Microbial Transformation and Biological Activities of the Prenylated Aromatic Compounds from Broussonetia kazinoki
Abstract
:1. Introduction
2. Results and Discussion
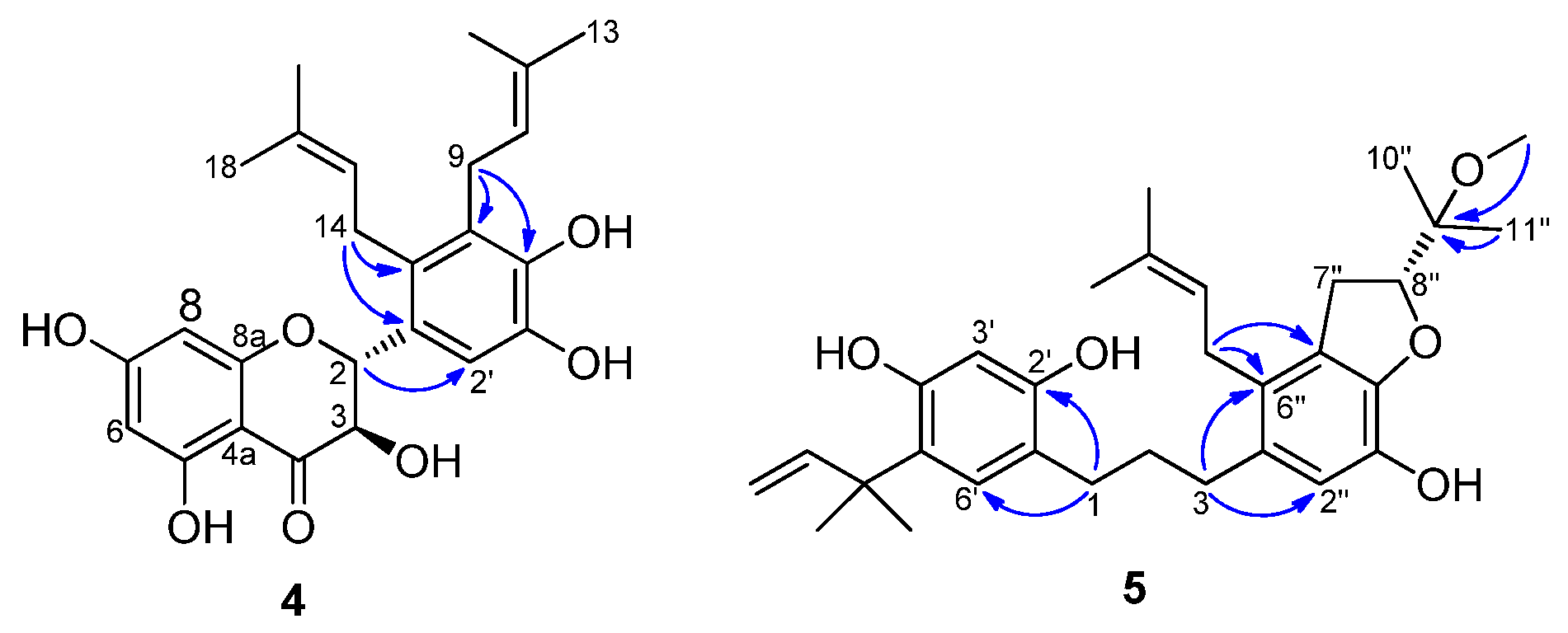
2.1. Structure Elucidation of Compounds from Broussonetia kazinoki
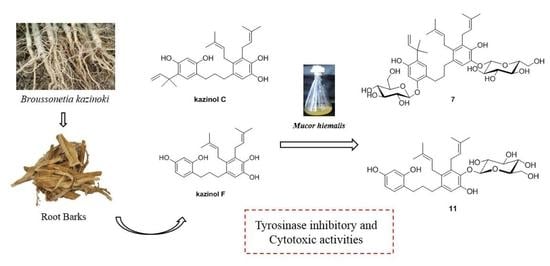
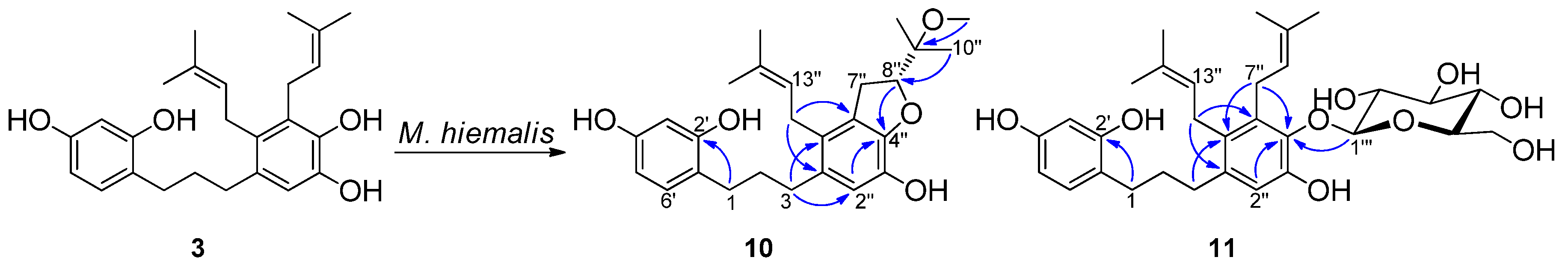
2.2. Microbial Transformation of Kazinols C and F by Mucor hiemalis
2.3. Tyrosinase Inhibitory Activity
2.4. Cytotoxic Activity
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Materials and Microorganisms
3.3. Isolation of Active Compounds from Broussonetia kazinoki
3.4. Microbial Screening Procedures
3.5. Scale-Up Fermentation of 1 and 3 with Mucor hiemalis
3.6. Mushroom Tyrosinase Activity
3.7. Cytotoxic Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Zhang, P.C.; Wang, S.; Wu, Y.; Chen, R.Y.; Yu, D.Q. Five new diprenylated flavonols from the leaves of Broussonetia kazinoki. J. Nat. Prod. 2001, 64, 1206–1209. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Lim, J.; Lee, D.Y.; Ryu, J.H.; Lim, J.S. Kazinol C from Broussonetia kazinoki activates AMP-activated protein kinase to induce antitumorigenic effects in HT-29 colon cancer cells. Oncol. Rep. 2015, 33, 223–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vu, N.K.; Ha, M.T.; Kim, C.S.; Gal, M.; Kim, J.A.; Woo, M.H.; Lee, J.H.; Min, B.S. Structural characterization of prenylated compounds from Broussonetia kazinoki and their antiosteoclastogenic activity. Phytochemistry 2021, 188, 112791. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Kim, H.J.; Cha, S.H.; Jun, H.S. Protective effects of Broussonetia kazinoki Siebold fruit extract against palmitate-induced lipotoxicity in mesangial cells. Evid.-Based Complement. Altern. Med. 2019, 2019, 4509403. [Google Scholar] [CrossRef]
- Lee, J.K.; Ha, H.; Lee, H.Y.; Park, S.J.; Jeong, S.I.; Choi, Y.J.; Shin, H.Y. Inhibitory effects of heartwood extracts of Broussonetia kazinoki Sieb on the development of atopic dermatitis in NC/Nga mice. Biosci. Biotechnol. Biochem. 2010, 74, 1802–1806. [Google Scholar] [CrossRef]
- Hwang, J.; Lee, S.J.; Yoo, M.; Go, G.Y.; Lee, D.Y.; Kim, Y.K.; Seo, D.W.; Kang, J.S.; Ryu, J.H.; Bae, G.U. Kazinol-P from Broussonetia kazinoki enhances skeletal muscle differentiation via p38MAPK and MyoD. Biochem. Biophys. Res. Commun. 2015, 456, 471–475. [Google Scholar] [CrossRef]
- Lee, H.J.; Park, J.H.; Jang, D.I.; Ryu, J.H. Antioxidant Components from Broussonetia kazinoki. Yakhak Hoeji 1997, 41, 439–443. [Google Scholar]
- Baek, Y.S.; Ryu, Y.B.; Curtis-Long, M.J.; Ha, T.J.; Rengasamy, R.; Yang, M.S.; Park, K.H. Tyrosinase inhibitory effects of 1,3-diphenylpropanes from Broussonetia kazinoki. Bioorg. Med. Chem. 2009, 17, 35–41. [Google Scholar] [CrossRef]
- El-Nashar, H.A.; El-Din, M.I.; Hritcu, L.; Eldahshan, O.A. Insights on the inhibitory power of flavonoids on tyrosinase activity: A survey from 2016 to 2021. Molecules 2021, 26, 7546. [Google Scholar] [CrossRef]
- Sari, S.; Barut, B.; Özel, A.; Şöhretoğlu, D. Tyrosinase inhibitory effects of Vinca major and its secondary metabolites: Enzyme kinetics and in silico inhibition model of the metabolites validated by pharmacophore modeling. Bioorg. Chem. 2019, 92, 103259. [Google Scholar] [CrossRef]
- Khan, M.T.H. Molecular design of tyrosinase inhibitors: A critical review of promising novel inhibitors from synthetic origins. Pure Appl. Chem. 2007, 79, 2277–2295. [Google Scholar] [CrossRef] [Green Version]
- Uyen, L.D.P.; Nguyen, D.H.; Kim, E.K. Mechanism of skin pigmentation. Biotechnol. Bioprocess Eng. 2008, 13, 383–395. [Google Scholar] [CrossRef]
- Carballo-Carbajal, I.; Laguna, A.; Romero-Giménez, J.; Cuadros, T.; Bové, J.; Martinez-Vicente, M.; Parent, A.; Gonzalez-Sepulveda, M.; Peñuelas, N.; Torra, A.; et al. Brain tyrosinase overexpression implicates age-dependent neuromelanin production in Parkinson’s disease pathogenesis. Nat. Commun. 2019, 10, 973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Souza, P.M.; Elias, S.T.; Simeoni, L.A.; de Paula, J.E.; Gomes, S.M.; Guerra, E.N.S.; Fonseca, Y.M. Plants from Brazilian Cerrado with potent tyrosinase inhibitory activity. PLoS ONE 2012, 7, e48589. [Google Scholar] [CrossRef] [Green Version]
- Bianchini, L.F.; Arruda, M.F.C.; Vieira, S.R.; Campelo, P.M.S.; Grégio, A.M.T.; Rosa, E.A.R. Microbial biotransformation to obtain new antifungals. Front. Microbiol. 2015, 6, 1433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cano-Flores, A.; Gómez, J.; Escalona-Torres, I.S.; Velasco-Bejarano, B. Microorganisms as biocatalysts and enzyme sources. In Microorganisms; Blumenberg, M., Shaaban, M., Elgaml, A., Eds.; IntechOpen: London, UK, 2020. [Google Scholar]
- Han, F.; Lee, I.-S. Microbial transformation of the antimalarial sesquiterpene endoperoxide dihydroartemisinin. Nat. Prod. Res. 2017, 31, 883–889. [Google Scholar] [CrossRef] [PubMed]
- Han, F.; Lee, I.-S. Microbial transformation of bavachin by Absidia coerulea. Phytochem. Lett. 2016, 18, 136–139. [Google Scholar] [CrossRef]
- Xiao, Y.; Lee, I.-S. Microbial metabolism of prenylated apigenin derivatives by Mucor hiemalis. Phytochem. Lett. 2016, 16, 197–202. [Google Scholar] [CrossRef]
- Ikuta, J.; Hano, Y.; Nomura, T.; Kawakami, Y.; Sato, T. Components of Broussonetia kazinoki SIEB. I. Structures of two new isoprenylated flavans and five new isoprenylated 1,3-diphenylpropane derivatives. Chem. Pharm. Bull. 1986, 34, 1968–1979. [Google Scholar]
- Ni, G.; Zhang, Q.J.; Wang, Y.H.; Chen, R.Y.; Zheng, Z.F.; Yu, D.Q. Chemical constituents of the stem bark of Morus cathayana. J. Asian Nat. Prod. Res. 2010, 12, 505–515. [Google Scholar] [CrossRef]
- Ryu, H.W.; Park, M.H.; Kwon, O.K.; Kim, D.Y.; Hwang, J.Y.; Jo, Y.H.; Ahn, K.S.; Hwang, B.Y.; Oh, S.R. Anti-inflammatory flavonoids from root bark of Broussonetia papyrifera in LPS-stimulated RAW264.7 cells. Bioorg. Chem. 2019, 92, 103233. [Google Scholar] [CrossRef]
- Tahara, S.; Ingham, J.L.; Mizutani, J. Stereochemical studies on dihydrofurano-isoflavones. Agric. Biol. Chem. 1987, 51, 211–216. [Google Scholar]
- Han, F.; Xiao, Y.; Lee, I.-S. Microbial transformation of galangin derivatives and cytotoxicity evaluation of their metabolites. Catalysts 2021, 11, 1020. [Google Scholar] [CrossRef]
- Han, F.; Xiao, Y.; Lee, I.-S. Microbial transformation of prenylquercetins by Mucor hiemalis. Molecules 2020, 25, 528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.J.; Yim, S.H.; Han, F.; Kang, B.Y.; Choi, H.J.; Jung, D.W.; Williams, D.R.; Gustafson, K.R.; Kennelly, E.J.; Lee, I.-S. Biotransformed metabolites of the hop prenylflavanone isoxanthohumol. Molecules 2019, 24, 394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, J.L.; Liu, T.L.; Xue, J.J.; Hong, W.; Zhang, Y.; Zhang, D.X.; Cui, C.C.; Liu, M.C.; Niu, S.L. Flavanoids derivatives from the root bark of Broussonetia papyrifera as a tyrosinase inhibitor. Ind. Crop. Prod. 2019, 138, 111445. [Google Scholar] [CrossRef]
- Ko, H.H.; Yen, M.H.; Wu, R.R.; Won, S.J.; Lin, C.N. Cytotoxic isoprenylated flavans of Broussonetia kazinoki. J. Nat. Prod. 1999, 62, 162–166. [Google Scholar] [CrossRef]


| Compound | Anti-Tyrosinase | Cell Line | ||
|---|---|---|---|---|
| A375P | B16F10 | B16F1 | ||
| 1 | >80 | 12.13 ± 0.24 | 26.01 ± 0.33 | 17.57 ± 0.33 |
| 2 | >80 | 11.46 ± 0.24 | 12.13 ± 0.87 | 13.35 ± 0.22 |
| 3 | 2.12 ± 0.21 | 18.16 ± 0.09 | 19.89 ± 0.73 | 13.77 ± 0.88 |
| 4 | 24.11 ± 0.30 | 13.77 ± 1.00 | 27.79 ± 0.63 | 24.38 ± 1.82 |
| 5 | >80 | 45.87 ± 0.65 | 43.24 ± 0.71 | 17.32 ± 0.22 |
| 6 | >80 | >80 | >80 | >80 |
| 7 | >80 | 23.63 ± 0.52 | 24.22 ± 1.26 | 17.80 ± 1.65 |
| 8 | >80 | >80 | >80 | >80 |
| 9 | >80 | >80 | >80 | >80 |
| 10 | 3.36 ± 0.21 | >80 | >80 | >80 |
| 11 | 0.71 ± 0.01 | >80 | >80 | >80 |
| Kojic acid | 42.67 ± 1.44 | - | - | - |
| 5-FU | - | 13.13 ± 1.55 | 4.61 ± 0.16 | 12.82 ± 0.16 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, E.; Han, F.; Park, J.; Lee, I.-S. Microbial Transformation and Biological Activities of the Prenylated Aromatic Compounds from Broussonetia kazinoki. Molecules 2022, 27, 1879. https://doi.org/10.3390/molecules27061879
Choi E, Han F, Park J, Lee I-S. Microbial Transformation and Biological Activities of the Prenylated Aromatic Compounds from Broussonetia kazinoki. Molecules. 2022; 27(6):1879. https://doi.org/10.3390/molecules27061879
Chicago/Turabian StyleChoi, EunA, Fubo Han, Jisu Park, and Ik-Soo Lee. 2022. "Microbial Transformation and Biological Activities of the Prenylated Aromatic Compounds from Broussonetia kazinoki" Molecules 27, no. 6: 1879. https://doi.org/10.3390/molecules27061879
APA StyleChoi, E., Han, F., Park, J., & Lee, I. -S. (2022). Microbial Transformation and Biological Activities of the Prenylated Aromatic Compounds from Broussonetia kazinoki. Molecules, 27(6), 1879. https://doi.org/10.3390/molecules27061879