Analysis of Staphylococcus aureus Molecules in Non-Treated Blood Using Mercury Immobilized Carbon Nanotube Sensor
Abstract
:1. Introduction
2. Objectives
3. Study Design
3.1. Instrument, Reagent and MCN Preparation
3.2. Cyclic Properties of the GC and MCN
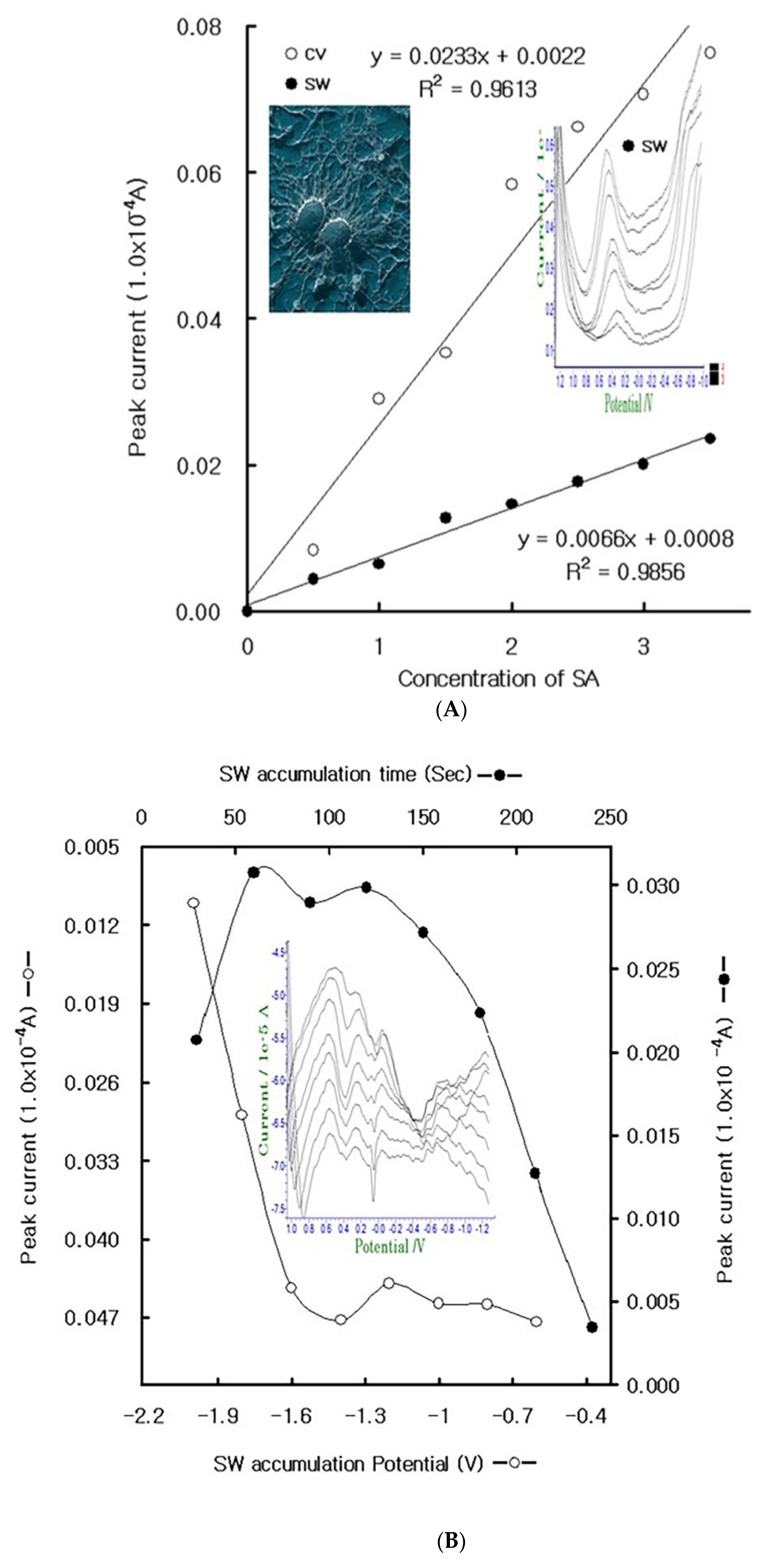
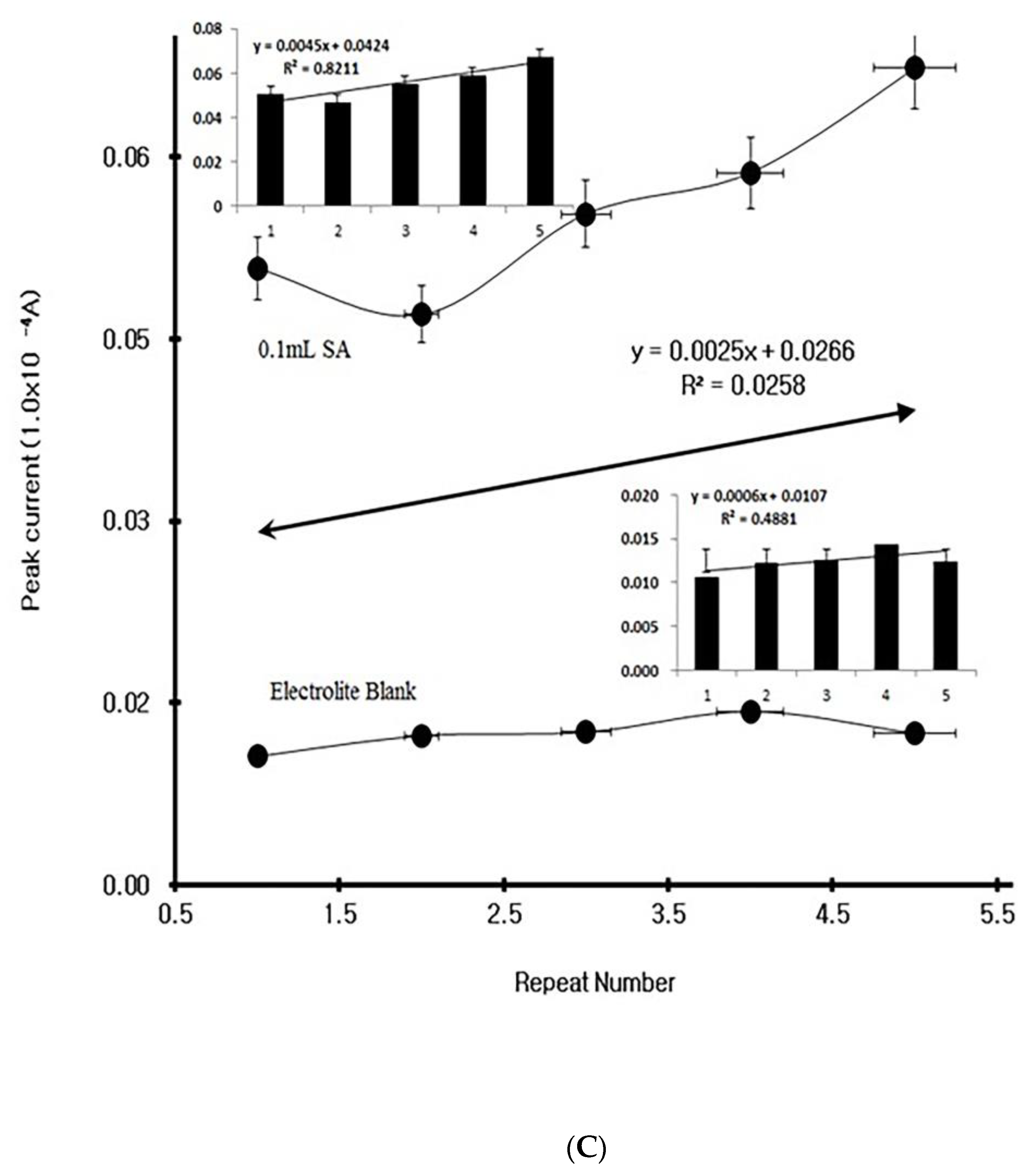
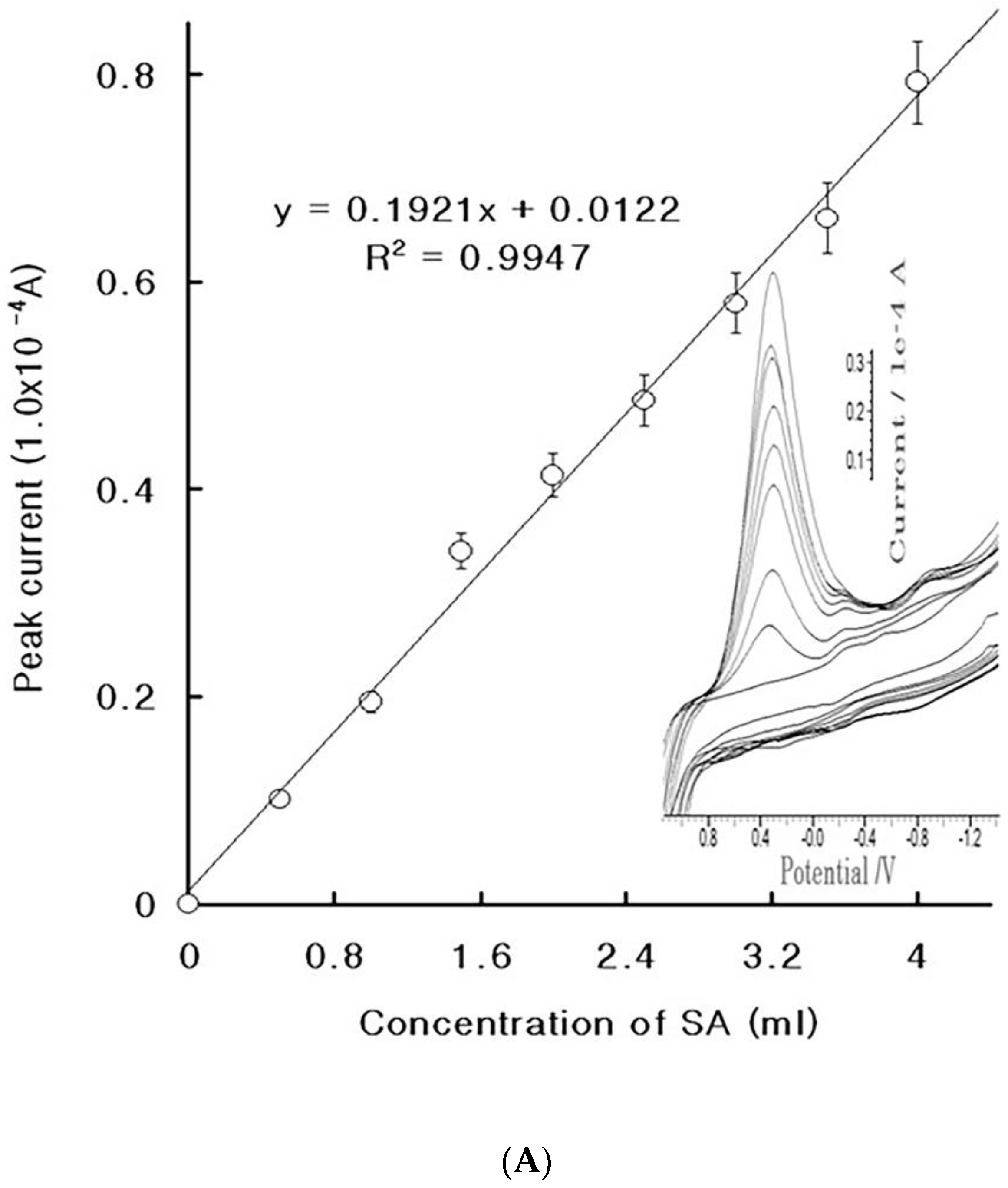
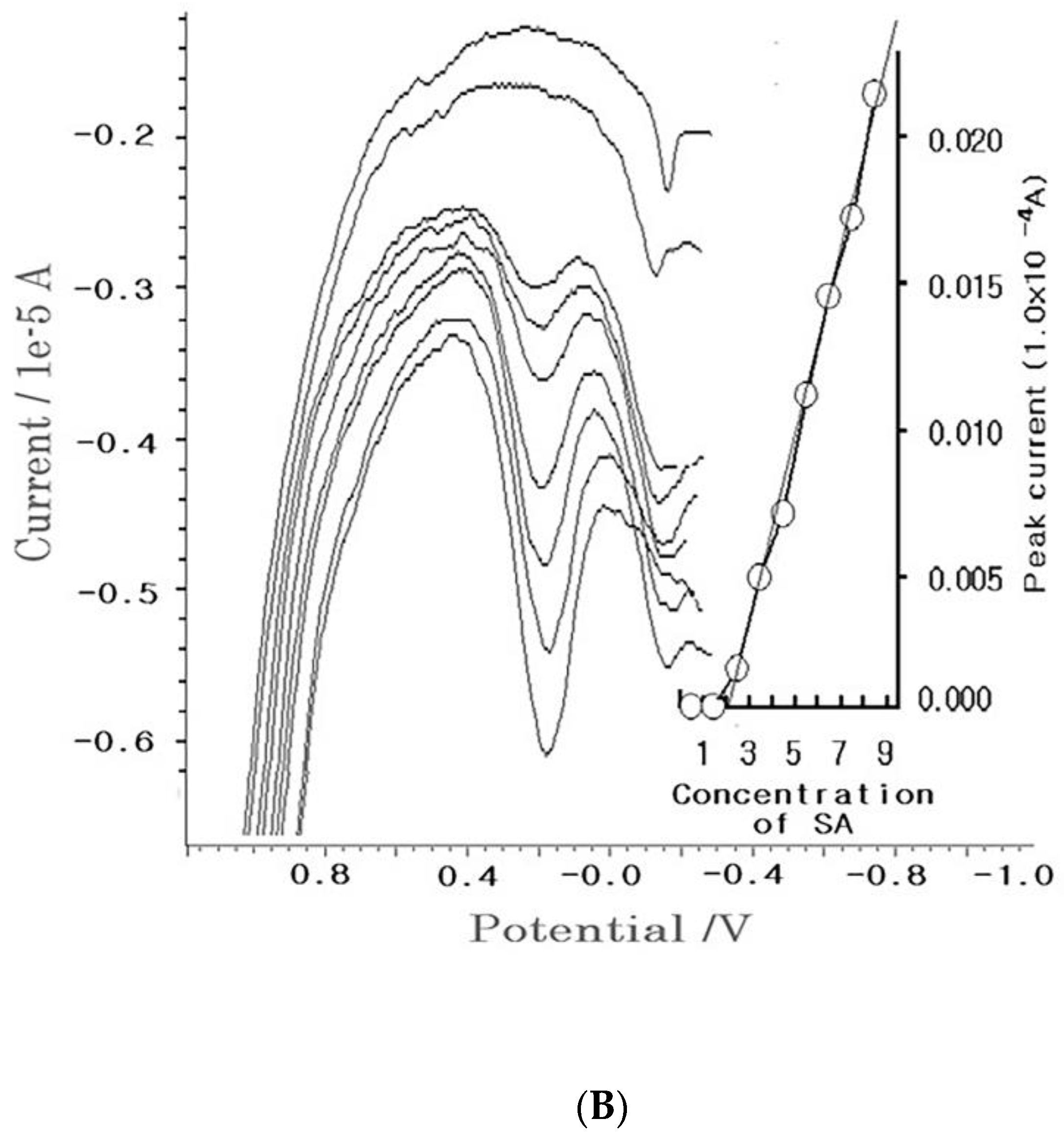
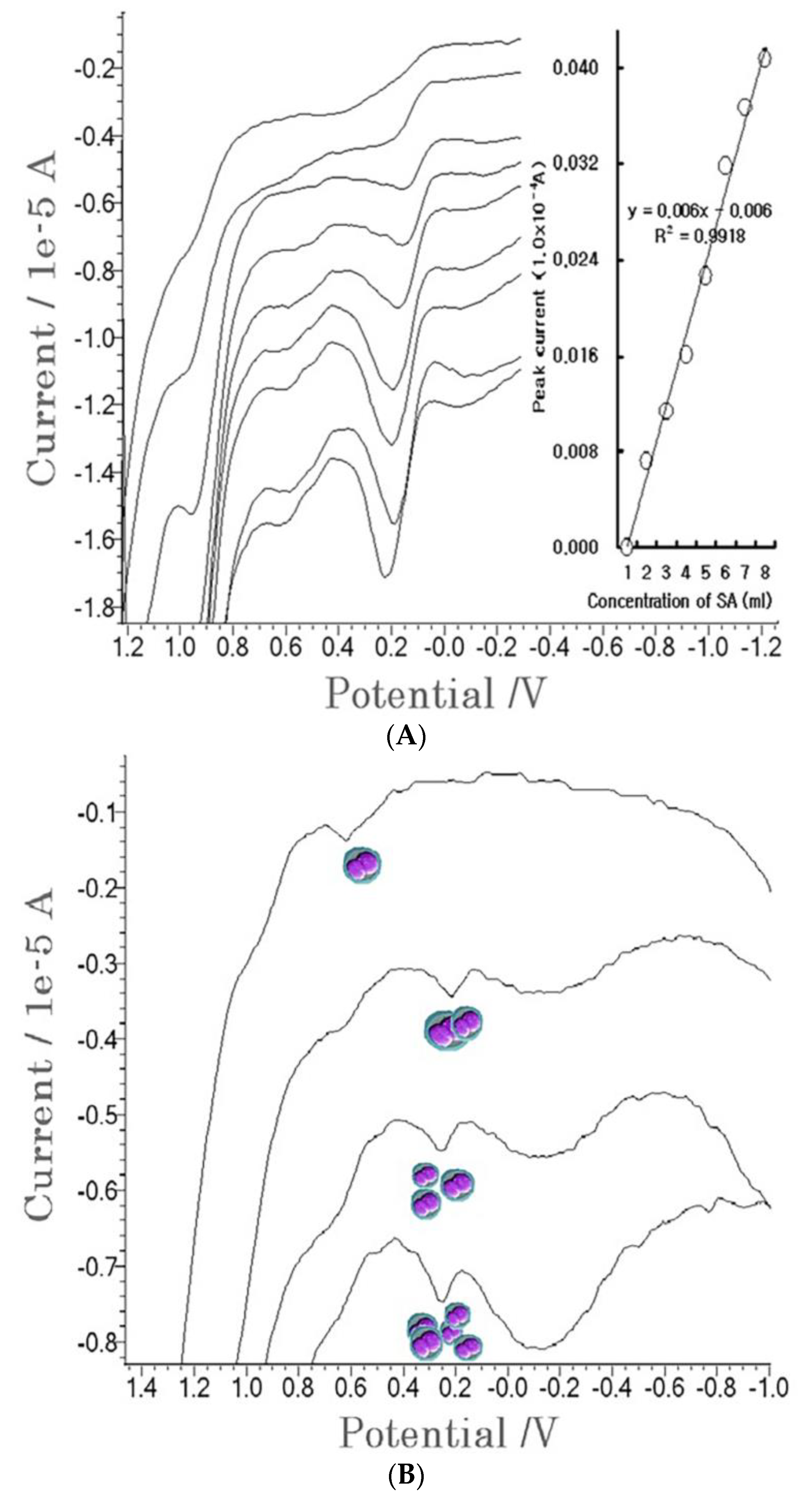
4. Results
4.1. Stripping Voltammetric SA and Positive Patient Sera Using MCN
4.2. Diagnostic Working Ranges of SA
4.3. Patient Diagnosis
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SA | Staphylococcus aureus |
| MCN | mercury immobilized on a carbon nanotube sensor |
| SW | stripping voltammetry |
| PCR | polymerase chain reaction |
| GC | graphite pencil electrode |
| RPLA | reversed passive latex agglutination |
| CFU | colony-forming unit |
| CVD | chemical vapor deposition |
| CV | Cyclic voltammetry |
References
- Cremonesi, P.; Perez, G.; Pisoni, G.; Moroni, P.; Morandi, S.; Luzzana, M.; Brasca, M.; Castiglioni, B. Detection of enterotoxigenic Staphylococcus aureus isolates in raw milk cheese. Lett. Appl. Microbiol. 2007, 45, 586–591. [Google Scholar] [CrossRef] [PubMed]
- Bernardo, K.; Fleer, S.; Pakulat, N.; Krut, O.; Hunger, F.; Kronke, M. Identification of Staphylococcus aureus exotoxins by combined sodium dodecyl sulfate gelelectrophoresis and matrix-assisted laser desorption/ionization-time of flight mass spectrometry. Proteomics 2002, 2, 740–746. [Google Scholar] [CrossRef]
- Zorgani, A.; Essery, S.D.; Madani, O.A.; Bentley, A.J.; James, V.S.; MacKenzie, D.A.C.; Keeling, J.W.; Rambaud, C.; Hilton, J.; Blackwell, C.C.; et al. Detection of pyrogenic toxins of Staphylococcus aureus in sudden infant death syndrome. FEMS Immunol. Med. Mic. 1999, 25, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.H.; Wei, H.C.; Lee, Y.C. One step immunochromatographic assay for the detection of Staphylococcus aureus. Food Control 2007, 18, 893–897. [Google Scholar] [CrossRef]
- Huang, S.H. Gold nanoparticle-based immunochromatographic assay for the detection of Staphylococcus aureus. Sens. Actuat. B Chem. 2007, 127, 335–340. [Google Scholar] [CrossRef]
- Grady, J.O.; Lacey, K.; Glynn, B.; Smith, T.J.; Barry, T.; Maher, M. tmRNA a novel high copy number RNA diagnostic target its application for Staphylococcus aureus detection using real time NASBA. FEMS Microbiol. Lett. 2009, 301, 218–223. [Google Scholar] [CrossRef] [Green Version]
- Wada, M.; Lkhagvadorj, E.; Bian, L.; Wang, C.; Chiba, Y.; Nagata, S.; Shimizu, T.; Yamashiro, Y.; Asahara, T.; Nomoto, K. Quantitative reverse transcription-PCR assay for the rapid detection of methicillin-resistant Staphylococcus aureus. J. Appl. Microbiol. 2010, 108, 779–788. [Google Scholar] [CrossRef]
- Cattoir, V.; Merabet, L.; Djibo, N.; Rioux, C.; Legrand, P.; Girou, E.; Lesprit, P. Clinical impact of a real-time PCR assay for rapid identification of Staphylococcus aureus and determination of methicillin resistance from positive blood cultures. Clin. Microbiol. Infect. 2011, 17, 425–431. [Google Scholar] [CrossRef] [Green Version]
- Fox, L.K.; Adams, D.S. The ability of the enzyme-linked immunosorbent assay to detect antibody against Staphylococcus aureus in milk following experimental intramammary infection. J. Vet. Med. B 2000, 47, 517–526. [Google Scholar] [CrossRef]
- Zouharova, M.; Rysanek, D. Multiplex PCR and RPLA identification of Staphylococcus aureus enterotoxigenic strains from bulk tank milk. Zoonoses Public Health 2008, 55, 313–319. [Google Scholar] [CrossRef]
- Ly, S.Y. Real-time voltammetric assay of cadmium ions in plant tissue and fish brain core. Bull. Korean Chem. Soc. 2006, 10, 1613–1615. [Google Scholar]
- Ly, S.Y. Diagnosis of copper ions in vascular tracts using a fluorine doped carbon nanotube sensor. Talanta 2008, 74, 1635–1641. [Google Scholar] [CrossRef] [PubMed]
- Ly, S.Y.; Cho, N.S. Diagnosis of human hepatitis B virus in non-treated blood by the bovine IgG DNA linked carbon nanotube biosensor. J. Clin. Virol. 2009, 44, 43–47. [Google Scholar] [CrossRef] [PubMed]
- Ly, S.Y.; Lee, J.H. Human-urine diabetes assay and in vivo rat bladder assay using a fluorine doped carbon nanotube catheter sensor. Ann. Biomed. Engl. 2009, 37, 2028–2033. [Google Scholar] [CrossRef]
- Thangamani, G.; Deshmukh, J.K.; Sadasivuni, K.K.; Chidambaram, K.; Ahamed, M.; Ponnamma, D.; AlMaadeed, M.A.A.; Pasha, S.K.K. Recent advances in electrochemical biosensor and gas sensors based on graphene and carbon nanotubes (CNT)-A. Adv. Mater. Lett. 2017, 8, 196–205. [Google Scholar] [CrossRef]
- Lenka, L.; Sadasivuni, K.K.; Kasak, P.; Tkac, J. Ti3C2 MXene-based nanobiosensors for detection of cancer biomarkers. In Novel Nanomaterials; IntechOpen: London, UK, 2020. [Google Scholar]
- Sultan, Ş.; Ünlü, C.; Trabzon, L. Affinity biosensors developed with quantum dots in microfluidic systems. Emerg. Mater. 2021, 4, 187–209. [Google Scholar]
- Amene, S.; Rezaei, N. Biosensing surfaces and therapeutic biomaterials for the central nervous system in COVID-19. Emerg. Mater. 2021, 1, 293–312. [Google Scholar]
- Manjunatha, J.G.; Swamy, B.E.K.; Mamatha, G.P.; Shankar, S.S.; Gilbert, O.; Chandrashekar, B.N.; Sherigara, B.S. Electrochemical response of dopamine at phthalic acid and triton X-100 modified carbon paste electrode: A cyclic voltammetry study. Int. J. Electrochem. Sci. 2009, 4, 1469–1478. [Google Scholar]
- Hareesha, N.; Manjunatha, J.G. Electro-oxidation of formoterol fumarate on the surface of novel poly(thiazole yellow-G) layered multi-walled carbon nanotube paste electrode. Sci. Rep. 2021, 17, 12797. [Google Scholar] [CrossRef]
- David, P.K.; Kimani, C.N.; Katabazi, F.A.; Okeng, A.; Okee, M.S.; Nanteza, A.; Joloba, M.L.; Najjuka, F.C. Identification of Staphylococcus aureus: DNase and Mannitol salt agar improve the efficiency of the tube coagulase test. Ann. Clin. Microbiol. Antimicrob. 2010, 9, 23. [Google Scholar]
- Wu, S.; Duan, N.; Gu, H.; Hao, L.; Ye, H.; Gong, W.; Wang, Z. A review of the methods for detection of Staphylococcus aureus enterotoxins. Toxins 2016, 8, 176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonçalves, E.; Carvalhal, R.; Mesquita, R.; Azevedo, J.; Coelho, M.J.; Magalhães, R.; Pia Ferraz, M.; Manso, M.C.; Gavinha, S.; Pina, C.; et al. Detection of Staphylococcus aureus (MRSA/MSSA) in surfaces of dental medicine equipment. Saudi J. Biol. Sci. 2020, 27, 1003–1008. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cho, I.H.; Choi, K.J.; Kim, J.H.; Lee, K.; Ly, S.Y. Analysis of Staphylococcus aureus Molecules in Non-Treated Blood Using Mercury Immobilized Carbon Nanotube Sensor. Molecules 2022, 27, 1837. https://doi.org/10.3390/molecules27061837
Cho IH, Choi KJ, Kim JH, Lee K, Ly SY. Analysis of Staphylococcus aureus Molecules in Non-Treated Blood Using Mercury Immobilized Carbon Nanotube Sensor. Molecules. 2022; 27(6):1837. https://doi.org/10.3390/molecules27061837
Chicago/Turabian StyleCho, In Hea, Kwang Jin Choi, Ji Hyun Kim, Kyung Lee, and Suw Young Ly. 2022. "Analysis of Staphylococcus aureus Molecules in Non-Treated Blood Using Mercury Immobilized Carbon Nanotube Sensor" Molecules 27, no. 6: 1837. https://doi.org/10.3390/molecules27061837




