bacteria protect against food allergen sensitization. Proc. Natl. Acad. Sci. USA 2014, 111, 13145–13150. [Google Scholar] [CrossRef]
- Rogier, R.; Evans-Marin, H.; Manasson, J.; van der Kraan, P.M.; Walgreen, B.; Helsen, M.M.; van den Bersselaar, L.A.; van de Loo, F.A.; Van Lent, P.L.; Abramson, S.B. Alteration of the intestinal microbiome characterizes preclinical inflammatory arthritis in mice and its modulation attenuates established arthritis. Sci. Rep. 2017, 7, 1–12. [Google Scholar]
- Biagi, G.; Cipollini, I.; Pompei, A.; Zaghini, G.; Matteuzzi, D. Effect of a Lactobacillus animalis strain on composition and metabolism of the intestinal microflora in adult dogs. Vet. Microbiol. 2007, 124, 160–165. [Google Scholar] [CrossRef]
- Niccoli, A.A.; Artesi, A.L.; Candio, F.; Ceccarelli, S.; Cozzali, R.; Ferraro, L.; Fiumana, D.; Mencacci, M.; Morlupo, M.; Pazzelli, P. Preliminary results on clinical effects of probiotic Lactobacillus salivarius LS01 in children affected by atopic dermatitis. J. Clin. Gastroenterol. 2014, 48, S34–S36. [Google Scholar] [CrossRef]
- Yun, X.; Shang, Y.; Li, M. Effect of Lactobacillus salivarius on Th1/Th2 cytokines and the number of spleen CD4+ CD25+ Foxp3+ Treg in asthma Balb/c mouse. Int. J. Clin. Exp. Pathol. 2015, 8, 7661. [Google Scholar]


| Isolate | Strain | Similarity (%) |
|---|---|---|
| MRS1 | Lactobacillus murinus strain NBRC 14221 | 100 |
| MRS2 | Lactobacillus animalis strain KCTC 3501 | 100 |
| MRS3 | Lactobacillus apodemi strain DSM 16634 | 99 |
| MRS4 | Lactobacillus salivarius strain JCM 1231 | 99 |
| MRS5 | Lactobacillus faecis strain AFL13-2 | 100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, C.-H.; Pan, C.-L.; Tsai, G.-J.; Chang, C.-J.; Tsai, W.-C.; Lu, S.-Y. Anti-Allergic Diarrhea Effect of Diosgenin Occurs via Improving Gut Dysbiosis in a Murine Model of Food Allergy. Molecules 2021, 26, 2471. https://doi.org/10.3390/molecules26092471
Huang C-H, Pan C-L, Tsai G-J, Chang C-J, Tsai W-C, Lu S-Y. Anti-Allergic Diarrhea Effect of Diosgenin Occurs via Improving Gut Dysbiosis in a Murine Model of Food Allergy. Molecules. 2021; 26(9):2471. https://doi.org/10.3390/molecules26092471
Chicago/Turabian StyleHuang, Chung-Hsiung, Chorng-Liang Pan, Guo-Jane Tsai, Chun-Ju Chang, Wei-Chung Tsai, and Shueh-Yu Lu. 2021. "Anti-Allergic Diarrhea Effect of Diosgenin Occurs via Improving Gut Dysbiosis in a Murine Model of Food Allergy" Molecules 26, no. 9: 2471. https://doi.org/10.3390/molecules26092471
APA StyleHuang, C.-H., Pan, C.-L., Tsai, G.-J., Chang, C.-J., Tsai, W.-C., & Lu, S.-Y. (2021). Anti-Allergic Diarrhea Effect of Diosgenin Occurs via Improving Gut Dysbiosis in a Murine Model of Food Allergy. Molecules, 26(9), 2471. https://doi.org/10.3390/molecules26092471







