A Comprehensive View on the Quercetin Impact on Colorectal Cancer
Abstract
:1. Epidemiology and Aetiology of Colorectal Cancer
2. Phytochemicals—Potential Benefits
3. Quercetin
3.1. Quercetin Biosynthesis in Plants
3.2. Regulation of Quercetin Biosynthesis in Plants
3.3. Quercetin Metabolism in Humans
4. Quercetin and Its Derivatives—Mechanisms of Action in CRC
5. Crucial Signal Transduction Pathways in CRC
5.1. Wnt/β-Catenin Signaling in CRC
5.2. PI3K/AKT-mTOR Signaling in CRC
5.3. MAPK Cascades in CRC
5.3.1. MAPK/ERK Signaling in CRC
5.3.2. MAPK/JNK Signaling in CRC
5.3.3. MAPK/ p38 Signaling in CRC
5.4. p53 Signaling in CRC
5.5. NF-κB Signaling in CRC
6. Quercetin Impacts the Growth and Proliferation in CRC
7. Quercetin Impacts the Cell Cycle in CRC
| Cell Cycle Arrest Phase and/or Molecular Targets | Testing System | Reference |
|---|---|---|
| At G0/G1 phase | In vivo: HCT-116 Xenograft mouse model | [146] |
| ? | ||
| At G0/G1 phase | In vitro: HT-29 cell culture | [131] |
| ? | ||
| At G1 or G2 | In vitro: HCT-116 cell culture | [138] |
| ? | ||
| At G2/M | In vitro: HT-29, HCT116 and SW480 cell cultures | [147] |
| ↓ p-AKT | ||
| ↑ Cyclin B1 | ||
| At G2/M | In vitro: RKO cell culture | [148] |
| ↓ CDK1, CDC25c, Cyclin B1 | ||
| ↑ p21 | ||
| At G2/M | In vitro: SW620 cell culture | [149] |
| ↑ p21, p58 | ||
| ↓ CDC6, CDK4, Cyclin D1 | In vitro: Caco-2 cell culture | [151] |
| ↓ Ki67 | In vitro: SW480 cell culture | [152] |
| ↓ Bcl-2 | In vitro: HT-29 cell culture | [131] |
| ↑ Bax, p53, Caspase-3 |
8. Quercetin Impacts Apoptosis in CRC
9. Quercetin Impacts Tumor Size in CRC
10. Quercetin Impacts Tumor Nodule Number in CRC
11. Quercetin Impacts Migration and Invasion in CRC
12. Quercetin Impacts Inflammation in CRC
13. Quercetin Impacts Oxidative Stress in CRC
14. Quercetin Impacts Chemoresistance in CRC
15. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Douaiher, J.; Ravipati, A.; Grams, B.; Chowdhury, S.; Alatise, O.; Are, C. Colorectal cancer-global burden, trends, and geographical variations. J. Surg. Oncol. 2017, 115, 619–630. [Google Scholar] [CrossRef] [PubMed]
- Al-Sohaily, S.; Biankin, A.; Leong, R.; Kohonen-Corish, M.; Warusavitarne, J. Molecular pathways in colorectal cancer. J. Gastroenterol. Hepatol. 2012, 27, 1403–1431. [Google Scholar] [CrossRef] [PubMed]
- Li, S.K.H.; Martin, A. Mismatch Repair and Colon Cancer: Mechanisms and Therapies Explored. Trends Mol. Med. 2016, 22, 274–289. [Google Scholar] [CrossRef]
- Rattray, N.J.W.; Charkoftaki, G.; Rattray, Z.; Hansen, J.E.; Vasiliou, V.; Johnson, C.H. Environmental influences in the etiology of colorectal cancer: The premise of metabolomics. Curr. Pharmacol. Rep. 2017, 3, 114–125. [Google Scholar] [CrossRef] [PubMed]
- Afrin, S.; Giampieri, F.; Gasparrini, M.; Forbes-Hernandez, T.Y.; Cianciosi, D.; Reboredo-Rodriguez, P.; Zhang, J.; Manna, P.P.; Daglia, M.; Atanasov, A.G.; et al. Dietary phytochemicals in colorectal cancer prevention and treatment: A focus on the molecular mechanisms involved. Biotechnol. Adv. 2020, 38, 107322. [Google Scholar] [CrossRef] [PubMed]
- Yamagishi, H.; Kuroda, H.; Imai, Y.; Hiraishi, H. Molecular pathogenesis of sporadic colorectal cancers. Chin. J. Cancer. 2016, 35, 26738600. [Google Scholar] [CrossRef] [Green Version]
- Rustgi, A.K. The genetics of hereditary colon cancer. Genes. Dev. 2007, 21, 2525–2538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fung, T.T.; Hu, F.B.; Wu, K.; Chiuve, S.E.; Fuchs, C.S.; Giovannucci, E. The Mediterranean and Dietary Approaches to Stop Hypertension (DASH) diets and colorectal cancer. Am. J. Clin. Nutr. 2010, 92, 1409–1435. [Google Scholar] [CrossRef] [Green Version]
- Nomura, A.M.Y.; Wilkens, L.R.; Murphy, S.P.; Hankin, J.H.; Henderson, B.E.; Pike, M.C.; Kolonel, L.N. Association of vegetable, fruit, and grain intakes with colorectal cancer: The Multiethnic Cohort Study. Am. J. Clin. Nutr. 2008, 88, 730–737. [Google Scholar] [CrossRef] [Green Version]
- Luo, W.P.; Fang, Y.J.; Lu, M.S.; Zhong, X.; Chen, Y.M.; Zhang, C.X. High consumption of vegetable and fruit colour groups is inversely associated with the risk of colorectal cancer: A case-control study. Br. J. Nutr. 2015, 113, 1129–1138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, C.M.; Wei, C.; Ensor, J.E.; Smolenski, D.J.; Amos, C.I.; Levin, B.; Berry, D.A. Meta-analyses of colorectal cancer risk factors. Cancer Causes Control 2013, 24, 1207–1222. [Google Scholar] [CrossRef] [PubMed]
- Ben, Q.; Zhong, J.; Liu, J.; Wang, L.; Sun, Y.; Yv, L.; Yuan, Y. Association Between Consumption of Fruits and Vegetables and Risk of Colorectal Adenoma: A PRISMA-Compliant Meta-Analysis of Observational Studies. Medicine 2015, 94, e1599. [Google Scholar] [CrossRef] [PubMed]
- AL-Ishaq, R.K.; Overy, A.J.; Büsselberg, D. Phytochemicals and Gastrointestinal Cancer: Cellular Mechanisms and Effects to Change Cancer Progression. Biomolecules 2020, 10, 105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González-Vallinas, M.; González-Castejón, M.; Rodríguez-Casado, A.; Ramírez de Molina, A. Dietary phytochemicals in cancer prevention and therapy: A complementary approach with promising perspectives. Nutr. Rev. 2013, 71, 585–599. [Google Scholar] [CrossRef] [PubMed]
- Johnson, I.T. Phytochemicals and cancer. Proc. Nutr. Soc. 2007, 66, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Forni, C.; Facchiano, F.; Bartoli, M.; Pieretti, S.; Facchiano, A.; D’Arcangelo, D.; Norelli, S.; Valle, G.; Nisini, R.; Beninati, S.; et al. Beneficial Role of Phytochemicals on Oxidative Stress and Age-Related Diseases. BioMed. Res. Int. 2019, 2019, 8748253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ho, J.W.S.; Cheung, M.W.M. Combination of phytochemicals as adjuvants for cancer therapy. Recent Pat. Anticancer. Drug Discov. 2014, 9, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.J.; Gan, R.Y.; Li, S.; Zhou, Y.; Li, A.N.; Xu, D.P.; Li, H.B. Antioxidant Phytochemicals for the Prevention and Treatment of Chronic Diseases. Molecules 2015, 20, 21138–21156. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Virgous, C.; Si, H. Synergistic anti-inflammatory effects and mechanisms of combined phytochemicals. J. Nutr. Biochem. 2019, 69, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Hu, M.J.; Wang, Y.Q.; Cui, Y.L. Antioxidant Activities of Quercetin and Its Complexes for Medicinal Application. Molecules 2019, 24, 1123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, P.; Arif, Y.; Bajguz, A.; Hayat, S. The role of quercetin in plants. Plant Physiol. Biochem. 2021, 166, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Hastings, J.; Owen, G.; Dekker, A.; Ennis, M.; Kale, N.; Muthukrishnan, V.; Turner, S.; Swainston, N.; Mendes, P.; Steinbeck, C. ChEBI in 2016: Improved services and an expanding collection of metabolites. Nucleic Acids Res. 2015, 44, D1214–D1219. [Google Scholar] [CrossRef] [PubMed]
- Law, V.; Knox, C.; Djoumbou, Y.; Jewison, T.; Guo, A.C.; Liu, Y.; Maciejewski, A.; Arndt, D.; Wilson, M.; Neveu, V.; et al. DrugBank 4.0: Shedding new light on drug metabolism. Nucleic Acids Res. 2014, 42, D1091–D1097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- National Center for Biotechnology Information. PubChem Compound Summary for CID 5280343, Quercetin. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Quercetin (accessed on 12 February 2022).
- Reyes-Farias, M.; Carrasco-Pozo, C. The anti-cancer effect of quercetin: Molecular implications in cancer metabolism. Int. J. Mol. Sci. 2019, 20, 3177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stavric, B. Quercetin in our diet: From potent mutagen to probable anticarcinogen. Clin. Biochem. 1994, 27, 245–248. [Google Scholar] [CrossRef]
- Nishimuro, H.; Ohnishi, H.; Sato, M.; Ohnishi-Kameyama, M.; Matsunaga, I.; Naito, S.; Kobori, M. Estimated daily intake and seasonal food sources of quercetin in Japan. Nutrients 2015, 7, 2345–2358. [Google Scholar] [CrossRef]
- Perez-Jimenez, J.; Fezeu, L.; Touvier, M.; Arnault, N.; Manach, C.; Hercberg, S.; Galan, P.; Scalbert, A. Dietary intake of 337 polyphenols in French adults. Am. J. Clin. Nutr. 2011, 93, 1220–1228. [Google Scholar] [CrossRef] [Green Version]
- Ovaskainen, M.L.; Torronen, R.; Koponen, J.M.; Sinkko, H.; Hellstrom, J.; Reinivuo, H.; Mattila, P. Dietary intake and major food sources of polyphenols in Finnish adults. J. Nutr. 2008, 138, 562–566. [Google Scholar] [CrossRef]
- Dabeek, W.M.; Marra, M.V. Dietary quercetin and kaempferol: Bioavailability and potential cardiovascular-related bioactivity in humans. Nutrients 2019, 11, 2288. [Google Scholar] [CrossRef] [Green Version]
- Bhagwat, S.; Haytowitz, D.B. USDA Database for the Flavonoid Content of Selected Foods. Release 3.2 (November 2015). Nutrient Data Laboratory, Beltsville Human Nutrition Research Center, ARS, USDA. Available online: https://data.nal.usda.gov/dataset/usda-database-flavonoid-content-selected-foods-release-32-november-2015 (accessed on 15 February 2022). [CrossRef]
- Nabavi, S.M.; Samec, D.; Tomczyk, M.; Milella, L.; Russo, D.; Habtemariam, S.; Shirooie, S. Flavonoid biosynthetic pathways in plants: Versatile targets for metabolic engineering. Biotechnol. Adv. 2020, 38, 107316. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Kong, D.; Fu, Y.; Sussman, M.R.; Wu, H. The effect of developmental and environmental factors on secondary metabolites in medicinal plants. Plant Physiol. Biochem. 2020, 148, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Sachan, A.; Ghosh, S.; Sen, S.K.; Mitra, A. Co-production of caffeic acid and p-hydroxybenzoic acid from p-coumaric acid by Streptomyces caeruleus MTCC 6638. Appl. Microbiol. Biotechnol. 2006, 71, 720–727. [Google Scholar] [CrossRef] [PubMed]
- Alrawaiq, N.S.; Abdullah, A. A review of flavonoid quercetin: Metabolism, bioactivity and antioxidant properties. Int. J. Pharmtech. Res. 2014, 6, 933–941. [Google Scholar]
- Lakhanpal, P.; Rai, D.K. Quercetin: A versatile flavonoid. Internet J. Med. Update 2007, 2, 20–35. [Google Scholar] [CrossRef] [Green Version]
- Wohl, J.; Petersen, M. Functional expression and characterization of cinnamic acid 4-hydroxylase from the hornwort Anthoceros agrestis in Physcomitrella patens. Plant Cell Rep. 2020, 39, 597–607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magar, R.T.; Sohng, J.K. A review on structure, modifications and structure-activity relation of quercetin and its derivatives. J. Microbiol. Biotechnol. 2020, 30, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Lesjak, M.; Beara, I.; Simin, N.; Pintac, D.; Majkic, T.; Bekvalac, K.; Mimica-Dukic, N. Antioxidant and anti-inflammatory activities of quercetin and its derivatives. J. Funct. Foods 2018, 40, 68–75. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information. PubChem Compound Summary. Available online: https://pubchem.ncbi.nlm.nih.gov (accessed on 12 February 2022).
- Liu, W.; Feng, Y.; Yu, S.; Fan, Z.; Li, X.; Li, J.; Yin, H. The Flavonoid Biosynthesis Network in Plants. Int. J. Mol. Sci. 2021, 22, 12824. [Google Scholar] [CrossRef] [PubMed]
- Petroni, K.; Tonelli, C. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 2011, 181, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Liang, D.; Xia, H.; Lin, L.J.; Wang, J.; Lv, X.L. Lignin and quercetin synthesis underlies berry russeting in ‘sunshine muscat’grape. Biomolecules 2020, 10, 690. [Google Scholar] [CrossRef] [PubMed]
- Becker, C.; Klaering, H.-P.; Schreiner, M.; Kroh, L.W.; Krumbein, A. Unlike quercetin glycosides, cyanidin glycoside in red leaf lettuce responds more sensitively to increasing low radiation intensity before than after head formation has started. J. Agric. Food Chem. 2014, 62, 6911–6917. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Yao, J.; Zhao, Y.; Xie, D.; Jiang, X.; Xu, Z. Efficient rutin and quercetin biosynthesis through flavonoids-related gene expression in Fagopyrum tataricum Gaertn. Hairy root cultures with UV-B irradiation. Front. Plant Sci. 2016, 7, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parvin, K.; Hasanuzzaman, M.; Bhuyan, M.H.M.; Mohsin, S.M.; Fujita, M. Quercetin mediated salt tolerance in tomato through the enhancement of plant antioxidant defense and glyoxalase systems. Plants 2019, 8, 247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Z.; Zhou, J.; Ren, T.; Du, H.; Liu, H.; Li, Y.; Zhang, C. Salt stress decreases seedling growth and development but increases quercetin and kaempferol content in Apocynum venetum. Plant Biol. 2020, 22, 813–821. [Google Scholar] [CrossRef] [PubMed]
- Almeida, A.F.; Borge, G.I.A.; Piskula, M.; Tudose, A.; Tudoreanu, L.; Valentová, K.; Williamson, G.; Santos, C.N. Bioavailability of Quercetin in Humans with a Focus on Interindividual Variation. Compr. Rev. Food Sci. Food Saf. 2018, 17, 714–731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ader, P.; Wessmann, A.; Wolffram, S. Bioavailability and metabolism of the flavonol quercetin in the pig. Free Rad. Biol. Med. 2000, 28, 1056–1067. [Google Scholar] [CrossRef]
- Erlund, I.; Kosonen, T.; Alfthan, G.; Maenpaa, J.; Perttunen, K.; Kenraali, J.; Aro, A. Pharmacokinetics of quercetin from quercetin aglycone and rutin in healthy volunteers. Eur. J. Clin. Pharmacol. 2000, 56, 545–553. [Google Scholar] [CrossRef]
- Graefe, E.U.; Derendorf, H.; Veit, M. Pharmacokinetics and bioavailability of the flavonol quercetin in humans. Int. J. Clin. Pharmacol. Ther. 1999, 37, 219–233. [Google Scholar]
- Crespy, V.; Morand, C.; Besson, C.; Manach, C.; Demigne, C.; Remesy, C. Quercetin, but not its glycosides, is absorbed from the rat stomach. J. Agric. Food Chem. 2002, 50, 618–621. [Google Scholar] [CrossRef]
- Day, A.J.; Canada, F.J.; Diaz, J.C.; Kroon, P.A.; McLauchlan, W.R.; Faulds, C.B.; Williamson, G. Dietary flavonoid and isoflavone glycosides are hydrolysed by the lactase site of lactase phlorizin hydrolase. FEBS Lett. 2000, 468, 166–170. [Google Scholar] [CrossRef] [Green Version]
- Hollman, P.C.; van Trijp, J.M.; Mengelers, M.J.; de Vries, J.H.; Katan, M.B. Bioavailability of the dietary antioxidant flavonol quercetin in man. Cancer Lett. 1997, 114, 139–140. [Google Scholar] [CrossRef]
- Hollman, P.C.; Devries, J.H.; Vanleeuwen, S.D.; Mengelers, M.J.; Katan, M.B. Absorption of dietary quercetin glycosides and quercetin in healthy ileostomy volunteers. Am. J. Clin. Nutr. 1995, 62, 1276–1282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hollman, P.C.; vander Gaag, M.; Mengelers, M.J.; van Trijp, J.M.; de Vries, J.H.; Katan, M.B. Absorption and disposition kinetics of the dietary antioxidant quercetin in man. Free Rad. Biol. Med. 1996, 21, 703–707. [Google Scholar] [CrossRef]
- Arts, I.C.W.; Sesink, A.L.A.; Faassen-Peters, M.; Hollman, P.C.H. The type of sugar moiety is a major determinant of the small intestinal uptake and subsequent biliary excretion of dietary quercetin glycosides. Br. J. Nutr. 2004, 91, 841–847. [Google Scholar] [CrossRef] [Green Version]
- Cermak, R.; Landgraf, S.; Wolffram, S. The bioavailability of quercetin in pigs depends on the glycoside moiety and on dietary factors. J. Nutr. 2003, 133, 2802–2807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reinboth, M.; Wolffram, S.; Abraham, G.; Ungemach, F.R.; Cermak, R. Oral bioavailability of quercetin from different quercetin glycosides in dogs. Br. J. Nutr. 2010, 104, 198–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russo, M.; Spagnuolo, C.; Tedesco, I.; Bilotto, S.; Russo, G.L. The flavonoid quercetin in disease prevention and therapy: Facts and fancies. Biochem. Pharmacol. 2012, 83, 6–15. [Google Scholar] [CrossRef]
- Vacek, J.; Papouskova, B.; Kosina, P.; Vrba, J.; Kren, V.; Ulrichova, J. Biotransformation of flavonols and taxifolin in hepatocyte in vitro systems as determined by liquid chromatography with various stationary phases and electrospray ionization-quadrupole time-of-flight mass spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2012, 899, 109–115. [Google Scholar] [CrossRef]
- Wolffram, S.; Block, M.; Ader, P. Quercetin-3-Glucoside Is Transported by the Glucose Carrier SGLT1 across the Brush Border Membrane of Rat Small Intestine. J. Nutr. 2002, 132, 630–635. [Google Scholar] [CrossRef] [Green Version]
- Lesser, S.; Cermak, R.; Wolffram, S. Bioavailability of quercetin in pigs is influenced by the dietary fat content. J. Nutr. 2004, 134, 1508–1511. [Google Scholar] [CrossRef] [PubMed]
- Menendez, C.; Duenas, M.; Galindo, P.; Gonzalez-Manzano, S.; Jimenez, R.; Moreno, L.; Perez-Vizcaino, F. Vascular deconjugation of quercetin glucuronide: The flavonoid paradox revealed? Molec. Nutr. Food Res. 2011, 55, 1780–1790. [Google Scholar] [CrossRef] [PubMed]
- Salehi, B.; Machin, L.; Monzote, L.; Sharifi-Rad, J.; Ezzat, S.M.; Salem, M.A.; Cho, W.C. Therapeutic potential of quercetin: New insights and perspectives for human health. ACS Omega 2020, 5, 11849–11872. [Google Scholar] [CrossRef] [PubMed]
- Shimoi, K.; Saka, N.; Nozawa, R.; Sato, M.; Amano, I.; Nakayama, T.; Kinae, N. Deglucuronidation of a flavonoid, luteolin monoglucuronide, during inflammation. Drug Metab. Dispos. 2001, 29, 1521–1524. [Google Scholar] [PubMed]
- Kawai, Y. β-Glucuronidase activity and mitochondrial dysfunction: The sites where flavonoid glucuronides act as anti-inflammatory agents. J. Clin. Biochem. Nutr. 2014, 54, 145–150. [Google Scholar] [CrossRef] [Green Version]
- Araújo, K.C.F.; de M.B. Costa, E.M.; Pazini, F.; Valadares, M.C.; de Oliveira, V. Bioconversion of quercetin and rutin and the cytotoxicity activities of the transformed products. Food Chem. Toxicol. 2013, 51, 93–96. [Google Scholar] [CrossRef] [PubMed]
- Beekmann, K.; Actis-Goretta, L.; van Bladeren, P.J.; Dionisi, F.; Destaillats, F.; Rietjens, I.M.C.M. A state-of-the-art overview of the effect of metabolic conjugation on the biological activity of flavonoids. Food Funct. 2012, 3, 1008–1018. [Google Scholar] [CrossRef]
- Lodi, F.; Jimenez, R.; Moreno, L.; Kroon, P.A.; Needs, P.W.; Hughes, D.A.; Perez-Vizcaino, F. Glucuronidated and sulfated metabolites of the flavonoid quercetin prevent endothelial dysfunction but lack direct vasorelaxant effects in rat aorta. Atherosclerosis 2009, 204, 34–39. [Google Scholar] [CrossRef]
- Tribolo, S.; Lodib, F.; Connor, C.; Suri, S.; Wilson, V.G.; Taylor, M.A.; Hughes, D.A. Comparative effects of quercetin and its predominant human metabolites on adhesion molecule expression in activated human vascular endothelial cells. Atherosclerosis 2008, 197, 50–56. [Google Scholar] [CrossRef]
- Williamson, G.; Barron, D.; Shimoi, K.; Terao, J. In vitro biological properties of flavonoid conjugates found in vivo. Free Rad. Res. 2005, 39, 457–469. [Google Scholar] [CrossRef]
- Cruz–Correa, M.; Shoskes, D.A.; Sanchez, P.; Zhao, R.; Hylind, L.M.; Wexner, S.D.; Giardiello, F.M. Combination treatment with curcumin and quercetin of adenomas in familial adenomatous polyposis. Clin. Gastroenterol. Hepatol. 2006, 4, 1035–1038. [Google Scholar] [CrossRef] [PubMed]
- Bobe, G.; Albert, P.S.; Sansbury, L.B.; Lanza, E.; Schatzkin, A.; Colburn, N.H.; Cross, A.J. Interleukin-6 as a potential indicator for prevention of high-risk adenoma recurrence by dietary flavonols in the polyp prevention trial. Cancer Prev. Res. 2010, 3, 764–775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Lee, I.M.; Zhang, S.M.; Blumberg, J.B.; Buring, J.E.; Sesso, H.D. Dietary intake of selected flavonols, flavones, and flavonoid-rich foods and risk of cancer in middle-aged and older women. Am. J. Clin. Nutr. 2009, 89, 905–912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossi, M.; Negri, E.; Talamini, R.; Bosetti, C.; Parpinel, M.; Gnagnarella, P.; La Vecchia, C. Flavonoids and colorectal cancer in Italy. Cancer Epidemiol. Prev. Biomark. 2006, 15, 1555–1558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prossomariti, A.; Piazzi, G.; Alquati, C.; Ricciardiello, L. Are Wnt/β-Catenin and PI3K/AKT/mTORC1 distinct pathways in colorectal cancer? Cell. Mol. Gastroenterol. Hepatol. 2020, 10, 491–506. [Google Scholar] [CrossRef] [PubMed]
- Stamos, J.L.; Weis, W.I. The β-catenin destruction complex. Cold Spring Harb. Perspect. Biol. 2013, 5, a007898. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Semenov, M.; Han, C.; Baeg, G.-H.; Tan, Y.; Zhang, Z.; Lin, X.; He, X. Control of β-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell 2002, 108, 837–847. [Google Scholar] [CrossRef] [Green Version]
- Aberle, H.; Bauer, A.; Stappert, J.; Kispert, A.; Kemler, R. β-catenin is a target for the ubiquitin–proteasome pathway. EMBO J. 1997, 16, 3797–3804. [Google Scholar] [CrossRef] [Green Version]
- Dann, C.E.; Hsieh, J.C.; Rattner, A.; Sharma, D.; Nathans, J.; Leahy, D.J. Insights into Wnt binding and signalling from the structures of two Frizzled cysteine-rich domains. Nature 2001, 412, 86–90. [Google Scholar] [CrossRef]
- Tamai, K.; Semenov, M.; Kato, Y.; Spokony, R.; Liu, C.; Katsuyama, Y.; He, X. LDL-receptor-related proteins in Wnt signal transduction. Nature 2000, 407, 530–535. [Google Scholar] [CrossRef]
- Zeng, X.; Tamai, K.; Doble, B.; Li, S.; Huang, H.; Habas, R.; He, X. A dual-kinase mechanism for Wnt co-receptor phosphorylation and activation. Nature 2005, 438, 873–877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molenaar, M.; Van De Wetering, M.; Oosterwegel, M.; Peterson-Maduro, J.; Godsave, S.; Korinek, V.; Clevers, H. XTcf-3 transcription factor mediates β-catenin-induced axis formation in Xenopus embryos. Cell 1996, 86, 391–399. [Google Scholar] [CrossRef] [Green Version]
- He, T.C.; Sparks, A.B.; Rago, C.; Hermeking, H.; Zawel, L.; Da Costa, L.T.; Kinzler, K.W. Identification of c-MYC as a target of the APC pathway. Science 1998, 281, 1509–1512. [Google Scholar] [CrossRef] [PubMed]
- Leung, J.Y.; Kolligs, F.T.; Wu, R.; Zhai, Y.; Kuick, R.; Hanash, S.; Fearon, E.R. Activation of AXIN2 expression by β-catenin-T cell factor: A feedback repressor pathway regulating Wnt signaling. J. Biol. Chem. 2002, 277, 21657–21665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jhanwar-Uniyal, M.; Wainwright, J.V.; Mohan, A.L.; Tobias, M.E.; Murali, R.; Gandhi, C.D.; Schmidt, M.H. Diverse signaling mechanisms of mTOR complexes: mTORC1 and mTORC2 in forming a formidable relationship. Adv. Biol. Reg. 2019, 72, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Sarbassov, D.D.; Ali, S.M.; King, J.E.; Latek, R.R.; Erdjument-Bromage, H.; Sabatini, D.M. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 2002, 110, 163–175. [Google Scholar] [CrossRef] [Green Version]
- Haar, E.V.; Lee, S.I.; Bandhakavi, S.; Griffin, T.J.; Kim, D.H. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat. Cell Biol. 2007, 9, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Sarbassov, D.D.; Ali, S.M.; Kim, D.H.; Guertin, D.A.; Latek, R.R.; Erdjument-Bromage, H.; Sabatini, D.M. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 2004, 14, 1296–1302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pearce, L.R.; Sommer, E.M.; Sakamoto, K.; Wullschleger, S.; Alessi, D.R. Protor-1 is required for efficient mTORC2-mediated activation of SGK1 in the kidney. Biochem. J. 2011, 436, 169–179. [Google Scholar] [CrossRef]
- Frias, M.A.; Thoreen, C.C.; Jaffe, J.D.; Schroder, W.; Sculley, T.; Carr, S.A.; Sabatini, D.M. mSin1 is necessary for Akt/PKB phosphorylation, and its isoforms define three distinct mTORC2s. Curr. Biol. 2006, 16, 1865–1870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Roock, W.; Claes, B.; Bernasconi, D.; De Schutter, J.; Biesmans, B.; Fountzilas, G.; Tejpar, S. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: A retrospective consortium analysis. Lancet Oncol. 2010, 11, 753–762. [Google Scholar] [CrossRef]
- Day, F.L.; Jorissen, R.N.; Lipton, L.; Mouradov, D.; Sakthianandeswaren, A.; Christie, M.; Sieber, O.M. PIK3CA and PTEN gene and exon mutation-specific clinicopathologic and molecular associations in colorectal cancer. Clin. Cancer Res. 2013, 19, 3285–3296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, S.M.; Kim, K.M.; Kim, S.Y.; Shin, M.S.; Na, E.Y.; Lee, S.H.; Lee, J.Y. Frequent somatic mutations in serine/threonine kinase 11/Peutz-Jeghers syndrome gene in left-sided colon cancer. Cancer Res. 1998, 58, 3787–3790. [Google Scholar]
- Carpten, J.D.; Faber, A.L.; Horn, C.; Donoho, G.P.; Briggs, S.L.; Robbins, C.M.; Thomas, J.E. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature 2007, 448, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Memmott, R.M.; Dennis, P.A. Akt-dependent and-independent mechanisms of mTOR regulation in cancer. Cell. Signal. 2009, 21, 656–664. [Google Scholar] [CrossRef] [Green Version]
- Alessi, D.R.; James, S.R.; Downes, C.P.; Holmes, A.B.; Gaffney, P.R.; Reese, C.B.; Cohen, P. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Bα. Curr. Biol. 1997, 7, 261–269. [Google Scholar] [CrossRef] [Green Version]
- Beretta, L.; Gingras, A.C.; Svitkin, Y.V.; Hall, M.N.; Sonenberg, N. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J. 1996, 15, 658–664. [Google Scholar] [CrossRef]
- Gingras, A.C.; Kennedy, S.G.; O’Leary, M.A.; Sonenberg, N.; Hay, N. 4E-BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the Akt (PKB) signaling pathway. Genes Dev. 1998, 12, 502–513. [Google Scholar] [CrossRef]
- Holz, M.K.; Ballif, B.A.; Gygi, S.P.; Blenis, J. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell 2005, 123, 569–580. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, K. Identification of a member of the MAPKKK family as a potential mediator of TGF-beta signal transduction. Science 1995, 270, 2008–2011. [Google Scholar] [CrossRef]
- Cheruku, H.R.; Mohamedali, A.; Cantor, D.I.; Tan, S.H.; Nice, E.C.; Baker, M.S. Transforming growth factor-β, MAPK and Wnt signaling interactions in colorectal cancer. EuPA Open Proteom. 2015, 8, 104–115. [Google Scholar] [CrossRef] [Green Version]
- Fang, J.Y.; Richardson, B.C. The MAPK signalling pathways and colorectal cancer. Lancet Oncol. 2005, 6, 322–327. [Google Scholar] [CrossRef]
- Han, C.W.; Jeong, M.S.; Jang, S.B. Structure, signaling and the drug discovery of the Ras oncogene protein. BMB Rep. 2017, 50, 355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durrant, D.E.; Morrison, D.K. Targeting the Raf kinases in human cancer: The Raf dimer dilemma. Br. J. Cancer 2018, 118, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smalley, K.S.M. A pivotal role for ERK in the oncogenic behaviour of malignant melanoma? Int. J. Cancer 2003, 104, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Lavoie, J.N.; L’Allemain, G.; Brunet, A. Cyclin D1 expression is regulated positively by the p42/p44MAPK and negatively by the p38/HOGMAPK pathway. J. Biol. Chem. 1996, 271, 20608–20616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorrentino, A.; Thakur, N.; Grimsby, S.; Marcusson, A.; Von Bulow, V.; Schuster, N.; Landström, M. The type I TGF-β receptor engages TRAF6 to activate TAK1 in a receptor kinase-independent manner. Nat. Cell Biol. 2008, 10, 1199–1207. [Google Scholar] [CrossRef]
- Yamashita, M.; Fatyol, K.; Jin, C.; Wang, X.; Liu, Z.; Zhang, Y.E. TRAF6 mediates Smad-independent activation of JNK and p38 by TGF-β. Mol. Cell 2008, 31, 918–924. [Google Scholar] [CrossRef] [Green Version]
- Sundar, R.; Gudey, S.K.; Heldin, C.H.; Landström, M. TRAF6 promotes TGFβ-induced invasion and cell-cycle regulation via Lys63-linked polyubiquitination of Lys178 in TGFβ type I receptor. Cell Cycle 2015, 14, 554–565. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Zhang, W.; Font-Burgada, J.; Palmer, T.; Hamil, A.S.; Biswas, S.K.; Karin, M. Ubiquitin-conjugating enzyme Ubc13 controls breast cancer metastasis through a TAK1-p38 MAP kinase cascade. Proc. Natl. Acad. Sci. USA 2014, 111, 13870–13875. [Google Scholar] [CrossRef] [Green Version]
- Comes, F.; Matrone, A.; Lastella, P.; Nico, B.; Susca, F.C.; Bagnulo, R.; Simone, C. A novel cell type-specific role of p38α in the control of autophagy and cell death in colorectal cancer cells. Cell Death Differ. 2007, 14, 693–702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.Y.; Miah, A.; Sales, K.M.; Fuller, B.; Seifalian, A.M.; Winslet, M. Inhibition of the p38 MAPK pathway sensitises human colon cancer cells to 5-fluorouracil treatment. Int. J. Oncol. 2011, 38, 1695–1702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slattery, M.L.; Mullany, L.E.; Wolff, R.K.; Sakoda, L.C.; Samowitz, W.S.; Herrick, J.S. The p53-signaling pathway and colorectal cancer: Interactions between downstream p53 target genes and miRNAs. Genomics 2019, 111, 762–771. [Google Scholar] [CrossRef] [PubMed]
- Slattery, M.L.; Curtin, K.; Wolff, R.K.; Boucher, K.M.; Sweeney, C.; Edwards, S.; Samowitz, W. A comparison of colon and rectal somatic DNA alterations. Dis. Colon Rectum 2009, 52, 1304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russo, A.; Bazan, V.; Iacopetta, B.; Kerr, D.; Soussi, T.; Gebbia, N. The TP53 colorectal cancer international collaborative study on the prognostic and predictive significance of p53 mutation: Influence of tumor site, type of mutation, and adjuvant treatment. J. Clin. Oncol. 2005, 23, 7518–7528. [Google Scholar] [CrossRef] [PubMed]
- Mandinova, A.; Lee, S.W. The p53 pathway as a target in cancer therapeutics: Obstacles and promise. Sci. Transl. Med. 2011, 3, 64rv1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Lozano, G. Molecular pathways: Targeting Mdm2 and Mdm4 in cancer therapy. Clin. Cancer Res. 2013, 19, 34–41. [Google Scholar] [CrossRef] [Green Version]
- Kumar, M.; Lu, Z.; Takwi, A.A.L.; Chen, W.; Callander, N.S.; Ramos, K.S.; Li, Y. Negative regulation of the tumor suppressor p53 gene by microRNAs. Oncogene 2011, 30, 843–853. [Google Scholar] [CrossRef] [Green Version]
- Ryan, K.M.; Phillips, A.C.; Vousden, K.H. Regulation and function of the p53 tumor suppressor protein. Curr. Opin. Cell Biol. 2001, 13, 332–337. [Google Scholar] [CrossRef]
- Shen, J.; Vakifahmetoglu, H.; Stridh, H.; Zhivotovsky, B.; Wiman, K.G. PRIMA-1MET induces mitochondrial apoptosis through activation of caspase-2. Oncogene 2008, 27, 6571–6580. [Google Scholar] [CrossRef] [Green Version]
- Soleimani, A.; Rahmani, F.; Ferns, G.A.; Ryzhikov, M.; Avan, A.; Hassanian, S.M. Role of the NF-κB signaling pathway in the pathogenesis of colorectal cancer. Gene 2020, 726, 144132. [Google Scholar] [CrossRef]
- Wong, D.; Teixeira, A.; Oikonomopoulos, S.; Humburg, P.; Lone, I.N.; Saliba, D. Extensive characterization of NF-kappaB binding uncovers non-canonical motifs and advances the interpretation of genetic functional traits. Genome Biol. 2011, 12, R70. [Google Scholar] [CrossRef] [PubMed]
- Baeuerle, P.A.; Henkel, T. Function and activation of NF-kappa B in the immune system. Ann. Rev. Immunol. 1994, 12, 141–179. [Google Scholar] [CrossRef] [PubMed]
- Bonizzi, G.; Karin, M. The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 2004, 25, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Hassanzadeh, P. Colorectal cancer and NF-κB signaling pathway. Gastroenterol. Hepatol. Bed Bench. 2011, 4, 127–132. [Google Scholar] [PubMed]
- Sakamoto, K.; Maeda, S. Targeting NF-κB for colorectal cancer. Expert Opin. Ther. Targets 2010, 14, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Evertsson, S.; Sun, X.F. Protein expression of NF-kappaB in human colorectal adenocarcinoma. Int. J. Mol. Med. 2002, 10, 547–550. [Google Scholar] [PubMed]
- Yang, L.; Liu, Y.; Wang, M.; Qian, Y.; Dong, X.; Gu, H.; Wang, H.; Guo, S.; Hisamitsu, T. Quercetin-induced apoptosis of HT-29 colon cancer cells via inhibition of the Akt-CSN6-Myc signaling axis. Mol. Med. Rep. 2016, 14, 4559–4566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Refolo, M.G.; D’Alessandro, R.; Malerba, N.; Laezza, C.; Bifulco, M.; Messa, C.; Caruso, M.G.; Notarnicola, M.; Tutino, V. Anti Proliferative and Pro Apoptotic Effects of Flavonoid Quercetin Are Mediated by CB1 Receptor in Human Colon Cancer Cell Lines. J. Cell. Physiol. 2015, 230, 2973–2980. [Google Scholar] [CrossRef] [PubMed]
- Raja, S.B.; Rajendiran, V.; Kasinathan, N.K.; Amrithalakshmi, P.; Venkatabalasubramanian, S.; Murali, M.R.; Devaraj, H.; Devaraj, S.N. Differential cytotoxic activity of Quercetin on colonic cancer cells depends on ROS generation through COX-2 expression. Food Chem. Toxicol. 2017, 106, 92–106. [Google Scholar] [CrossRef] [PubMed]
- Shree, A.; Islam, J.; Sultana, S. Quercetin ameliorates reactive oxygen species generation, inflammation, mucus depletion, goblet disintegration, and tumor multiplicity in colon cancer: Probable role of adenomatous polyposis coli, β-catenin. Phytother. Res. 2021, 35, 2171–2184. [Google Scholar] [CrossRef] [PubMed]
- Dihal, A.A.; Van der Woude, H.; Hendriksen, P.J.M.; Charif, H.; Dekker, L.J.; Ijsselstijn, L.; de Boer, V.C.J.; Alink, G.M.; Burgers, P.C.; Rietjens, I.M.C.M.; et al. Transcriptome and proteome profiling of colon mucosa from quercetin fed F344 rats point to tumor preventive mechanisms, increased mitochondrial fatty acid degradation and decreased glycolysis. Proteomics. 2008, 8, 45–61. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Zhang, Z.; Li, W.; Kong, F.; Yi, P.; Huang, J.; Zhang, S. Network pharmacology-based prediction and verification of the active ingredients and potential targets of zuojinwan for treating colorectal cancer. Drug Des. Dev. Ther. 2020, 14, 2725. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wang, T.; Chen, D.; Ma, Q.; Zheng, Y.; Liao, S.; Zhang, J. Quercetin preferentially induces apoptosis in KRAS-mutant colorectal cancer cells via JNK signaling pathways. Cell Biol. Int. 2019, 43, 117–124. [Google Scholar] [CrossRef]
- Al-Ghamdi, M.A.; AL-Enazy, A.; Huwait, E.A.; Albukhari, A.; Harakeh, S.; Moselhy, S.S. Aumento da anexina V em resposta à combinação de galato de epigalocatequina e quercetina como uma potente parada do ciclo celular do câncer colorretal. Braz. J. Biol. 2021, 83, e248746. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, B.; Xu, P.; Yang, B. Integrated whole transcriptome profiling and bioinformatics analysis for revealing regulatory pathways associated with quercetin-induced apoptosis in HCT-116 cells. Front. Pharmacol. 2019, 10, 798. [Google Scholar] [CrossRef] [Green Version]
- Catalán, M.; Ferreira, J.; Carrasco-Pozo, C. The microbiota-derived metabolite of quercetin, 3, 4-dihydroxyphenylacetic acid prevents malignant transformation and mitochondrial dysfunction induced by hemin in colon cancer and normal colon epithelia cell lines. Molecules 2020, 25, 4138. [Google Scholar] [CrossRef]
- Dihal, A.A.; de Boer, V.C.; van der Woude, H.; Tilburgs, C.; Bruijntjes, J.P.; Alink, G.M.; Stierum, R.H. Quercetin, but not its glycosidated conjugate rutin, inhibits azoxymethane-induced colorectal carcinogenesis in F344 rats. J. Nutr. 2006, 136, 2862–2867. [Google Scholar] [CrossRef]
- Thakar, T.; Leung, W.; Nicolae, C.M. Ubiquitinated-PCNA protects replication forks from DNA2-mediated degradation by regulating Okazaki fragment maturation and chromatin assembly. Nat. Commun. 2020, 11, 2147. [Google Scholar] [CrossRef]
- Maga, G.; Hubscher, U. Proliferating cell nuclear antigen (PCNA): A dancer with many partners. J. Cell Sci. 2003, 116, 3051–3060. [Google Scholar] [CrossRef] [Green Version]
- Guo, C.; Liu, S.; Sun, M.Z. Potential role of Anxa1 in cancer. Future Oncol. 2013, 9, 1773–1793. [Google Scholar] [CrossRef]
- Su, N.; Xu, X.-Y.; Chen, H. Increased expression of annexin A1 is correlated with K-ras mutation in colorectal cancer. Tohoku J. Exp. Med. 2010, 222, 243–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.S.; Wannatung, T.; Lee, S.; Yang, W.K.; Chung, S.H.; Lim, J.S.; Choe, W.; Kang, I.; Kim, S.S.; Ha, J. Quercetin enhances hypoxia-mediated apoptosis via direct inhibition of AMPK activity in HCT116 colon cancer. Apoptosis 2012, 17, 938–949. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Yang, X.; Chen, C.; Cai, S.; Hu, J. Isorhamnetin suppresses colon cancer cell growth through the PI3K-Akt-mTOR pathway. Molec. Med. Rep. 2014, 9, 935–940. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Yi, J.; Wu, Y.; Chen, X.; Zeng, J.; Wu, J.; Peng, W. 3, 3′-dimethylquercetin inhibits the proliferation of human colon cancer RKO cells through Inducing G2/M cell cycle arrest and apoptosis. Anti Cancer Agents Med. Chem. 2019, 19, 402–409. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Guo, Y.; Wang, M.; Dong, H.; Zhang, J.; Zhang, L. Quercetrin from Toona sinensis leaves induces cell cycle arrest and apoptosis via enhancement of oxidative stress in human colorectal cancer SW620 cells. Oncol. Rep. 2017, 38, 3319–3326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clute, P.; Pines, J. Temporal and spatial control of cyclin B1 destruction in metaphase. Nat. Cell Biol. 1999, 1, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Darband, S.G.; Kaviani, M.; Yousefi, B.; Sadighparvar, S.; Pakdel, F.G.; Attari, J.A.; Mohebbi, I.; Naderi, S.; Majidinia, M. Quercetin: A functional dietary flavonoid with potential chemo-preventive properties in colorectal cancer. J. Cell. Physiol. 2018, 233, 6544–6560. [Google Scholar] [CrossRef] [PubMed]
- Na, S.; Ying, L.; Jun, C.; Ya, X.; Suifeng, Z.; Yuxi, H.; Jing, W.; Zonglang, L.; Xiajoun, Y.; Yue, W. Study on the molecular mechanism of nightshade in the treatment of colon cancer. Bioengineered 2022, 13, 1575–1589. [Google Scholar] [CrossRef] [PubMed]
- Kee, J.Y.; Han, Y.H.; Kim, D.S.; Mun, J.G.; Park, J.; Jeong, M.Y.; Um, J.Y.; Hong, S.H. Inhibitory effect of quercetin on colorectal lung metastasis through inducing apoptosis, and suppression of metastatic ability. Phytomedicine 2016, 23, 1680–1690. [Google Scholar] [CrossRef]
- Xavier, C.P.; Lima, C.F.; Rohde, M.; Pereira-Wilson, C. Quercetin enhances 5-fluorouracil-induced apoptosis in MSI colorectal cancer cells through p53 modulation. Cancer Chemother. Pharmacol. 2011, 68, 1449–1457. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.A.; Zhang, S.; Yin, Q.; Zhang, J. Quercetin induces human colon cancer cells apoptosis by inhibiting the nuclear factor-kappa B Pathway. Pharmacogn. Mag. 2015, 11, 404–409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cincin, Z.B.; Unlu, M.; Kiran, B.; Bireller, E.S.; Baran, Y.; Cakmakoglu, B. Apoptotic Effects of Quercitrin on DLD-1 Colon Cancer Cell Line. Pathol. Oncol. Res. 2015, 21, 333–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, G.T.; Lee, S.H.; Kim, J.I.; Kim, Y.M. Quercetin regulates the sestrin 2-AMPK- p38 MAPK signaling pathway and induces apoptosis by increasing the generation of intracellular ROS in a p53-independent manner. Int. J. Mol. Med. 2014, 33, 863–869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, G.T.; Lee, S.H.; Kim, Y.M. Quercetin Regulates Sestrin 2-AMPK-mTOR Signaling Pathway and Induces Apoptosis via Increased Intracellular ROS in HCT116 Colon Cancer Cells. J. Cancer. Prev. 2013, 18, 264–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, H.; Liu, Y.; Wu, K.; Luo, H.; Cui, L. AMPK activation overcomes anti-EGFR antibody resistance induced by KRAS mutation in colorectal cancer. Cell Comm. Sign. 2020, 18, 115. [Google Scholar] [CrossRef] [PubMed]
- Bulzomi, P.; Galluzzo, P.; Bolli, A.; Leone, S.; Acconcia, F.; Marino, M. The pro-apoptotic effect of quercetin in cancer cell lines requires ERβ-dependent signals. J. Cell. Physiol. 2012, 227, 1891–1898. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Pathak, S.; Jothimani, G.; Roy, S. Antiproliferative effects of combinational therapy of Lycopodium clavatum and quercetin in colon cancer cells. J. Basic Clin. Physiol. Pharmacol. 2020, 31, 20190193. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.; Piao, M.; Song, Y.; Liu, C. Quercetin suppresses AOM/DSS-induced colon carcinogenesis through its anti-inflammation effects in mice. J. Immunol. Res. 2020, 2020, 9242601. [Google Scholar] [CrossRef] [PubMed]
- Hashemzaei, M.; Delarami Far, A.; Yari, A.; Heravi, R.E.; Tabrizian, K.; Taghdisi, S.M.; Sadegh, S.E.; Tsarouhas, K.; Kouretas, D.; Tzanakakis, G.; et al. Anticancer and apoptosis-inducing effects of quercetin in vitro and in vivo. Oncol. Rep. 2017, 38, 819–828. [Google Scholar] [CrossRef] [Green Version]
- Benito, I.; Encío, I.J.; Milagro, F.I.; Alfaro, M.; Martínez-Peñuela, A.; Barajas, M.; Marzo, F. Microencapsulated Bifidobacterium bifidum and Lactobacillus gasseri in Combination with Quercetin Inhibit Colorectal Cancer Development in ApcMin/+ Mice. Int. J. Molec. Sci. 2021, 22, 4906. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Song, Y.; Zhang, X. Quercetin Suppresses the Migration and Invasion in Human Colon Cancer Caco-2 Cells Through Regulating Toll-like Receptor 4/Nuclear Factor-kappa B Pathway. Pharmacogn. Mag. 2016, 12, S237–S244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, J.; Song, D.; Jiang, S.; Yang, X.; Ding, T.; Zhang, H.; Yin, Q. Quercetin restrains TGF-β1-induced epithelial–mesenchymal transition by inhibiting Twist1 and regulating E-cadherin expression. Biochem. Biophys. Res. Comm. 2018, 498, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Mendonsa, A.M.; Na, T.Y.; Gumbiner, B.M. E-cadherin in contact inhibition and cancer. Oncogene 2018, 37, 4769–4780. [Google Scholar] [CrossRef] [PubMed]
- Damodharan, U.; Ganesan, R.; Radhakrishnan, U.C. Expression of MMP2 and MMP9 (Gelatinases A and B) in Human Colon Cancer Cells. Appl. Biochem. Biotechnol. 2011, 165, 1245–1252. [Google Scholar] [CrossRef] [PubMed]
- Sang-Oh, Y.; Soo-Jin, P.; Chang-Hyun, Y.; An-Sik, C. Roles of matrix metalloproteinases in tumor metastasis and angiogenesis. J. Biochem. Molec. Biol. 2003, 36, 128–137. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Zhang, J.; Wang, K.; Han, W.; Wang, X.; Gao, M.; Wang, Z.; Sun, Y.; Yan, H.; Zhang, H.; et al. Quercetin overcomes colon cancer cells resistance to chemotherapy by inhibiting solute carrier family 1, member 5 transporter. Eur. J. Pharmacol. 2020, 881, 178185. [Google Scholar] [CrossRef]
- Qi, J.; Yu, J.; Li, Y.; Luo, J.; Zhang, C.; Ou, S.; Peng, X. Alternating consumption of β-glucan and quercetin reduces mortality in mice with colorectal cancer. Food Sci. Nutr. 2019, 7, 3273–3285. [Google Scholar] [CrossRef] [PubMed]
- Papuc, C.; Goran, G.V.; Predescu, C.N.; Nicorescu, V.; Stefan, G. Plant polyphenols as antioxidant and antibacterial agents for shelf-life extension of meat and meat products: Classification, structures, sources, and action mechanisms. Comprehens. Rev. Food Sci. Food Saf. 2017, 16, 1243–1268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scalise, M.; Pochini, L.; Console, L.; Losso, M.A.; Indiveri, C. The human SLC1A5 (ASCT2) amino acid transporter: From function to structure and role in cell biology. Front. Cell Dev. Biol. 2018, 96. [Google Scholar] [CrossRef] [PubMed]
- Al Obeed, O.A.; Alkhayal, K.A.; Al Sheikh, A.; Zubaidi, A.M.; Vaali-Mohammed, M.A.; Boushey, R.; Abdulla, M.H. Increased expression of tumor necrosis factor-α is associated with advanced colorectal cancer stages. World J. Gastroenterol. 2014, 20, 18390. [Google Scholar] [CrossRef]
- Reed, J.C. Apoptosis-targeted therapies for cancer. Cancer Cell 2003, 3, 17–22. [Google Scholar] [CrossRef] [Green Version]
- Lawrence, T.; Willoughby, D.A.; Gilroy, D.W. Anti-inflammatory lipid mediators and insights into the resolution of inflammation. Nat. Rev. Immunol. 2002, 2, 787–795. [Google Scholar] [CrossRef]
- Kobori, M.; Takahashi, Y.; Akimoto, Y.; Sakurai, M.; Matsunaga, I.; Nishimuro, H.; Ippoushi, K.; Oike, H.; Ohnishi-Kameyama, M. Chronic high intake of quercetin reduces oxidative stress and induces expression of the antioxidant enzymes in the liver and visceral adipose tissues in mice. J. Funct. Foods. 2015, 15, 551–560. [Google Scholar] [CrossRef]
- Granado-Serrano, A.B.; Martín, M.A.; Bravo, L.; Goya, L.; Ramos, S. Quercetin modulates Nrf2 and glutathione-related defenses in HepG2 cells: Involvement of p38. Chem. Biol. Interact. 2012, 195, 154–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawamura, K.; Qi, F.; Kobayashi, J. Potential relationship between the biological effects of low-dose irradiation and mitochondrial ROS production. J. Radiat. Res. 2018, 59 (Suppl. 2), ii91–ii97. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Hu, X.; Liu, Y.; Dong, S.; Wen, Z.; He, W.; Zhang, S.; Huang, Q.; Shi, M. ROS signaling under metabolic stress: Cross-talk between AMPK and AKT pathway. Mol. Cancer. 2017, 16, 79. [Google Scholar] [CrossRef] [Green Version]
- Jalmi, S.K.; Sinha, A.K. ROS mediated MAPK signaling in abiotic and biotic stress-striking similarities and differences. Front. Plant Sci. 2015, 6, 769. [Google Scholar] [CrossRef] [Green Version]
- Stoiber, W.; Obermayer, A.; Steinbacher, P.; Krautgartner, W.D. The role of reactive oxygen species (ROS) in the formation of extracellular traps (ETs) in humans. Biomolecules 2015, 5, 702–723. [Google Scholar] [CrossRef] [Green Version]
- Vurusaner, B.; Poli, G.; Basaga, H. Tumor suppressor genes and ROS: Complex networks of interactions. Free Radic. Biol. Med. 2012, 52, 7–18. [Google Scholar] [CrossRef]
- Aires, V.; Limagne, E.; Cotte, A.K.; Latruffe, N.; Ghiringhelli, F.; Delmas, D. Resveratrol metabolites inhibit human metastatic colon cancer cells progression and synergize with chemotherapeutic drugs to induce cell death. Molec. Nutr. Food Res. 2013, 57, 1170–1181. [Google Scholar] [CrossRef] [PubMed]
- Shakibaei, M.; Mobasheri, A.; Lueders, C.; Busch, F.; Shayan, P.; Goel, A. Curcumin enhances the effect of chemotherapy against colorectal cancer cells by inhibition of NF-κB and Src protein kinase signaling pathways. PLoS ONE 2013, 8, e57218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, B.; Jiang, L.; Xia, Q.; Zhong, L. Synergistic inhibitory effects of curcumin and 5-fluorouracil on the growth of the human colon cancer cell line HT-29. Chemotherapy 2005, 52, 23–28. [Google Scholar] [CrossRef]
- Hwang, J.-T.; Ha, J.; Park, O.J. Combination of 5-fluorouracil and genistein induces apoptosis synergistically in chemo-resistant cancer cells through the modulation of AMPK and COX-2 signaling pathways. Biochem. Biophys. Res. Comm. 2005, 332, 433–440. [Google Scholar] [CrossRef] [PubMed]

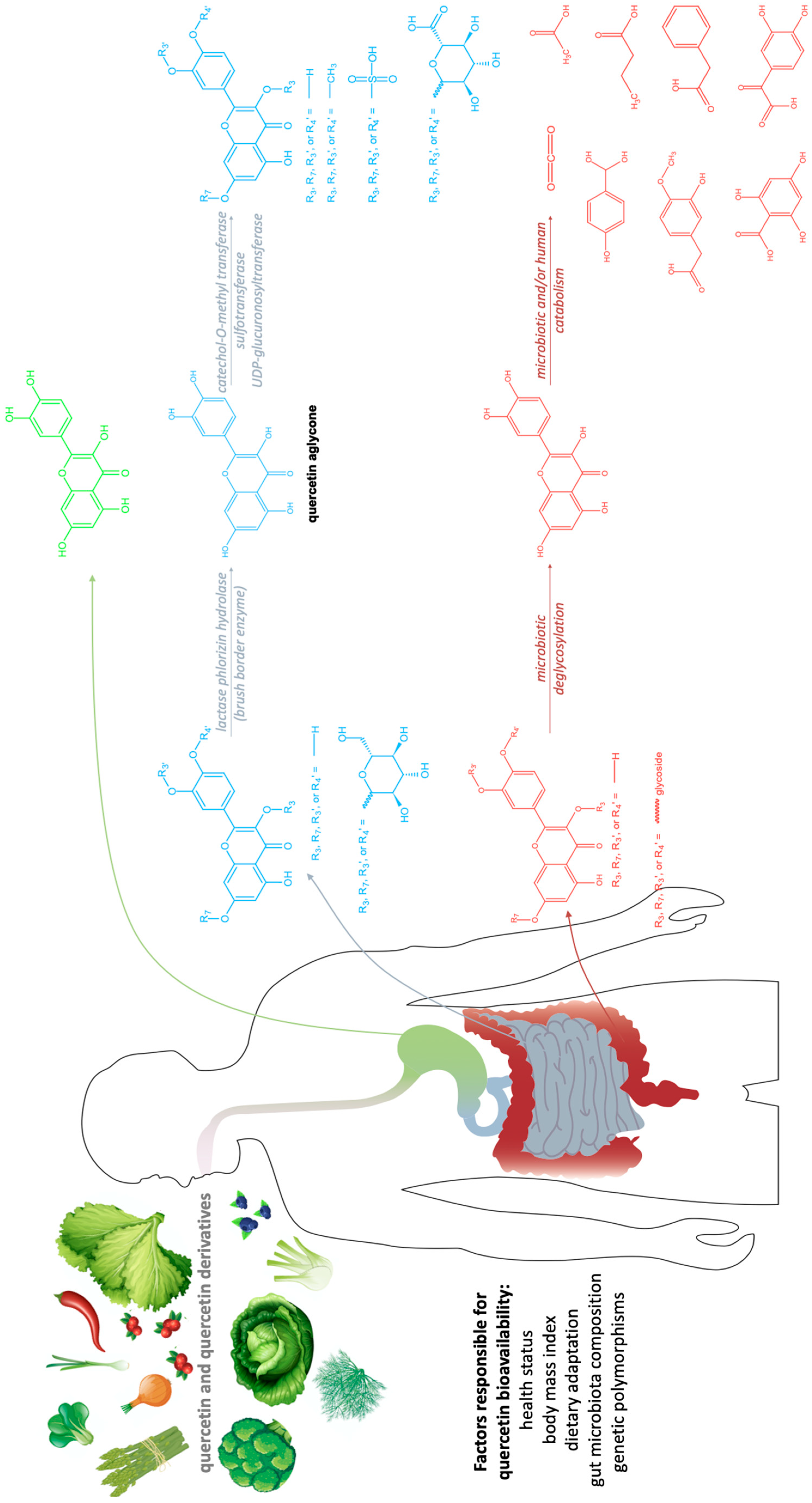

| Plant | Quercetin Concentration |
|---|---|
| (mg/100 g Fresh Weight) | |
| Dill | 79.0 |
| Fennel leaves | 46.8 |
| Onion | 45.0 |
| Oregano | 42.0 |
| Chili pepper | 32.6 |
| Spinach | 27.2 |
| Cranberry | 25.0 |
| Kale | 22.6 |
| Cherry | 17.4 |
| Lettuce | 14.7 |
| Blueberry | 14.6 |
| Asparagus | 14.0 |
| Broccoli | 13.7 |
| Chives | 10.4 |
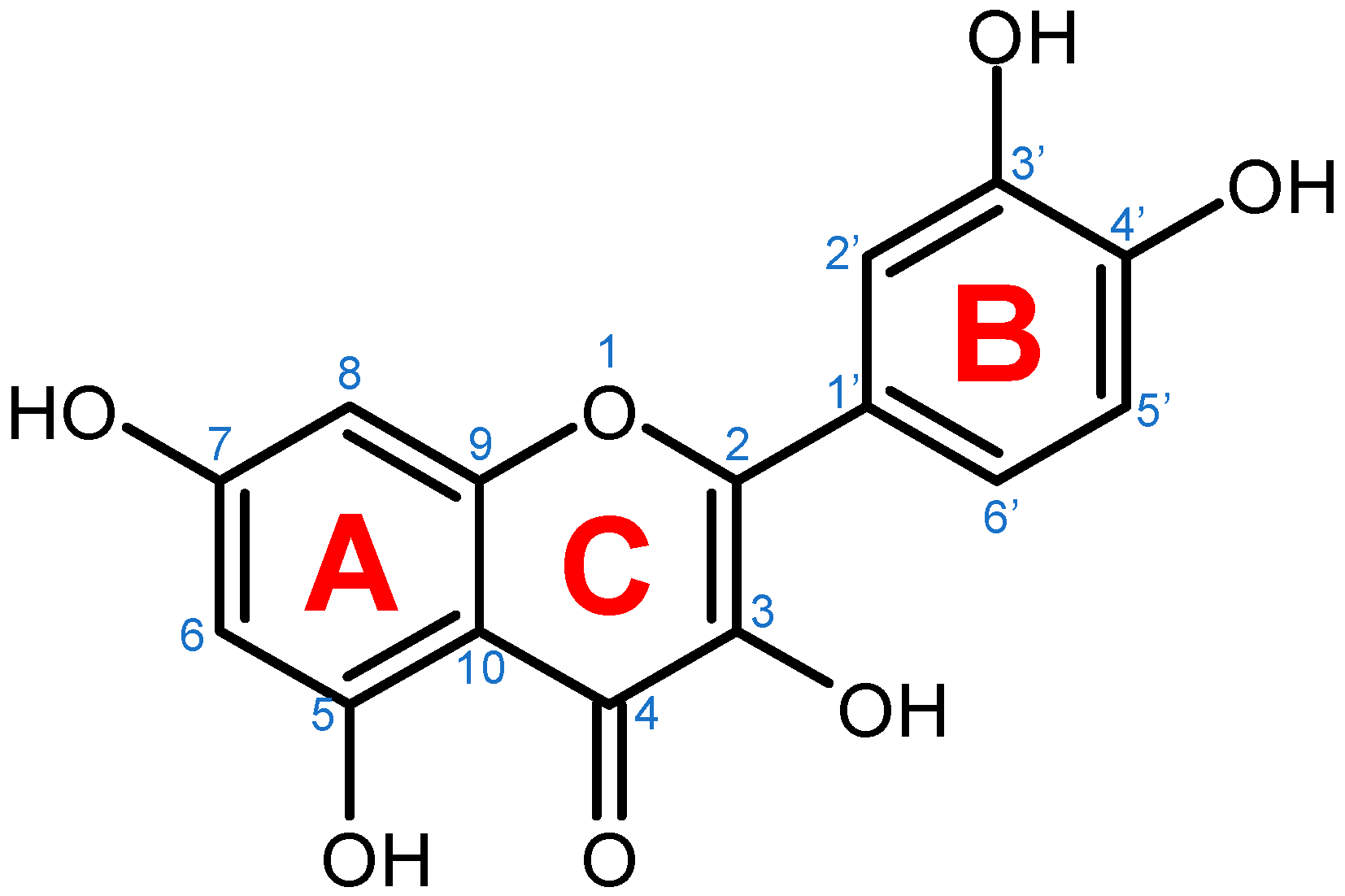
| Selected Quercetin Derivative | Chemical Structure | Modification on A-Ring | Modification on B-Ring | Modification on C-Ring |
|---|---|---|---|---|
| Quercetin |  | - | - | 3-OH to 3-O- glucoside |
| 3-O-glucoside | ||||
| (Isoquercetin) | ||||
| Quercetin |  | - | - | 3-OH to 3-O- galactoside |
| 3-O-galactoside | ||||
| (Hyperoside) | ||||
| Quercetin |  | - | - | 3-OH to 3-O- rhamnoside |
| 3-O-rhamnoside | ||||
| (Quercitrin) | ||||
| Quercetin |  | 7-OH to 7-O-glucoside | - | - |
| 7-O-glucoside | ||||
| (Quercimeritrin) | ||||
| Quercetin |  | - | - | 3-OH to 3-O- rutinoside |
| 3-O-rutinoside | ||||
| (Rutin) | ||||
| Quercetin |  | - | 3′-OH to 3′-methyl ether | - |
| 3-methyl ether | ||||
| (Isorhamnetin) | ||||
| Isorhamnetin |  | - | 3′-OH to 3′-methyl ether 4′-OH to 4′-O-glucoside | 3-OH to 3-O- rutinoside |
| 3-O-rutinoside- | ||||
| 4′-O-glucoside | ||||
| Isorhamnetin |  | 7-OH to 7-O-glucoside | 3′-OH to 3′-methyl ether | 3-OH to 3-O- rutinoside |
| 3-O-rutinoside- | ||||
| 7-O-glucoside | ||||
| Quercetin |  | - | 3′-OH to 3′-methyl ether | 3-OH to 3-methyl ether |
| 3,3′-dimethyl ether | ||||
| Quercetin |  | - | 4′-OH to 4′-O-glucoside | - |
| 4′-O-glucoside | ||||
| (Spiraeoside) | ||||
| Quercetin |  | 7-OH to 7-O-rhamnoside | - | - |
| 7-O-rhamnoside | ||||
| Quercetin |  | 7-OH to 7-O-rhamnoside | - | 3-OH to 3-O- glucoside |
| 3-O-glucoside- | ||||
| 7-O-rhamnoside | ||||
| (VincetoxicosideA) | ||||
| 4′-O-methyl |  | - | 4′-OH to 4′- methyl ether | - |
| quercetin | ||||
| (Tamarixetin) | ||||
| 7-O-methyl |  | 7-OH to 7-methyl ether | - | - |
| quercetin | ||||
| (Rhamnetin) | ||||
| 3′, 7-dimethyl quercetin |  | 7-OH to 7-methyl ether | 3′-OH to 3′-methyl ether | - |
| (Rhamnazin) |
| Molecular Targets | Testing System | Reference |
|---|---|---|
| ↓ p-AKT, MYC | In vitro: HT-29 cell culture | [131] |
| ↓ CB1 receptor, Wnt/β-catenin, p-GSK3β, | In vitro: Caco-2 and DLD-1 cell cultures | [132] |
| p-PI3K, p-AKT, p-S6, p-4E-BP1, p-STAT3 | ||
| ↓ p-AKT, p-GSK3β, Cyclin D1 | In vitro: HT-29 and HCT-15 cell cultures | [133] |
| ↓ PCNA | In vivo: Wistar rats | [134] |
| ↓ ANXA1 | In vivo: F344 rats | [135] |
| ? | In vitro: HCT-116 and HT-29 cell cultures | [136] |
| ? | In vitro: DLD-1KRASG13D, DLD-1KRASWT, SW480KRASG12V, HCT-116KRASG13D, Colo205KRASWT, WIDRKRASWT, and HT-29 KRASWT cell cultures | [137] |
| ? | In vitro: HCT-116 cell culture | [138] |
| ? | In vitro: HCT15 and CO115 cell cultures | [135] |
| ? | In vitro: HCT-116 cell culture | [139] |
| ? | In vitro: RKO and CCD841 cell cultures | [140] |
| ? | In vivo: F344 AOM treated rats | [141] |
| Molecular Targets | Testing System | Reference |
|---|---|---|
| ↑ JNK, c-Jun | In vitro: Caco-2 and DLD-1 cell cultures | [132] |
| ↑ COX-2 | In vitro: HT-29 and HCT-15 cell cultures | [133] |
| ↑ Caspase-3, Cytochrome-c | ||
| ↑ Bax, PARP, APC | In vivo: Wistar rats | [134] |
| ↓ Bcl-2, β-catenin | ||
| ↓ p-ERK, KRAS | In vitro: HCT-15 cell culture | [135] |
| ↓ TSC22 domain family 3 | In vivo: F344 rats | [135] |
| ↓ p-AKT, KRAS | In vitro: CO115 cell culture | [135] |
| ↓ PI3K, AKT, p-AKT, Bcl-2 | In vitro: HCT-116 and HT29 cell cultures | [136] |
| ↑ Bax | ||
| ↑ Caspase-3, p-JNK | In vitro: DLD-1KRASG13D and DLD-1KRASWT cell cultures | [137] |
| ↓ p-AKT | ||
| ↑ Caspase-3, Cytochrome-c | In vitro: RKO and CCD841 cell cultures | [140] |
| ↓ AMPK, HIF-1 | In vitro: HCT-116 | [146] |
| ↓ Bcl-2 | In vitro: RKO cell culture | [148] |
| ↑ Bax, cleaved-Caspase-3, cleaved-Caspase-9 | ||
| ↑ Bax, Cytochrome-c, Caspase-9, Apaf-1, Caspase-3 | In vitro: SW620 cell culture | [149] |
| ↓ GPx, Catalase | ||
| ↓ PI3K, AKT ↑ Caspase-3, Bax | In vitro: SW480 cell culture | [152] |
| ↑ PARP, cleaved-Caspase-3, cleaved-Caspase-9 | In vitro: CT-26 cell culture | [153] |
| ↓ Bcl-2, Bcl-xL | ||
| ↓ MMP-2, MMP-9, N-cadherin, β-catenin, Snail | In vivo: mouse model of CRC lung metastasis | [153] |
| ↑ E-cadherin | ||
| ↑ p53, BAX, p-p38 ↓ Bcl-2 | In vitro: HCT-15 cell culture | [154] |
| ↑ p53, cleaved-Caspase 3, cleaved-Caspase 9, PARP, cleaved-PARP ↓ Bcl-2 | In vitro: CO115 cell culture | [154] |
| ↑ Bax, Caspase-3, Caspase-9 | In vitro: Caco-2 and SW-620 cell cultures | [155] |
| ↓ Bcl-2, NF-κB | ||
| ↓ MMP | In vitro: DLD-1 cell culture | [156] |
| ↓ MMP | In vitro: HCT-116 cell culture | [157] |
| ↑ SIRT-2, p-AMPK, p-p38 | In vitro: HCT-116 cell culture | [158] |
| ↓ p-mTOR | ||
| ↑ Caspase-3, cleaved-PARP, p-p38 | In vitro: DLD-1 cell culture | [160] |
| ↓ Bcl-2, Cyclin D1, | In vitro: Colo320 cell culture | [161] |
| ↑ Bax, Caspase-3, Wnt1, Catalase | ||
| ? | In vivo: AOM/DSS-treated wild-type C57BL/6J mice | [162] |
| ? | In vitro: HCT-116 cell culture | [138] |
| ? | In vitro: HCT-116 cell culture | [141] |
| ? | In vitro: HCT-116p53-wt, HCT-116p53-null, HCT-15KRAS-mutated cell culture | [154] |
| ? | In vitro: CT-26 cell culture | [163] |
| Molecular Targets | Testing System | Reference |
|---|---|---|
| ↓ PI3K, AKT | In vivo: SPF grade BALB/C nude mice | [152] |
| ↑ caspase-3, Bax | ||
| ↑ p-Erk, p-JNK, p-p38 | In vivo: mouse model of CRC lung metastasis | [153] |
| ? | In vivo: AOM/DSS-treated wild-type C57BL/6J mice | [162] |
| ? | In vivo: F344 AOM-treated rats | [139] |
| Molecular Targets | Testing System | Reference |
|---|---|---|
| ? | In vivo: F344 AOM-treated rats | [139] |
| ? | In vivo: AOM/DSS-treated wild-type C57BL/6J mice | [162] |
| ? | In vivo: CT-26 mouse Xenograft model | [163] |
| ? | In vivo: ApcMin/+ mice | [164] |
| Molecular Targets | Testing System | Reference |
|---|---|---|
| ↑ E-cadherin | In vitro: Caco-2 cell culture | [165] |
| ↓ MMP-2, MMP-9, TLR4, NF-ҡB, TNF-α, COX-2,IL-6 | ||
| ↑ E-cadherin ↓ Twist1, Vimentin | In vitro: SW-480 cell culture [165] | [165] |
| Molecular Targets | Testing System | Reference |
|---|---|---|
| ↓ COX-2, iNOS, NF-κB | In vivo: Wistar rats | [134] |
| ↓ SLC1A5 glutamine transporter | In vitro: SW620/Ad300 cell culture | [170] |
| ↓ TNF-α | In vivo: AOM/DSS-treated wild-type C57BL/6J mice | [171] |
| ? | In vivo: AOM/DSS-treated wild-type C57BL/6J mice | [162] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neamtu, A.-A.; Maghiar, T.-A.; Alaya, A.; Olah, N.-K.; Turcus, V.; Pelea, D.; Totolici, B.D.; Neamtu, C.; Maghiar, A.M.; Mathe, E. A Comprehensive View on the Quercetin Impact on Colorectal Cancer. Molecules 2022, 27, 1873. https://doi.org/10.3390/molecules27061873
Neamtu A-A, Maghiar T-A, Alaya A, Olah N-K, Turcus V, Pelea D, Totolici BD, Neamtu C, Maghiar AM, Mathe E. A Comprehensive View on the Quercetin Impact on Colorectal Cancer. Molecules. 2022; 27(6):1873. https://doi.org/10.3390/molecules27061873
Chicago/Turabian StyleNeamtu, Andreea-Adriana, Teodor-Andrei Maghiar, Amina Alaya, Neli-Kinga Olah, Violeta Turcus, Diana Pelea, Bogdan Dan Totolici, Carmen Neamtu, Adrian Marius Maghiar, and Endre Mathe. 2022. "A Comprehensive View on the Quercetin Impact on Colorectal Cancer" Molecules 27, no. 6: 1873. https://doi.org/10.3390/molecules27061873
APA StyleNeamtu, A.-A., Maghiar, T.-A., Alaya, A., Olah, N.-K., Turcus, V., Pelea, D., Totolici, B. D., Neamtu, C., Maghiar, A. M., & Mathe, E. (2022). A Comprehensive View on the Quercetin Impact on Colorectal Cancer. Molecules, 27(6), 1873. https://doi.org/10.3390/molecules27061873







