Synthesis of Phenylboronic Acid-Functionalized Magnetic Nanoparticles for Sensitive Soil Enzyme Assays
Abstract
:1. Introduction
2. Results
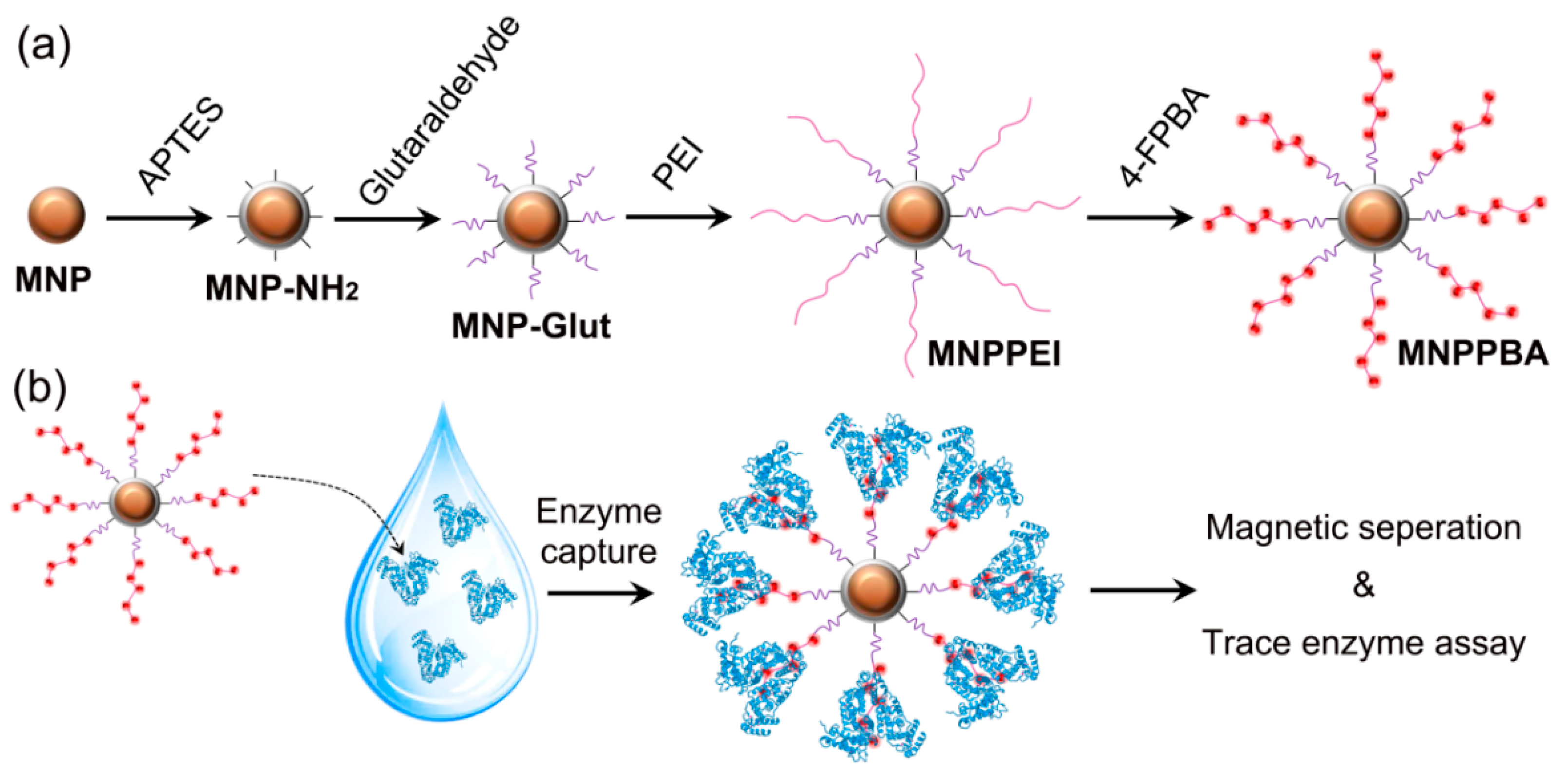
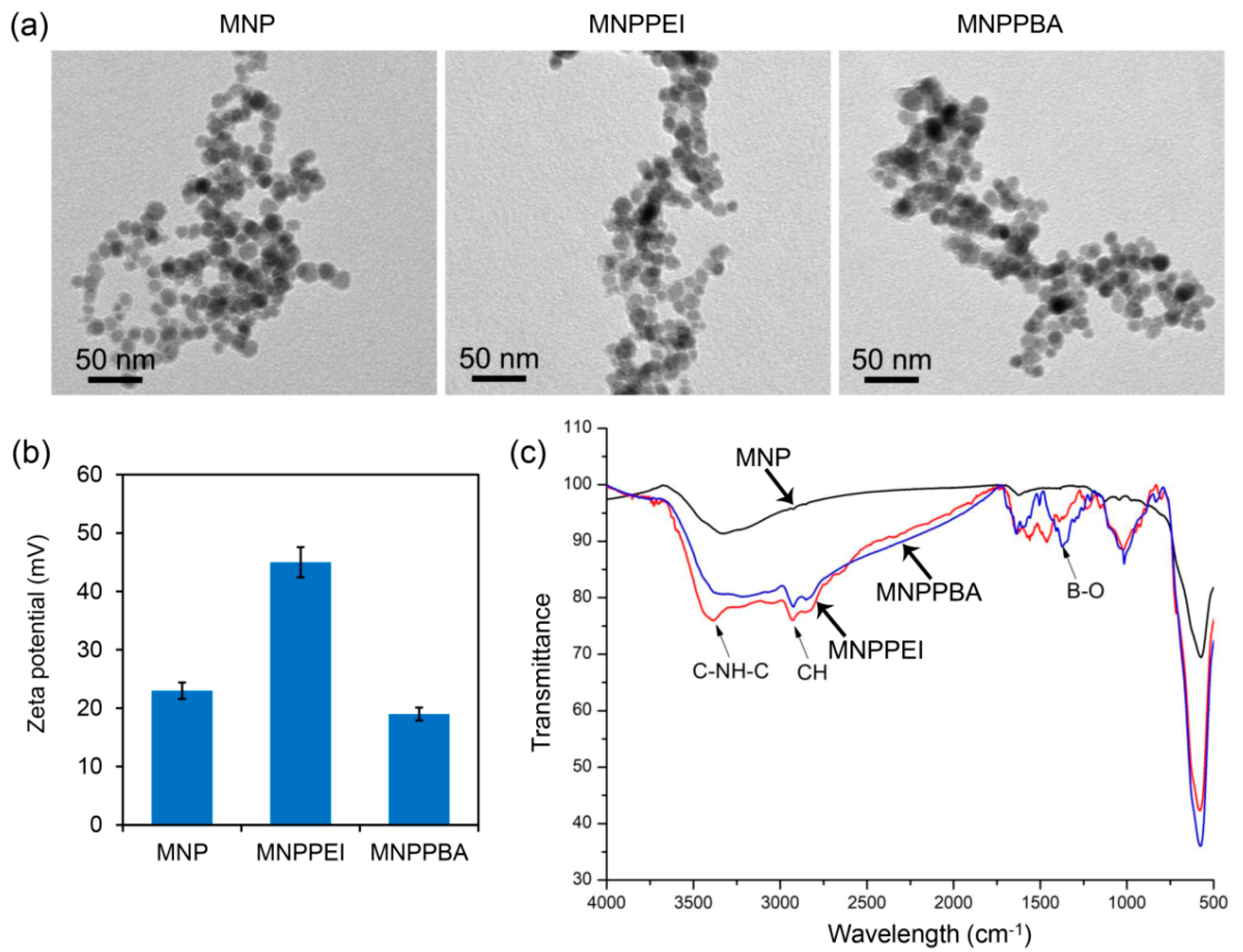
2.1. Synthesis of the Phenylboronic Acid-Grafted Magnetic Nanoparticles
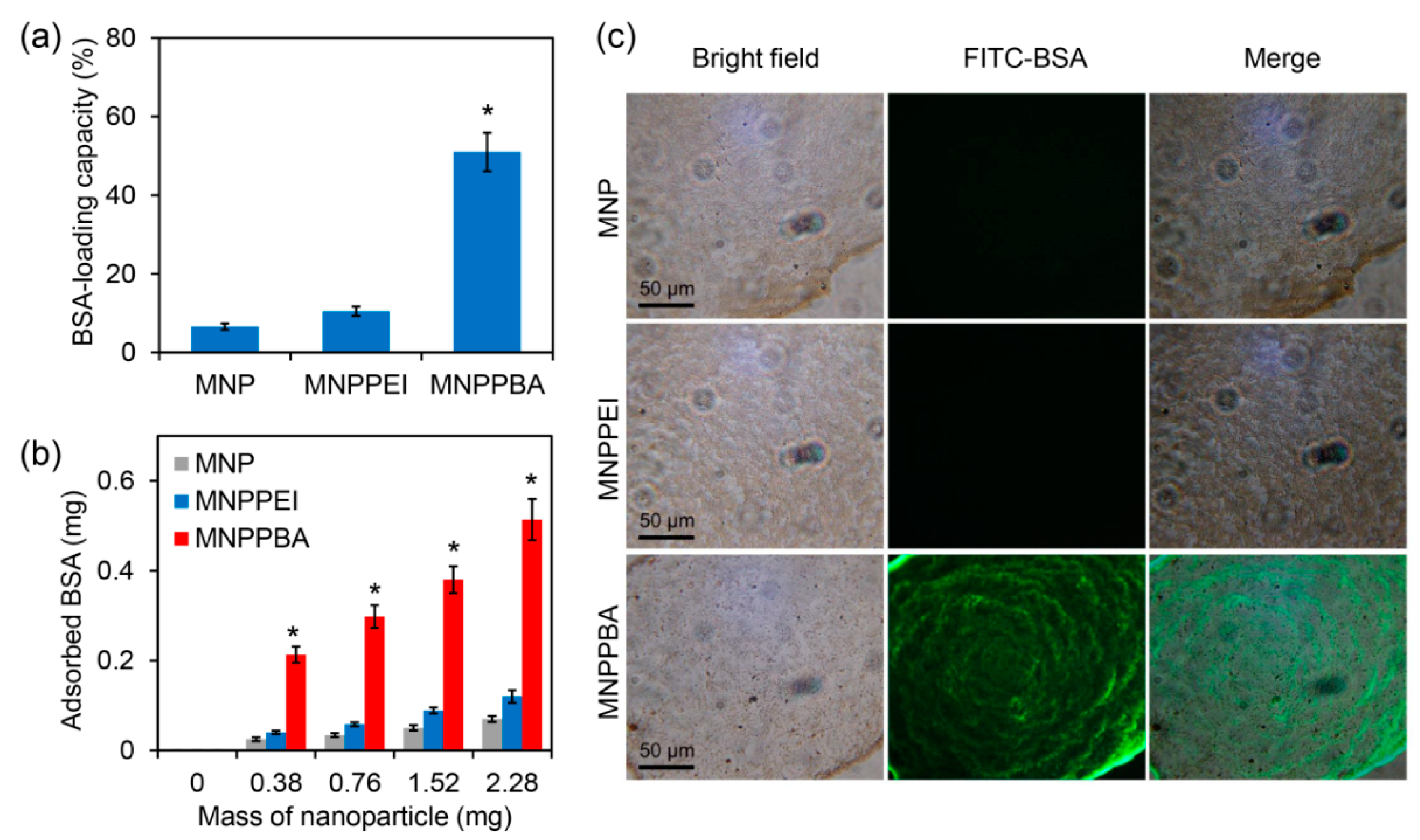
2.2. Efficient Protein Capture by the Synthesized Magnetic Nanoparticles
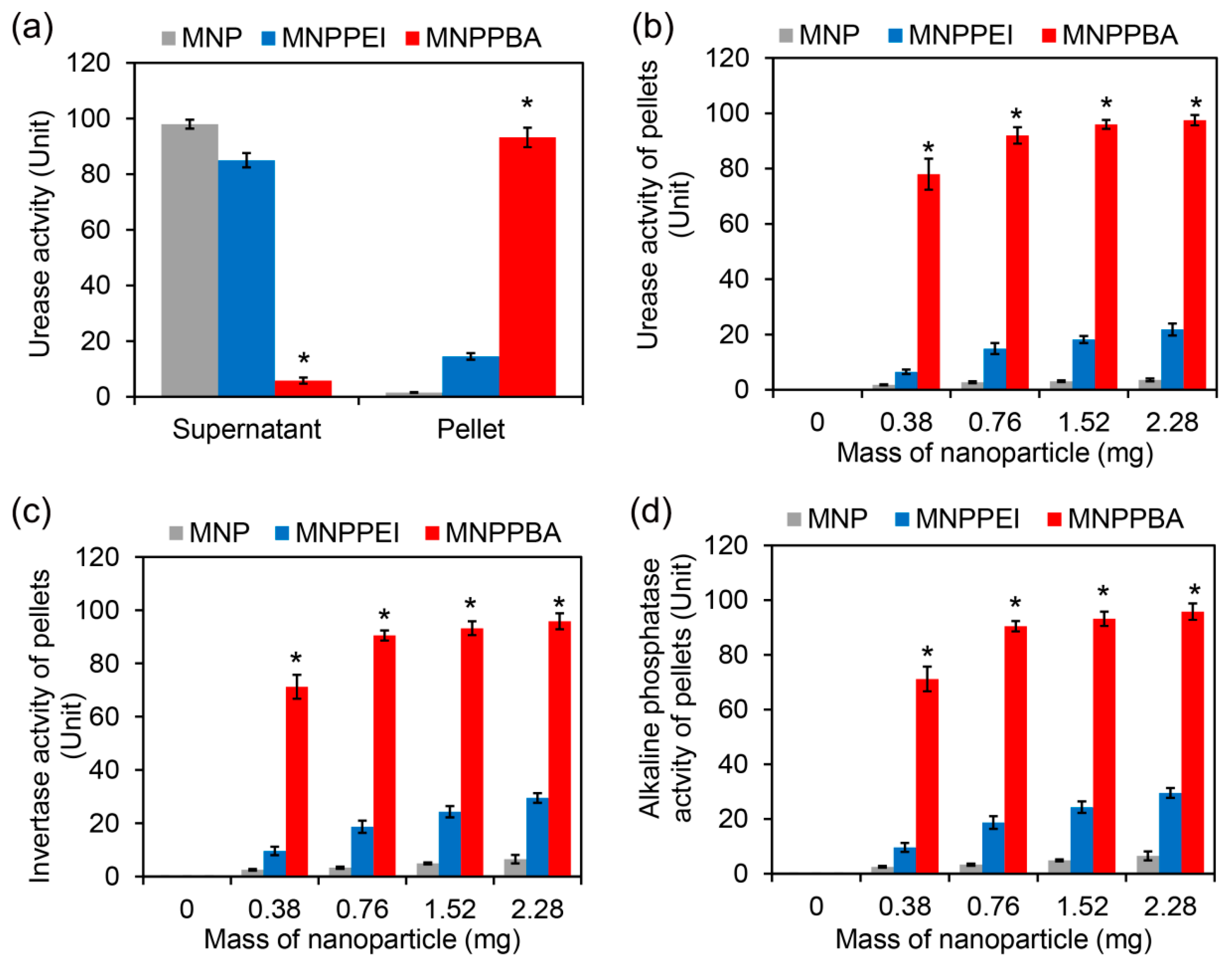
2.3. Efficient Enrichment of Enzymes in Rhizosphere Soil Supernatants by the Synthesized Magnetic Nanoparticles
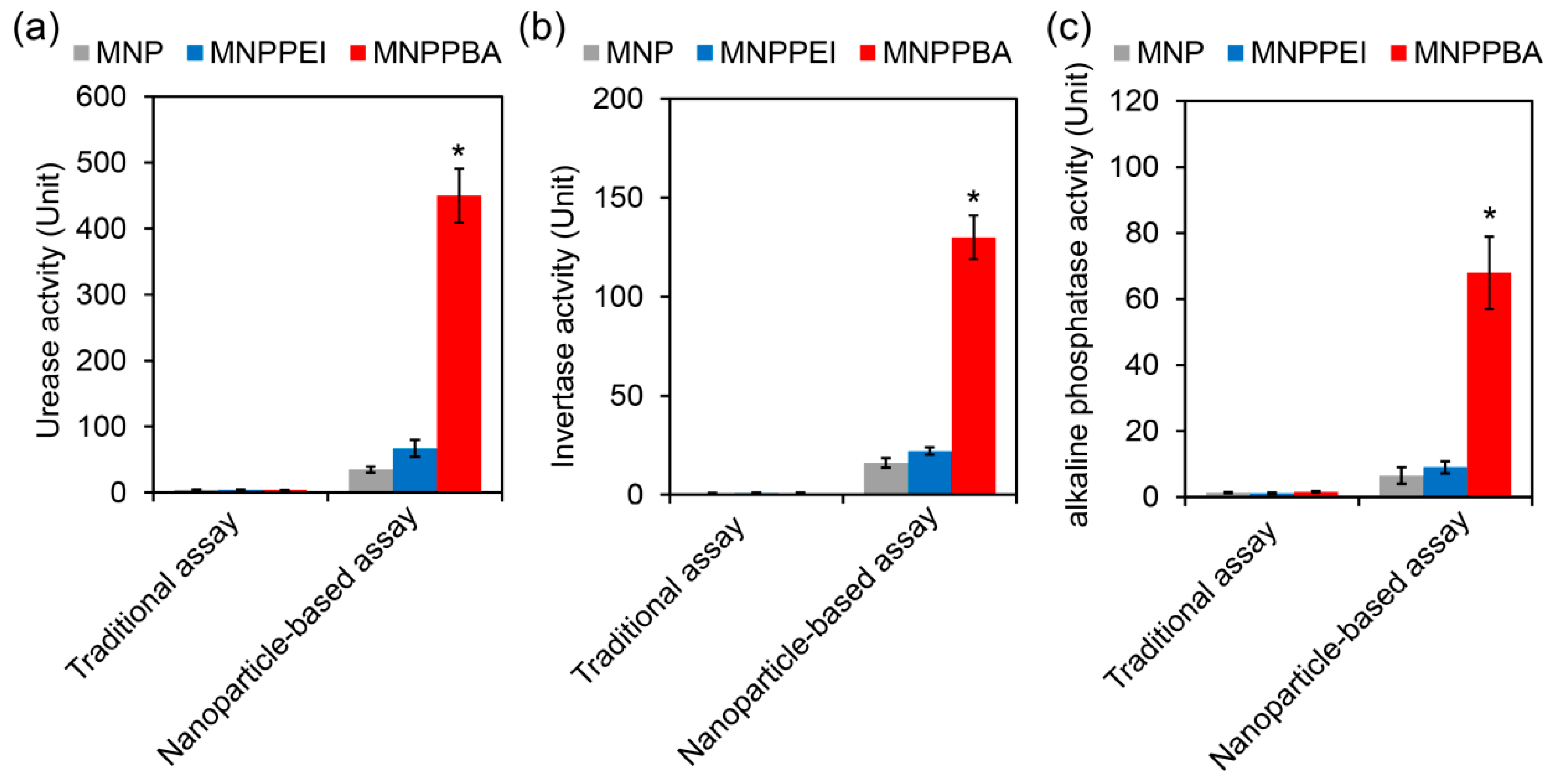
2.4. Sensitive Rhizosphere Enzyme Assay Based on the Synthesized Magnetic Nanoparticles
3. Materials and Methods
3.1. Chemical Agents
3.2. Synthesis of the Magnetic Nanoparticles
3.3. Characterization of the Magnetic Nanoparticles
3.4. Protein-Capturing Assays
3.5. Enzyme-Capturing Assays
3.6. Enrichment of Enzymes in Rhizosphere Soil Supernatants and Enzyme Assays
3.7. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Wang, Y.; Yan, D.; Wang, J.; Ding, Y.; Song, X. Effects of elevated CO2 and drought on plant physiology, soil carbon and soil enzyme activities. Pedosphere 2017, 27, 846–855. [Google Scholar] [CrossRef]
- Liu, X.; Li, C.; Meng, M.; Zhai, L.; Zhang, B.; Jia, Z.; Gu, Z.; Liu, Q.; Zhang, Y.; Zhang, J. Comparative effects of the recovery from sulfuric and nitric acid rain on the soil enzyme activities and metabolic functions of soil microbial communities. Sci. Total Environ. 2020, 714, 136788. [Google Scholar] [CrossRef]
- Ndossi, E.M.; Becker, J.N.; Hemp, A.; Dippold, M.A.; Kuzyakov, Y.; Razavi, B.S. Effects of land use and elevation on the functional characteristics of soil enzymes at Mt. Kilimanjaro. Eur. J. Soil Biol. 2020, 97, 103167. [Google Scholar] [CrossRef]
- Wahsha, M.; Nadimi-Goki, M.; Fornasier, F.; Al-Jawasreh, R.; Hussein, E.I.; Bini, C. Microbial enzymes as an early warning management tool for monitoring mining site soils. Catena 2017, 148, 40–45. [Google Scholar] [CrossRef]
- Zhou, Y.; Staver, A.C. Enhanced activity of soil nutrient-releasing enzymes after plant invasion: A meta-analysis. Ecology 2019, 100, e02830. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, W.A.; Nodeh, H.R.; Sanagi, M.M. Graphene-based materials as solid phase extraction sorbent for trace metal ions, organic compounds, and biological sample preparation. Crit. Rev. Anal. Chem. 2016, 46, 267–283. [Google Scholar] [CrossRef]
- Deng, S.; Popova, I.E.; Dick, L.; Dick, R. Bench scale and microplate format assay of soil enzyme activities using spectroscopic and fluorometric approaches. Appl. Soil Ecol. 2013, 64, 84–90. [Google Scholar] [CrossRef]
- Khosravi-Darani, K.; Vasheghani-Farahani, E. Application of supercritical fluid extraction in biotechnology. Crit. Rev. Biotechnol. 2005, 25, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Guo, Z.; Wang, X.; Qiu, C. Sample preparation. J. Chromatogr. 2008, 1184, 191–219. [Google Scholar] [CrossRef]
- Ding, H.; Fazelinia, H.; Spruce, L.A.; Weiss, D.A.; Zderic, S.A.; Seeholzer, S.H. Urine proteomics: Evaluation of different sample preparation workflows for quantitative, reproducible, and improved depth of analysis. J. Proteome Res. 2020, 19, 1857–1862. [Google Scholar] [CrossRef] [PubMed]
- Yin, P.; Sun, N.; Deng, C.; Li, Y.; Zhang, X.; Yang, P. Facile preparation of magnetic graphene double-sided mesoporous composites for the selective enrichment and analysis of endogenous peptides. Proteomics 2013, 13, 2243–2250. [Google Scholar] [CrossRef]
- Xue, S.; Wang, Y.; Wu, D.; Shen, J.; Wei, Y.; Wang, C. Core-shell structured magnetic mesoporous carbon nanospheres derived from metal-polyphenol coordination polymer-coated Fe3O4 and its application in the enrichment of phthalates from water samples. J. Sep. Sci. 2019, 42, 3512–3520. [Google Scholar] [CrossRef]
- Li, Q.; Zhou, X.; Xing, D. Rapid and highly sensitive detection of mercury ion (Hg2+) by magnetic beads-based electrochemiluminescence assay. Biosens. Bioelectron. 2010, 26, 859–862. [Google Scholar] [CrossRef]
- Zhou, J.; Liang, Y.; He, X.; Chen, L.; Zhang, Y. Dual-functionalized magnetic metal–organic framework for highly specific enrichment of phosphopeptides. ACS Sustain. Chem. Eng. 2017, 5, 11413–11421. [Google Scholar] [CrossRef]
- Guan, G.; Yang, L.; Mei, Q.; Zhang, K.; Zhang, Z.; Han, M.Y. Chemiluminescence switching on peroxidase-like Fe3O4 nanoparticles for selective detection and simultaneous determination of various pesticides. Anal. Chem. 2012, 84, 9492–9497. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, M.; Shen, H.; Che, G.; Qiao, Y.; Liu, B.; Wang, L. Recyclable multifunctional magnetic mesoporous silica nanocomposite for ratiometric detection, rapid adsorption, and efficient removal of Hg(II). ACS Sustain. Chem. Eng. 2018, 6, 1744–1752. [Google Scholar] [CrossRef]
- El-Sheikh, A.H.; Qawariq, R.F.; Abdelghani, J.I. Adsorption and magnetic solid-phase extraction of NSAIDs from pharmaceutical wastewater using magnetic carbon nanotubes: Effect of sorbent dimensions, magnetite loading and competitive adsorption study. Environ. Technol. Innov. 2019, 16, 100496. [Google Scholar] [CrossRef]
- Liu, C.; Liao, Y.; Huang, X.; Chen, L.; Li, Y. Fabrication of N,N-dimethyldodecylamine functionalized magnetic adsorbent for efficient enrichment of flavonoids. Talanta 2019, 194, 771–777. [Google Scholar] [CrossRef]
- Zhang, Y.; Ma, W.; Li, D.; Yu, M.; Guo, J.; Wang, C. Benzoboroxole-functionalized magnetic core/shell microspheres for highly specific enrichment of glycoproteins under physiological conditions. Small 2014, 10, 1379–1386. [Google Scholar] [CrossRef]
- Zhu, M.; Liu, W.; Liu, H.; Liao, Y.; Wei, J.; Zhou, X.; Xing, D. Construction of Fe3O4/Vancomycin/PEG Magnetic Nanocarrier for Highly Efficient Pathogen Enrichment and Gene Sensing. ACS Appl. Mater. Interfaces 2015, 7, 12873–12881. [Google Scholar] [CrossRef]
- Zarghani, M.; Akhlaghinia, B. Fe3O4 magnetic nanoparticles (MNPs) as an efficient catalyst for selective oxidation of benzylic and allylic C–H bonds to carbonyl compounds with tert-butyl hydroperoxide. RSC Adv. 2016, 6, 38592–38601. [Google Scholar] [CrossRef]
- Shabanian, M.; Khoobi, M.; Hemati, F.; Khonakdar, H.A.; ebrahimi, S.e.S.; Wagenknecht, U.; Shafiee, A. New PLA/PEI-functionalized Fe3O4 nanocomposite: Preparation and characterization. J. Ind. Eng. Chem. 2015, 24, 211–218. [Google Scholar] [CrossRef]
- Ghasemi, A.; Sohrabi, M.R.; Motiee, F. Preparation and characterization of a new sawdust/MNP/PEI nanocomposite and its applications for removing Pb (II) ions from aqueous solution. Water Sci. Technol. 2018, 78, 2469–2480. [Google Scholar] [CrossRef]
- Zhang, J.; Feng, X.; Wang, J.; Fang, G.; Liu, J.; Wang, S. Nano-crystalline cellulose-coated magnetic nanoparticles for affinity adsorption of glycoproteins. Analyst 2020, 145, 3407–3413. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, X.; He, X.; Chen, L.; Zhang, Y. A self-assembled polydopamine film on the surface of magnetic nanoparticles for specific capture of protein. Nanoscale 2012, 4, 3141–3147. [Google Scholar] [CrossRef]
- Ma, M.; Zhang, Y.; Gong, H.; Li, F.; Gu, N. Silica-coated magnetite nanoparticles labeled by nimotuzumab, a humanised monoclonal antibody to epidermal growth factor receptor: Preparations, specific targeting and bioimaging. J. Nanosci. Nanotechnol. 2013, 13, 6541–6545. [Google Scholar] [CrossRef]
- Xu, W.; Peng, J.; Ni, D.; Zhang, W.; Wu, H.; Mu, W. Preparation, characterization and application of levan/montmorillonite biocomposite and levan/BSA nanoparticle. Carbohydr. Polym. 2020, 234, 115921. [Google Scholar] [CrossRef]
- Zhang, M.; Qiao, J.; Qi, L. Dual-functional polymer-modified magnetic nanoparticles for isolation of lysozyme. Anal. Chim. Acta 2018, 1035, 70–76. [Google Scholar] [CrossRef]
- Yunoki, T.; Kimura, Y.; Fujimori, A. Maintenance properties of enzyme molecule stereostructure at high temperature by adsorption on organo-modified magnetic nanoparticle layer template. Bull. Chem. Soc. Jpn. 2019, 92, 1662–1671. [Google Scholar] [CrossRef]
- Coutinho, T.C.; Malafatti, J.O.D.; Paris, E.C.; Tardioli, P.W.; Farinas, C.S. Hydroxyapatite-CoFe2O4 magnetic nanoparticle composites for industrial enzyme immobilization, use, and recovery. ACS Appl. Nano Mater. 2020, 3, 12334–12345. [Google Scholar] [CrossRef]
- Johnson, P.A.; Park, H.J.; Driscoll, A.J. Enzyme nanoparticle fabrication: Magnetic nanoparticle synthesis and enzyme immobilization. Methods Mol. Biol. 2011, 679, 183–191. [Google Scholar] [PubMed]
- Hepziba Suganthi, S.; Swathi, K.V.; Biswas, R.; Basker, S.; Ramani, K. Co-immobilization of multiple enzymes onto surface-functionalized magnetic nanoparticle for the simultaneous hydrolysis of multiple substrates containing industrial wastes. Appl. Nanosci. 2019, 9, 1439–1457. [Google Scholar] [CrossRef]
- You, T.; Liu, D.; Chen, J.; Yang, Z.; Dou, R.; Gao, X.; Wang, L. Effects of metal oxide nanoparticles on soil enzyme activities and bacterial communities in two different soil types. J. Soils Sediments 2017, 18, 211–221. [Google Scholar] [CrossRef]
- Mishra, P.; Xue, Y.; Eivazi, F.; Afrasiabi, Z. Size, concentration, coating, and exposure time effects of silver nanoparticles on the activities of selected soil enzymes. Geoderma 2021, 381, 114682. [Google Scholar] [CrossRef]
- Vranish, J.N.; Ancona, M.G.; Oh, E.; Susumu, K.; Lasarte Aragones, G.; Breger, J.C.; Walper, S.A.; Medintz, I.L. Enhancing coupled enzymatic activity by colocalization on nanoparticle surfaces: Kinetic evidence for directed channeling of intermediates. ACS Nano 2018, 12, 7911–7926. [Google Scholar] [CrossRef]
- Dai, Y.; Liu, R.; Zhou, Y.; Li, N.; Hou, L.; Ma, Q.; Gao, B. Fire Phoenix facilitates phytoremediation of PAH-Cd co-contaminated soil through promotion of beneficial rhizosphere bacterial communities. Environ. Int. 2020, 136, 105421. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, C.; Shi, Z.; Cai, J.; Wang, P.; Wang, F.; Ju, M.; Liu, J.; Yu, Q. Synthesis of Phenylboronic Acid-Functionalized Magnetic Nanoparticles for Sensitive Soil Enzyme Assays. Molecules 2022, 27, 6883. https://doi.org/10.3390/molecules27206883
Li C, Shi Z, Cai J, Wang P, Wang F, Ju M, Liu J, Yu Q. Synthesis of Phenylboronic Acid-Functionalized Magnetic Nanoparticles for Sensitive Soil Enzyme Assays. Molecules. 2022; 27(20):6883. https://doi.org/10.3390/molecules27206883
Chicago/Turabian StyleLi, Can, Zhishang Shi, Jinxing Cai, Ping Wang, Fang Wang, Meiting Ju, Jinpeng Liu, and Qilin Yu. 2022. "Synthesis of Phenylboronic Acid-Functionalized Magnetic Nanoparticles for Sensitive Soil Enzyme Assays" Molecules 27, no. 20: 6883. https://doi.org/10.3390/molecules27206883





