Antiviral Peptides (AVPs) of Marine Origin as Propitious Therapeutic Drug Candidates for the Treatment of Human Viruses
Abstract
:1. Introduction
2. Biosynthesis of AVPs: A Brief Overview
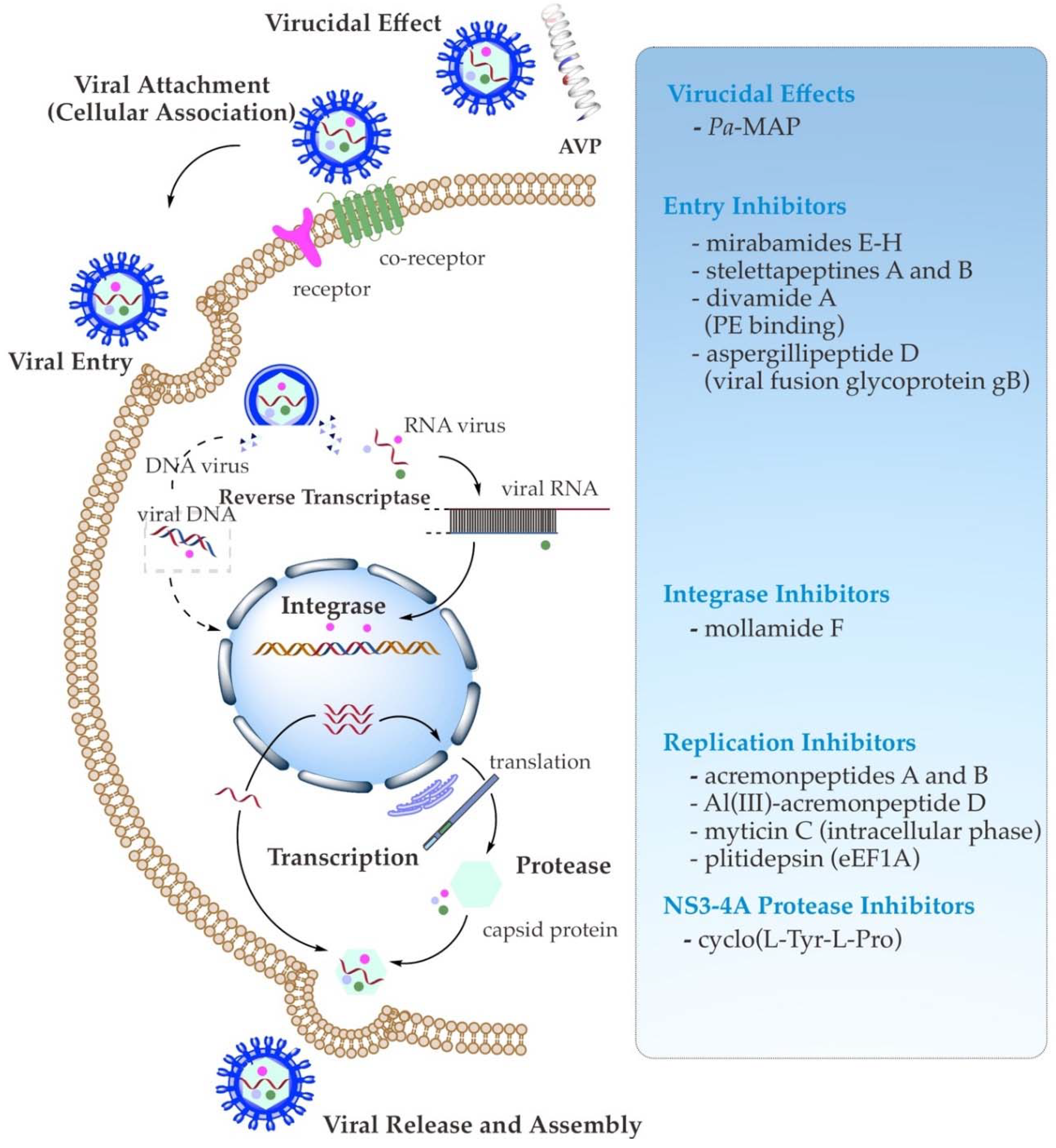
3. Antiviral Mechanism of Action of AVPs
3.1. Direct Binding Inhibition (Virucidal Effect)
3.2. Viral Attachment (Cellular Association) and Entry Inhibition
3.3. Viral Enzymes and Replication Inhibition
4. Anti-HIV (Human Immunodeficiency Virus) Marine AVPs
5. Anti-Influenza-Virus Marine AVPs
6. Anti-HSV (Human Simplex Virus) Marine AVPs
7. Anti-HCV (Hepatitis C Virus) Marine AVPs
8. Anti-SARS-CoV-2 Marine AVPs
9. Concluding Remarks and Future Perspective
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zumla, A.; Hui, D.S.C. Emerging and reemerging infectious diseases: Global overview. Infect. Dis. Clin. N. Am. 2019, 33, xiii–xix. [Google Scholar] [CrossRef] [PubMed]
- Global HIV and AIDS Statistics—Fact Sheet. Available online: https://www.unaids.org/en/resources/fact-sheet (accessed on 8 December 2021).
- World Health Organization. Global Progress Report on HIV, Viral Hepatitis and Sexually Transmitted Infections, 2021: Accountability for the Global Health Sector Strategies 2016–2021: Actions for Impact; World Health Organization: Geneva, Switzerland, 2021.
- Iuliano, A.D.; Roguski, K.M.; Chang, H.H.; Muscatello, D.J.; Palekar, R.; Tempia, S.; Cohen, C.; Gran, J.M.; Schanzer, D.; Cowling, B.; et al. Estimates of global seasonal influenza-associated respiratory mortality: A modelling study. Lancet 2018, 391, 1285–1300. [Google Scholar] [CrossRef]
- Chow, E.J.; Doyle, J.D.; Uyeki, T.M. Influenza virus-related critical illness: Prevention, diagnosis, treatment. Crit. Care 2019, 23, 214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lanini, S.; Ustianowski, A.; Pisapia, R.; Zumla, A.; Ippolito, G. Viral hepatitis: Etiology, epidemiology, transmission, diagnostics, treatment, and prevention. Infect. Dis. Clin. N. Am. 2019, 33, 1045–1062. [Google Scholar] [CrossRef]
- Martellucci, C.A.; Flacco, M.E.; Cappadona, R.; Bravi, F.; Mantovani, L.; Manzoli, L. SARS-CoV-2 pandemic: An overview. Adv. Biol. Regul. 2020, 77, 100736. [Google Scholar] [CrossRef]
- Morens, D.M.; Fauci, A.S. Emerging pandemic diseases: How we got to COVID-19. Cell 2020, 182, 1077–1092. [Google Scholar] [CrossRef]
- Mulder, K.C.L.; Lima, L.A.; Miranda, V.J.; Dias, S.C.; Franco, O.L. Current scenario of peptide-based drugs: The key roles of cationic antitumor and antiviral peptides. Front. Microbiol. 2013, 4, 321. [Google Scholar] [CrossRef] [Green Version]
- Sable, R.; Parajuli, P.; Jois, S. Peptides, peptidomimetics, and polypeptides from marine sources: A wealth of natural sources for pharmaceutical applications. Mar. Drugs 2017, 15, 124. [Google Scholar] [CrossRef] [Green Version]
- Mahlapuu, M.; Björn, C.; Ekblom, J. Antimicrobial peptides as therapeutic agents: Opportunities and challenges. Crit. Rev. Biotechnol. 2020, 40, 978–992. [Google Scholar] [CrossRef]
- Mahlapuu, M.; Håkansson, J.; Ringstad, L.; Björn, C. Antimicrobial peptides: An emerging category of therapeutic agents. Front. Cell. Infect. Microbiol. 2016, 6, 194. [Google Scholar] [CrossRef] [Green Version]
- Moravej, H.; Moravej, Z.; Yazdanparast, M.; Heiat, M.; Mirhosseini, A.; Moosazadeh Moghaddam, M.; Mirnejad, R. Antimicrobial peptides: Features, action, and their resistance mechanisms in bacteria. Microb. Drug Resist. 2018, 24, 747–767. [Google Scholar] [CrossRef] [PubMed]
- Vilas Boas, L.C.P.; Campos, M.L.; Berlanda, R.L.A.; de Carvalho Neves, N.; Franco, O.L. Antiviral peptides as promising therapeutic drugs. Cell. Mol. Life Sci. 2019, 76, 3525–3542. [Google Scholar] [CrossRef] [PubMed]
- Cappello, E.; Nieri, P. From life in the sea to the clinic: The marine drugs approved and under clinical trial. Life 2021, 11, 1390. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.W.; Alford, M.A.; Haney, E.F. Antibiofilm activity of host defence peptides: Complexity provides opportunities. Nat. Rev. Microbiol. 2021, 19, 786–797. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Separovic, F.; O’Brien-Simpson, N.M.; Wade, J.D. Chemically modified and conjugated antimicrobial peptides against superbugs. Chem. Soc. Rev. 2021, 50, 4932–4973. [Google Scholar] [CrossRef] [PubMed]
- Badani, H.; Garry, R.F.; Wimley, W.C. Peptide entry inhibitors of enveloped viruses: The importance of interfacial hydrophobicity. Biochim. Biomembr. 2014, 1838, 2180–2197. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.K.; Shih, L.Y.; Chang, K.Y. Large-scale analysis of antimicrobial activities in relation to amphipathicity and charge reveals novel characterization of antimicrobial peptides. Molecules 2017, 22, 2037. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Wang, G. APD: The antimicrobial peptide database. Nucleic Acids Res. 2004, 32, 590–592. [Google Scholar] [CrossRef] [Green Version]
- Mishra, B.; Wang, G. The importance of amino acid composition in natural amps: An evolutional, structural, and functional perspective. Front. Immunol. 2012, 3, 221. [Google Scholar] [CrossRef] [Green Version]
- Macedo, M.W.F.S.; da Cunha, N.B.; Carneiro, J.A.; da Costa, R.A.; de Alencar, S.A.; Cardoso, M.H.; Franco, O.L.; Dias, S.C. Marine organisms as a rich source of biologically active peptides. Front. Mar. Sci. 2021, 8, 667764. [Google Scholar] [CrossRef]
- Ovchinnikova, T.V. Marine peptides: Structure, bioactivities, and a new hope for therapeutic application. Mar. Drugs 2021, 19, 407. [Google Scholar] [CrossRef] [PubMed]
- Cheung, R.C.F.; Wong, J.H.; Pan, W.L.; Chan, Y.S.; Yin, C.M.; Dan, X.L.; Wang, H.X.; Fang, E.F.; Lam, S.K.; Ngai, P.H.K.; et al. Antifungal and antiviral products of marine organisms. Appl. Microbiol. Biotechnol. 2014, 98, 3475–3494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gogineni, V.; Schinazi, R.F.; Hamann, M.T. Role of marine natural products in the genesis of antiviral agents. Chem. Rev. 2015, 115, 9655–9706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, H.K.; Seo, C.H.; Park, Y. Marine peptides and their anti-infective activities. Mar. Drugs 2015, 13, 618–654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riccio, G.; Ruocco, N.; Mutalipassi, M.; Costantini, M.; Zupo, V.; Coppola, D.; de Pascale, D.; Lauritano, C. Ten-year research update review: Antiviral activities from marine organisms. Biomolecules 2020, 10, 1007. [Google Scholar] [CrossRef]
- Raihan, T.; Rabbee, M.F.; Roy, P.; Choudhury, S.; Baek, K.-H.; Azad, A.K. Microbial metabolites: The emerging hotspot of antiviral compounds as potential candidates to avert viral pandemic alike COVID-19. Front. Mol. Biosci. 2021, 8, 732256. [Google Scholar] [CrossRef]
- Murugan, N.A.; Raja, K.M.P.; Saraswathi, N.T. Peptide-based antiviral drugs. In Advances in Experimental Medicine and Biology; Liu, X., Zhan, P., Menéndez-Arias, L., Poongavanam, V., Eds.; Springer: Singapore, 2021; Volume 1322, pp. 261–284. [Google Scholar]
- Agarwal, G.; Gabrani, R. Antiviral peptides: Identification and validation. Int. J. Pept. Res. Ther. 2021, 27, 149–168. [Google Scholar] [CrossRef]
- Qureshi, A.; Thakur, N.; Tandon, H.; Kumar, M. AVPdb: A database of experimentally validated antiviral peptides targeting medically important viruses. Nucleic Acids Res. 2014, 42, 1147–1153. [Google Scholar] [CrossRef] [Green Version]
- Beltrán Lissabet, J.F.; Belén, L.H.; Farias, J.G. AntiVPP 1.0: A portable tool for prediction of antiviral peptides. Comput. Biol. Med. 2019, 107, 127–130. [Google Scholar] [CrossRef]
- Chowdhury, S.M.; Talukder, S.A.; Khan, A.M.; Afrin, N.; Ali, M.A.; Islam, R.; Parves, R.; Al Mamun, A.; Sufian, M.A.; Hossain, M.N.; et al. Antiviral peptides as promising therapeutics against SARS-CoV-2. J. Phys. Chem. B 2020, 124, 9785–9792. [Google Scholar] [CrossRef]
- McIntosh, J.A.; Donia, M.S.; Schmidt, E.W. Ribosomal peptide natural products: Bridging the ribosomal and nonribosomal worlds. Nat. Prod. Rep. 2009, 26, 537–559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dang, T.; Süssmuth, R.D. Bioactive peptide natural products as lead structures for medicinal use. Acc. Chem. Res. 2017, 50, 1566–1576. [Google Scholar] [CrossRef] [PubMed]
- Arnison, P.G.; Bibb, M.J.; Bierbaum, G.; Bowers, A.A.; Bugni, T.S.; Bulaj, G.; Camarero, J.A.; Campopiano, D.J.; Challis, G.L.; Clardy, J.; et al. Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013, 30, 108–160. [Google Scholar] [CrossRef] [PubMed]
- Payne, J.A.E.; Schoppet, M.; Hansen, M.H.; Cryle, M.J. Diversity of nature’s assembly lines-recent discoveries in non-ribosomal peptide synthesis. Mol. Bio Syst. 2017, 13, 9–22. [Google Scholar] [CrossRef]
- Süssmuth, R.D.; Mainz, A. Nonribosomal peptide synthesis—Principles and prospects. Angew. Chem. Int. Ed. 2017, 56, 3770–3821. [Google Scholar] [CrossRef]
- Lou, Z.; Sun, Y.; Rao, Z. Current progress in antiviral strategies. Trends Pharmacol. Sci. 2014, 35, 86–102. [Google Scholar] [CrossRef]
- Kausar, S.; Said Khan, F.; Ishaq Mujeeb Ur Rehman, M.; Akram, M.; Riaz, M.; Rasool, G.; Hamid Khan, A.; Saleem, I.; Shamim, S.; Malik, A. A review: Mechanism of action of antiviral drugs. Int. J. Immunopathol. Pharmacol. 2021, 35, 20587384211002621. [Google Scholar] [CrossRef]
- Skalickova, S.; Heger, Z.; Krejcova, L.; Pekarik, V.; Bastl, K.; Janda, J.; Kostolansky, F.; Vareckova, E.; Zitka, O.; Adam, V.; et al. Perspective of use of antiviral peptides against influenza virus. Viruses 2015, 7, 5428–5442. [Google Scholar] [CrossRef] [Green Version]
- Gao, B.; Zhao, D.; Li, L.; Cheng, Z.; Guo, Y. Antiviral peptides with in vivo activity: Development and modes of action. Chempluschem 2021, 86, 1547–1558. [Google Scholar] [CrossRef]
- Ford, P.W.; Gustafson, K.R.; McKee, T.C.; Shigematsu, N.; Maurizi, L.K.; Pannell, L.K.; Williams, D.E.; De Silva, E.D.; Lassota, P.; Allen, T.M.; et al. Papuamides A-D, HIV-inhibitory and cytotoxic depsipeptides from the sponges Theonella mirabilis and Thenonella swinhoei collected in Papua New Guinea. J. Am. Chem. Soc. 1999, 121, 5899–5909. [Google Scholar] [CrossRef]
- Andjelic, C.D.; Planelles, V.; Barrows, L.R. Characterizing the anti-HIV activity of papuamide A. Mar. Drugs 2008, 6, 528–549. [Google Scholar] [CrossRef]
- Demirkhanyan, L.H.; Marin, M.; Padilla-parra, S.; Zhan, C.; Miyauchi, K.; Jean-baptiste, M.; Novitskiy, G.; Lu, W.; Melikyan, G.B. Multifaceted mechanisms of HIV-1 entry inhibition by human α-defensin. J. Biol. Chem. 2012, 287, 28821–28838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, D.; Lu, W. Defensins : A double-edged sword in host immunity. Front. Immunol. 2020, 11, 764. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, T.; Sumi, M.; Kubota, H.; Taki, T.; Okahata, Y.; Sato, T. Inhibition of influenza virus infections by sialylgalactose-binding peptides selected from a phage library. J. Med. Chem. 2009, 52, 4247–4256. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, T.; Onishi, A.; Saito, T.; Shimada, A.; Inoue, H.; Taki, T.; Nagata, K.; Okahata, Y.; Sato, T. mSialic acid-mimic peptides as hemagglutinin inhibitors for anti-influenza therapy. J. Med. Chem. 2010, 53, 4441–4449. [Google Scholar] [CrossRef]
- Nicol, M.Q.; Ligertwood, Y.; Bacon, M.N.; Dutia, B.M.; Nash, A.A. A novel family of peptides with potent activity against influenza a viruses. J. Gen. Virol. 2012, 93, 980–986. [Google Scholar] [CrossRef]
- Hoffmann, J.; Schneider, C.; Heinbockel, L.; Brandenburg, K.; Reimer, R.; Gabriel, G. A new class of synthetic anti-lipopolysaccharide peptides inhibits influenza A virus replication by blocking cellular attachment. Antivir. Res. 2014, 104, 23–33. [Google Scholar] [CrossRef]
- Jenssen, H.; Andersen, J.H.; Uhlin-Hansen, L.; Gutteberg, T.J.; Rekdal, Ø. Anti-HSV activity of lactoferricin analogues is only partly related to their affinity for heparan sulfate. Antivir. Res. 2004, 61, 101–109. [Google Scholar] [CrossRef]
- Jiang, J.; Cun, W.; Wu, X.; Shi, Q.; Tang, H.; Luo, G. Hepatitis C virus attachment mediated by apolipoprotein E binding to cell surface heparan sulfate. J. Virol. 2012, 86, 7256–7267. [Google Scholar] [CrossRef] [Green Version]
- Leblanc, E.V.; Kim, Y.; Capicciotti, C.J.; Colpitts, C.C. Hepatitis C virus glycan-dependent interactions and the potential for novel preventative strategies. Pathogens 2021, 10, 685. [Google Scholar] [CrossRef]
- Chi, X.; Niu, Y.; Cheng, M.; Liu, X.; Feng, Y.; Zheng, F.; Fan, J.; Li, X.; Jin, Q.; Zhong, J.; et al. Identification of a potent and broad-spectrum hepatitis C virus fusion inhibitory peptide from the E2 stem domain. Sci. Rep. 2016, 6, 25224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Si, Y.; Liu, S.; Liu, X.; Jacobs, J.L.; Cheng, M.; Niu, Y.; Jin, Q.; Wang, T.; Yang, W. A human claudin-1-derived peptide inhibits hepatitis C virus entry. Hepatology 2012, 56, 507–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, X.J.; Zhu, Y.Z.; Zhao, P.; Qi, Z.T. Entry inhibitors: New advances in HCV treatment. Emerg. Microbes Infect. 2016, 5, e3. [Google Scholar] [CrossRef] [PubMed]
- Düzgüneş, N.; Fernandez-Fuentes, N.; Konopka, K. Inhibition of viral membrane fusion by peptides and approaches to peptide design. Pathogens 2021, 10, 1599. [Google Scholar] [CrossRef] [PubMed]
- Mänz, B.; Götz, V.; Wunderlich, K.; Eisel, J.; Kirchmair, J.; Stech, J.; Stech, O.; Chase, G.; Frank, R.; Schwemmle, M. Disruption of the viral polymerase complex assembly as a novel approach to attenuate influenza A virus. J. Biol. Chem. 2011, 286, 8414–8424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beran, R.K.F.; Pyle, A.M. Hepatitis C viral NS3-4A protease activity is enhanced by the NS3 helicase. J. Biol. Chem. 2008, 283, 29929–29937. [Google Scholar] [CrossRef] [Green Version]
- Chatel-Chaix, L.; Baril, M.; Lamarre, D. Hepatitis C virus NS3/4A protease inhibitors: A light at the end of the tunnel. Viruses 2010, 2, 1752–1765. [Google Scholar] [CrossRef] [Green Version]
- Tonk, M.; Růžek, D.; Vilcinskas, A. Compelling evidence for the activity of antiviral peptides against SARS-CoV-2. Viruses 2021, 13, 912. [Google Scholar] [CrossRef]
- Heydari, H.; Golmohammadi, R.; Mirnejad, R.; Tebyanian, H.; Fasihi-Ramandi, M.; Moosazadeh Moghaddam, M. Antiviral peptides against Coronaviridae family: A review. Peptides 2021, 139, 170526. [Google Scholar] [CrossRef]
- Bakovic, A.; Risner, K.; Bhalla, N.; Alem, F.; Chang, T.L.; Weston, W.K.; Harness, J.A.; Narayanan, A. Brilacidin demonstrates inhibition of SARS-CoV-2 in cell culture. Viruses 2021, 13, 271. [Google Scholar] [CrossRef]
- Xia, S.; Yan, L.; Xu, W.; Agrawal, A.S.; Algaissi, A.; Tseng, C.T.K.; Wang, Q.; Du, L.; Tan, W.; Wilson, I.A.; et al. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci. Adv. 2019, 5, eaav4580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, H.; To, K.K.W.; Sze, K.H.; Yung, T.T.M.; Bian, M.; Lam, H.; Yeung, M.L.; Li, C.; Chu, H.; Yuen, K.Y. A broad-spectrum virus- and host-targeting peptide against respiratory viruses including influenza virus and SARS-CoV-2. Nat. Commun. 2020, 11, 4252. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, S.; Li, D.; Wei, D.Q.; Zhao, J.; Wang, J. Human intestinal defensin 5 inhibits SARS-CoV-2 invasion by cloaking ACE2. Gastroenterology 2020, 159, 1145–1147. [Google Scholar] [CrossRef] [PubMed]
- Mahendran, A.S.K.; Lim, Y.S.; Fang, C.M.; Loh, H.S.; Le, C.F. The potential of antiviral peptides as COVID-19 therapeutics. Front. Pharmacol. 2020, 11, 575444. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Van Wagoner, R.M.; Harper, M.K.; Baker, H.L.; Hooper, J.N.A.; Bewley, C.A.; Ireland, C.M. Mirabamides E-H, HIV-inhibitory depsipeptides from the sponge Stelletta clavosa. J. Nat. Prod. 2011, 74, 185–193. [Google Scholar] [CrossRef] [Green Version]
- Shin, H.J.; Rashid, M.A.; Cartner, L.K.; Bokesch, H.R.; Wilson, J.A.; McMahon, J.B.; Gustafson, K.R. Stellettapeptins A and B, HIV-inhibitory cyclic depsipeptides from the marine sponge Stelletta sp. Tetrahedron Lett. 2015, 56, 4215–4219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Z.; Harper, M.K.; Pond, C.D.; Barrows, L.R.; Ireland, C.M.; Van Wagoner, R.M. Thiazoline peptides and a tris-phenethyl urea from Didemnum molle with anti-HIV activity. J. Nat. Prod. 2012, 75, 1436–1440. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Fang, W.; Tan, S.; Lin, X.; Xun, T.; Yang, B.; Liu, S.; Liu, Y. Aspernigrins with anti-HIV-1 activities from the marine-derived fungus Aspergillus niger SCSIO Jcsw6F30. Bioorg. Med. Chem. Lett. 2016, 26, 361–365. [Google Scholar] [CrossRef]
- Smith, T.E.; Pond, C.D.; Pierce, E.; Harmer, Z.P.; Kwan, J.; Zachariah, M.M.; Harper, M.K.; Wyche, T.P.; Matainaho, T.K.; Bugni, T.S.; et al. Accessing chemical diversity from the uncultivated symbionts of small marine animals. Nat. Chem. Biol. 2018, 14, 179–185. [Google Scholar] [CrossRef]
- He, F.; Bao, J.; Zhang, X.; Tu, Z.; Shi, Y.; Qi, S. Asperterrestide A, a cytotoxic cyclic tetrapeptide from the marine-derived fungus Aspergillus terreus SCSGAF0162. J. Nat. Prod. 2013, 76, 1182–1186. [Google Scholar] [CrossRef]
- Ma, X.; Nong, X.H.; Ren, Z.; Wang, J.; Liang, X.; Wang, L.; Qi, S.H. Antiviral peptides from marine gorgonian-derived fungus Aspergillus sp. SCSIO 41501. Tetrahedron Lett. 2017, 58, 1151–1155. [Google Scholar] [CrossRef]
- Liang, X.; Nong, X.H.; Huang, Z.H.; Qi, S.H. Antifungal and antiviral cyclic peptides from the deep-sea-derived fungus Simplicillium obclavatum EIODSF 020. J. Agric. Food Chem. 2017, 65, 5114–5121. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Zang, R.; Wang, X.; Chen, Z.; Song, X.; Ju, J.; Huang, H. Natural hydroxamate-containing siderophore acremonpeptides A-D and an aluminum complex of acremonpeptide D from the marine-derived Acremonium persicinum SCSIO 115. J. Nat. Prod. 2019, 82, 2594–2600. [Google Scholar] [CrossRef] [PubMed]
- Novoa, B.; Romero, A.; Álvarez, Á.L.; Moreira, R.; Pereiro, P.; Costa, M.M.; Dios, S.; Estepa, A.; Parra, F.; Figueras, A. Antiviral activity of myticin C peptide from mussel: An ancient defense against herpesviruses. J. Virol. 2016, 90, 7692–7702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vilas Boas, L.C.P.; de Lima, L.M.P.; Migliolo, L.; Mendes, G.d.S.; de Jesus, M.G.; Franco, O.L.; Silva, P.A. Linear antimicrobial peptides with activity against herpes simplex virus 1 and aichi virus. Pept. Sci. 2017, 108, e22871. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, E.F.; Rateb, M.E.; Abou El-Kassem, L.T.; Hawas, U.W. Anti-HCV protease of diketopiperazines produced by the Red Sea sponge-associated fungus Aspergillus versicolor. Appl. Biochem. 2017, 53, 101–106. [Google Scholar] [CrossRef] [Green Version]
- Wibowo, J.T.; Kellermann, M.Y.; Köck, M.; Putra, M.Y.; Murniasih, T.; Mohr, K.I.; Wink, J.; Praditya, D.F.; Steinmann, E.; Schupp, P.J. Anti-Infective and antiviral activity of valinomycin and its analogues from a sea cucumber-associated bacterium, Streptomyces sp. SV 21. Mar. Drugs 2021, 19, 81. [Google Scholar] [CrossRef]
- Singh, I.P.; Bodiwala, H.S. Recent advances in anti-HIV natural products. Nat. Prod. Rep. 2010, 27, 1781–1800. [Google Scholar] [CrossRef]
- Günaydın, G.; Edfeldt, G.; Garber, D.A.; Asghar, M.; Noel-Romas, L.; Burgener, A.; Wählby, C.; Wang, L.; Rohan, L.C.; Guenthner, P.; et al. Impact of Q-Griffithsin anti-HIV microbicide gel in non-human primates: In situ analyses of epithelial and immune cell markers in rectal mucosa. Sci. Rep. 2019, 9, 18120. [Google Scholar] [CrossRef] [Green Version]
- Lee, C. Griffithsin, a highly potent broad-spectrum antiviral lectin from red algae: From discovery to clinical application. Mar. Drugs 2019, 17, 567. [Google Scholar] [CrossRef] [Green Version]
- Vo, T.S.; Kim, S.K. Potential anti-HIV agents from marine resources: An overview. Mar. Drugs 2010, 8, 2871–2892. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Liu, J.; Yang, B.; Lin, X.; Yang, X.-W.; Liu, Y. Marine natural products with anti-HIV in the last decade. Curr. Med. Chem. 2013, 20, 953–973. [Google Scholar] [CrossRef] [PubMed]
- Sagar, S.; Kaur, M.; Minneman, K.P. Antiviral lead compounds from marine sponges. Mar. Drugs 2010, 8, 2619–2638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plaza, A.; Gustchina, E.; Baker, H.L.; Kelly, M.; Bewley, C.A. Mirabamides A–D, depsipeptides from the sponge Siliquariaspongia mirabilis that inhibit HIV-1 fusion. J. Nat. Prod. 2007, 70, 1753–1760. [Google Scholar] [CrossRef] [PubMed]
- Plaza, A.; Bifulco, G.; Masullo, M.; Lloyd, J.R.; Keffer, J.L.; Colin, P.L.; Hooper, J.N.A.; Bell, L.J.; Bewley, C.A. Mutremdamide A and koshikamides C-H, peptide inhibitors of HIV-1 entry from different theonella species. J. Org. Chem. 2010, 75, 4344–4355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donia, M.S.; Fricke, W.F.; Ravel, J.; Schmidt, E.W. Variation in tropical reef symbiont metagenomes defined by secondary metabolism. PLoS ONE 2011, 6, e17897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tianero, M.D.B.; Donia, M.S.; Young, T.S.; Schultz, P.G.; Schmidt, E.W. Ribosomal route to small-molecule diversity. J. Am. Chem. Soc. 2012, 134, 418–425. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, S.S.; Faulkner, D.J.; Rubins, K.; Bushman, F.D. Dolastatin 3 and two novel cyclic peptides from a Palauan collection of Lyngbya majuscula. J. Nat. Prod. 2000, 63, 279–282. [Google Scholar] [CrossRef] [Green Version]
- Craigie, R. HIV integrase, a brief overview from chemistry to therapeutics. J. Biol. Chem. 2001, 276, 23213–23216. [Google Scholar] [CrossRef] [Green Version]
- Youssef, F.S.; Ashour, M.L.; Singab, A.N.B.; Wink, M. A comprehensive review of bioactive peptides from marine fungi and their biological significance. Mar. Drugs 2019, 17, 559. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Gong, X.; Li, P.; Lai, D.; Zhou, L. Structural diversity and biological activities of cyclic depsipeptides from fungi. Molecules 2018, 23, 169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderegg, R.J.; Biemann, K.; Büchi, G.; Cushman, M. Malformin C, a new metabolite of Aspergillus niger. J. Am. Chem. Soc. 1976, 98, 3365–3370. [Google Scholar] [CrossRef] [PubMed]
- Kobbe, B.; Cushman, M.; Wogan, G.N.; Demain, A.L. Production and antibacterial activity of malformin C, a toxic metabolite of Aspergillus niger. Appl. Environ. Microbiol. 1977, 33, 996–997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kojima, Y.; Sunazuka, T.; Nagai, K.; Hirose, T.; Namatame, M.; Ishiyama, A.; Otoguro, K.; Omura, S. Solid-phase synthesis and biological activity of malformin C and its derivatives. J. Antibiot. 2009, 62, 681–686. [Google Scholar] [CrossRef]
- Hagimori, K.; Fukuda, T.; Hasegawa, Y.; Omura, S.; Tomoda, H. Fungal malformins inhibit bleomycin-induced G2 checkpoint in Jurkat cells. Biol. Pharm. Bull. 2007, 30, 1379–1383. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Jiang, Z.; Lam, W.; Gullen, E.A.; Yu, Z.; Wei, Y.; Wang, L.; Zeiss, C.; Beck, A.; Cheng, E.C.; et al. Study of malformin C, a fungal source cyclic pentapeptide, as an anti-cancer drug. PLoS ONE 2015, 10, e0140069. [Google Scholar] [CrossRef]
- Youssef, D.T.A.; Almagthali, H.; Shaala, L.A.; Schmidt, E.W. Secondary metabolites of the genus didemnum: A comprehensive review of chemical diversity and pharmacological properties. Mar. Drugs 2020, 18, 307. [Google Scholar] [CrossRef]
- Fu, Y.; Jaarsma, A.H.; Kuipers, O.P. Antiviral activities and applications of ribosomally synthesized and post-translationally modified peptides (RiPPs). Cell. Mol. Life Sci. 2021, 78, 3921–3940. [Google Scholar] [CrossRef]
- Zhao, M. Lantibiotics as probes for phosphatidylethanolamine. Amino Acids 2011, 41, 1071–1079. [Google Scholar] [CrossRef] [Green Version]
- Huarte, N.; Carravilla, P.; Cruz, A.; Lorizate, M.; Nieto-Garai, J.A.; Kräusslich, H.G.; Pérez-Gil, J.; Requejo-Isidro, J.; Nieva, J.L. Functional organization of the HIV lipid envelope. Sci. Rep. 2016, 6, 34190. [Google Scholar] [CrossRef] [Green Version]
- Sundquist, W.I.; Kra, H.-G. HIV-1 Assembly, Budding, and Maturation. Cold Spring Harb. Prespect. Med. 2012, 2, a006. [Google Scholar] [CrossRef]
- Lorizate, M.; Kräusslich, H.G. Role of lipids in virus replication. Cold Spring Harb. Prespect. Biol. 2011, 3, a004820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richard, A.S.; Zhang, A.; Park, S.J.; Farzan, M.; Zong, M.; Choe, H. Virion-associated phosphatidylethanolamine promotes TIM1-mediated infection by Ebola, dengue, and West Nile viruses. Proc. Nat. Acad. Sci. USA 2015, 112, 14682–14687. [Google Scholar] [CrossRef] [Green Version]
- Tabata, T.; Petitt, M.; Puerta-Guardo, H.; Michlmayr, D.; Wang, C.; Fang-Hoover, J.; Harris, E.; Pereira, L. Zika virus targets different primary human placental cells, suggesting two routes for vertical transmission. Cell Host Microbe 2016, 20, 155–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vestergaard, M.; Berglund, N.A.; Hsu, P.C.; Song, C.; Koldsø, H.; Schiøtt, B.; Sansom, M.S.P. Structure and dynamics of cinnamycin-lipid complexes: Mechanisms of selectivity for phosphatidylethanolamine lipids. ACS Omega 2019, 4, 18889–18899. [Google Scholar] [CrossRef] [PubMed]
- Naruse, N.; Tenmyo, O.; Tomita, K.; Konishi, M.; Miyaki, T.; Kawaguchi, H.; Fukase, K.; Wakamiya, T.; Shiba, T. Lanthiopeptin, a new peptide antibiotic. production, isolation and properties of lanthiopeptin. J. Antibiot. 1989, 42, 837–845. [Google Scholar] [CrossRef] [Green Version]
- Guex, N.; Peitsch, M.C.; Schwede, T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis 2009, 30, 162–173. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- Domeneghetti, S.; Franzoi, M.; Damiano, N.; Norante, R.; M. El Halfawy, N.; Mammi, S.; Marin, O.; Bellanda, M.; Venier, P. Structural and antimicrobial features of peptides related to myticin C, a special defense molecule from the Mediterranean mussel Mytilus galloprovincialis. J. Agric. Food Chem. 2015, 63, 9251–9259. [Google Scholar] [CrossRef]
- Bienert, S.; Waterhouse, A.; De Beer, T.A.P.; Tauriello, G.; Studer, G.; Bordoli, L.; Schwede, T. The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res. 2017, 45, D313–D319. [Google Scholar] [CrossRef] [Green Version]
- Liepinsh, E.; Otting, G.; Harding, M.M.; Ward, L.G.; Mackay, J.P.; Haymet, A.D.J. Solution structure of a hydrophobic analogue of the winter flounder antifreeze protein. Eur. J. Biochem. 2002, 269, 1259–1266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Jia, J.; Wang, L.; Li, F.; Wang, Y.; Jiang, Y.; Song, X.; Qin, S.; Zheng, K.; Ye, J.; et al. Anti-HSV-1 activity of aspergillipeptide D, a cyclic pentapeptide isolated from fungus Aspergillus sp. SCSIO 41501. Virol. J. 2020, 17, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flores-Holguín, N.; Frau, J.; Glossman-Mitnik, D. Computational peptidology approach to the study of the chemical reactivity and bioactivity properties of aspergillipeptide D, a cyclopentapeptide of marine origin. Sci. Rep. 2022, 12, 506. [Google Scholar] [CrossRef]
- Anjum, K.; Abbas, S.Q.; Shah, S.A.A.; Akhter, N.; Batool, S.; Hassan, S.S.U. Marine sponges as a drug treasure. Biomol. Ther. 2016, 24, 347–362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varijakzhan, D.; Loh, J.Y.; Yap, W.S.; Yusoff, K.; Seboussi, R.; Lim, S.H.E.; Lai, K.S.; Chong, C.M. Bioactive compounds from marine sponges: Fundamentals and applications. Mar. Drugs 2021, 19, 246. [Google Scholar] [CrossRef] [PubMed]
- Balseiro, P.; Falcó, A.; Romero, A.; Dios, S.; Martínez-López, A.; Figueras, A.; Estepa, A.; Novoa, B. Mytilus galloprovincialis myticin C: A chemotactic molecule with antiviral activity and immunoregulatory properties. PLoS ONE 2011, 6, e23140. [Google Scholar] [CrossRef] [Green Version]
- Gong, Z.; Ewart, K.V.; Hu, Z.; Fletcher, G.L.; Hew, C.L. Skin antifreeze protein genes of the winter flounder, Pleuronectes americanus, encode distinct and active polypeptides without the secretory signal and prosequences. J. Biol. Chem. 1996, 271, 4106–4112. [Google Scholar] [CrossRef] [Green Version]
- Migliolo, L.; Silva, O.N.; Silva, P.A.; Costa, M.P.; Costa, C.R.; Nolasco, D.O.; Barbosa, J.A.R.G.; Silva, M.R.R.; Bemquerer, M.P.; Lima, L.M.P.; et al. Structural and functional characterization of a multifunctional alanine-rich peptide analogue from Pleuronectes americanus. PLoS ONE 2012, 7, e47047. [Google Scholar] [CrossRef]
- Galdiero, S.; Falanga, A.; Cantisani, M.; Vitiello, M.; Morelli, G.; Galdiero, M. Peptide-lipid interactions: Experiments and applications. Int. J. Mol. Sci. 2013, 14, 18758–18789. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.Y.; Jan, J.T.; Ma, S.H.; Kuo, C.J.; Juan, H.F.; Cheng, Y.S.E.; Hsu, H.H.; Huang, H.C.; Wu, D.; Brik, A.; et al. Small molecules targeting severe acute respiratory syndrome human coronavirus. Proc. Nat. Acad. Sci. USA 2004, 101, 10012–10017. [Google Scholar] [CrossRef] [Green Version]
- Calland, N.; Albecka, A.; Belouzard, S.; Wychowski, C.; Duverlie, G.; Descamps, V.; Hober, D.; Dubuisson, J.; Rouillé, Y.; Séron, K. (−)-Epigallocatechin-3-gallate is a new inhibitor of hepatitis C virus entry. Hepatology 2012, 55, 720–729. [Google Scholar] [CrossRef] [PubMed]
- De Clercq, E. Potential antivirals and antiviral strategies against SARS coronavirus infections. Expert Rev. Anti-Infect. Ther. 2006, 4, 291–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mousavi Maleki, M.S.; Rostamian, M.; Madanchi, H. Antimicrobial peptides and other peptide-like therapeutics as promising candidates to combat SARS-CoV-2. Expert Rev. Anti Infect. Ther. 2021, 19, 1205–1217. [Google Scholar] [CrossRef] [PubMed]
- Mahmud, S.; Biswas, S.; Kumar Paul, G.; Mita, M.A.; Afrose, S.; Robiul Hasan, M.; Sharmin Sultana Shimu, M.; Uddin, M.A.R.; Salah Uddin, M.; Zaman, S.; et al. Antiviral peptides against the main protease of SARS-CoV-2: A molecular docking and dynamics study. Arab. J. Chem. 2021, 14, 103315. [Google Scholar] [CrossRef] [PubMed]
- Jang, W.D.; Jeon, S.; Kim, S.; Lee, S.Y. Drugs repurposed for COVID-19 by virtual screening of 6,218 drugs and cell-based assay. Proc. Nat. Acad. Sci. USA 2021, 118, e2024302118. [Google Scholar] [CrossRef]
- Sultana, J.; Crisafulli, S.; Gabbay, F.; Lynn, E.; Shakir, S.; Trifirò, G. Challenges for drug repurposing in the COVID-19 pandemic era. Front. Pharmacol. 2020, 11, 588654. [Google Scholar] [CrossRef]
- Gatti, M.; De Ponti, F. Drug repurposing in the COVID-19 era: Insights from case studies showing pharmaceutical peculiarities. Pharmaceutics 2021, 13, 302. [Google Scholar] [CrossRef]
- Ancy, I.; Sivanandam, M.; Kumaradhas, P. Possibility of HIV-1 protease inhibitors-clinical trial drugs as repurposed drugs for SARS-CoV-2 main protease: A molecular docking, molecular dynamics and binding free energy simulation study. J. Biomol. Struct. Dyn. 2020, 39, 5368–5375. [Google Scholar] [CrossRef]
- Jukič, M.; Kores, K.; Janežič, D.; Bren, U. Repurposing of drugs for SARS-CoV-2 using inverse docking fingerprints. Front. Chem. 2021, 9, 757826. [Google Scholar] [CrossRef]
- Mei, M.; Tan, X. Current strategies of antiviral drug discovery for COVID-19. Front. Mol. Biosci. 2021, 8, 671263. [Google Scholar] [CrossRef]
- Singh, T.U.; Parida, S.; Lingaraju, M.C.; Kesavan, M.; Kumar, D.; Singh, R.K. Drug repurposing approach to fight COVID-19. Pharmacol. Rep. 2020, 72, 1479–1508. [Google Scholar] [CrossRef] [PubMed]
- Lovato, E.C.W.; Barboza, L.N.; Wietzikoski, S.; de Souza, A.N.V.; Auth, P.A.; Junior, A.G.; dos Reis Lívero, F.A. Repurposing drugs for the management of patients with confirmed coronavirus disease 2019 (COVID-19). Curr. Pharm. Des. 2020, 27, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Taglialatela-Scafati, O. New hopes for drugs against COVID-19 come from the sea. Mar. Drugs 2021, 19, 104. [Google Scholar] [CrossRef] [PubMed]
- Papapanou, M.; Papoutsi, E.; Giannakas, T.; Katsaounou, P. Plitidepsin: Mechanisms and clinical profile of a promising antiviral agent against Covid-19. J. Pers. Med. 2021, 11, 668. [Google Scholar] [CrossRef] [PubMed]
- Martinez, M.A. Plitidepsin: A repurposed drug for the treatment of COVID-19. Antimicrob. Agents Chemother. 2021, 65, e00200-21. [Google Scholar] [CrossRef]
- Alonso-Álvarez, S.; Pardal, E.; Sánchez-Nieto, D.; Navarro, M.; Caballero, M.D.; Mateos, M.V.; Martin, A. Plitidepsin: Design, development and potential place in therapy. Drug Des. Dev. Ther. 2017, 11, 253–254. [Google Scholar] [CrossRef] [Green Version]
- Kalhotra, P.; Chittepu, V.C.S.R.; Osorio-Revilla, G.; Gallardo-Velazquez, T. Field-template, QSAR, ensemble molecular docking, and 3D-RISM solvation studies expose potential of FDA-approved marine drugs as SARS-CoVID-2 main protease inhibitors. Molecules 2021, 26, 936. [Google Scholar] [CrossRef]
- Vishvakarma, V.K.; Singh, M.B.; Jain, P.; Kumari, K.; Singh, P. Hunting the main protease of SARS-CoV-2 by plitidepsin: Molecular docking and temperature-dependent molecular dynamics simulations. Amino Acids 2022, 54, 205–213. [Google Scholar] [CrossRef]
- White, K.M.; Rosales, R.; Yildiz, S.; Kehrer, T.; Miorin, L.; Moreno, E.; Jangra, S.; Uccellini, M.B.; Rathnasinghe, R.; Coughlan, L.; et al. Plitidepsin has potent preclinical efficacy against SARS-CoV-2 by targeting the host protein eEF1A. Science 2021, 371, 926–931. [Google Scholar] [CrossRef]
- Varona, J.F.; Landete, P.; Lopez-Martin, J.A.; Estrada, V.; Paredes, R.; Guisado-Vasco, P.; Fernandez de Orueta, L.; Torralba, M.; Fortun, J.; Vates, R.; et al. Preclinical and randomized phase I studies of plitidepsin in adults hospitalized with COVID-19. Life Sci. Alliance 2022, 5, e202101200. [Google Scholar] [CrossRef]
- Guisado-Vasco, P.; Carralón-González, M.M.; Aguareles-Gorines, J.; Martí-Ballesteros, E.M.; Sánchez-Manzano, M.D.; Carnevali-Ruiz, D.; García-Coca, M.; Barrena-Puertas, R.; de Viedma, R.G.; Luque-Pinilla, J.M.; et al. Plitidepsin as a successful rescue treatment for prolonged viral SARS-CoV-2 replication in a patient with previous anti-CD20 monoclonal antibody-mediated B cell depletion and chronic lymphocytic leukemia. J. Hematol. Oncol. 2022, 15, 4. [Google Scholar] [CrossRef] [PubMed]
- Horton, D.A.; Bourne, G.T.; Smythe, M.L. Exploring privileged structures: The combinatorial synthesis of cyclic peptides. J. Comput. Aided Mol. Des. 2002, 16, 415–430. [Google Scholar] [CrossRef] [PubMed]
- Zorzi, A.; Deyle, K.; Heinis, C. Cyclic peptide therapeutics: Past, present and future. Curr. Opin. Chem. Biol. 2017, 38, 24–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdalla, M.A.; McGaw, L.J. Natural cyclic peptides as an attractive modality for therapeutics: A mini review. Molecules 2018, 23, 2080. [Google Scholar] [CrossRef] [Green Version]
- Bechara, C.; Sagan, S. Cell-penetrating peptides: 20 years later, where do we stand? FEBS Lett. 2013, 587, 1693–1702. [Google Scholar] [CrossRef] [PubMed]
- Vlieghe, P.; Lisowski, V.; Martinez, J.; Khrestchatisky, M. Synthetic therapeutic peptides: Science and market. Drug Discov. Today 2010, 15, 40–56. [Google Scholar] [CrossRef]
- Flores-Holguín, N.; Frau, J.; Glossman-Mitnik, D. Virtual screening of marine natural compounds by means of chemoinformatics and CDFT-based computational peptidology. Mar. Drugs 2020, 18, 478. [Google Scholar] [CrossRef]


| Targeted Virus | Peptide | Biosynthetic Class | Origin | IC50/EC50/SI/Infectivity | Mechanism of Antiviral Action (Target of Inhibition) | Reference |
|---|---|---|---|---|---|---|
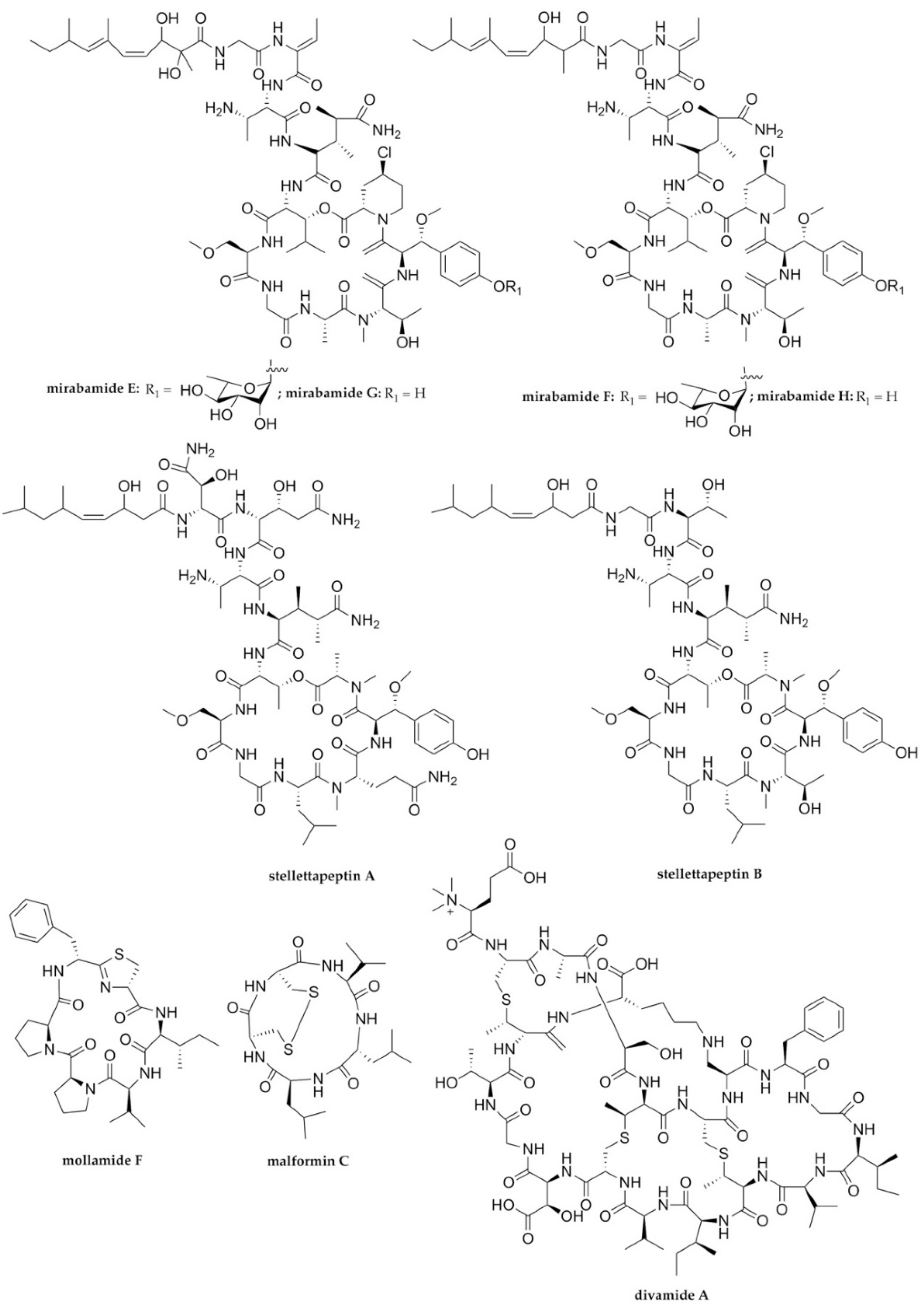
| HIV-1 | Mirabamides E–H | Cyclodepsipeptides/NRPs | Sponge Stelletta clavosa | 121, 62, 68, 41 nM | Viral fusion | [68] |
| HIV-1 | Stellettapeptines A and B | Cyclodepsipeptides/NRPs | Sponge Stelletta sp. | 23 and 27 nM | Viral entry (Viral membrane) | [69] |
| HIV-1 | Mollamide F | Cyclodepsipeptide/NRP | Tunicate Didemnum molle PNG07-2-050 | 78 μM (cytoprotective) 39 μM (HIV-integrase) | Viral integrase | [70] |
| HIV-1 | Malformin C | Cyclopeptide/NRP | Endophytic fungus Aspergillus niger SCSIO Jscw6F30 | 1.4 μM | ND * | [71] |
| HIV-1 | Divamide A | Lanthipeptide/ribosomal peptide | Tunicate Didemnum molle E11-036 | 0.225 μM | PE binding | [72] |
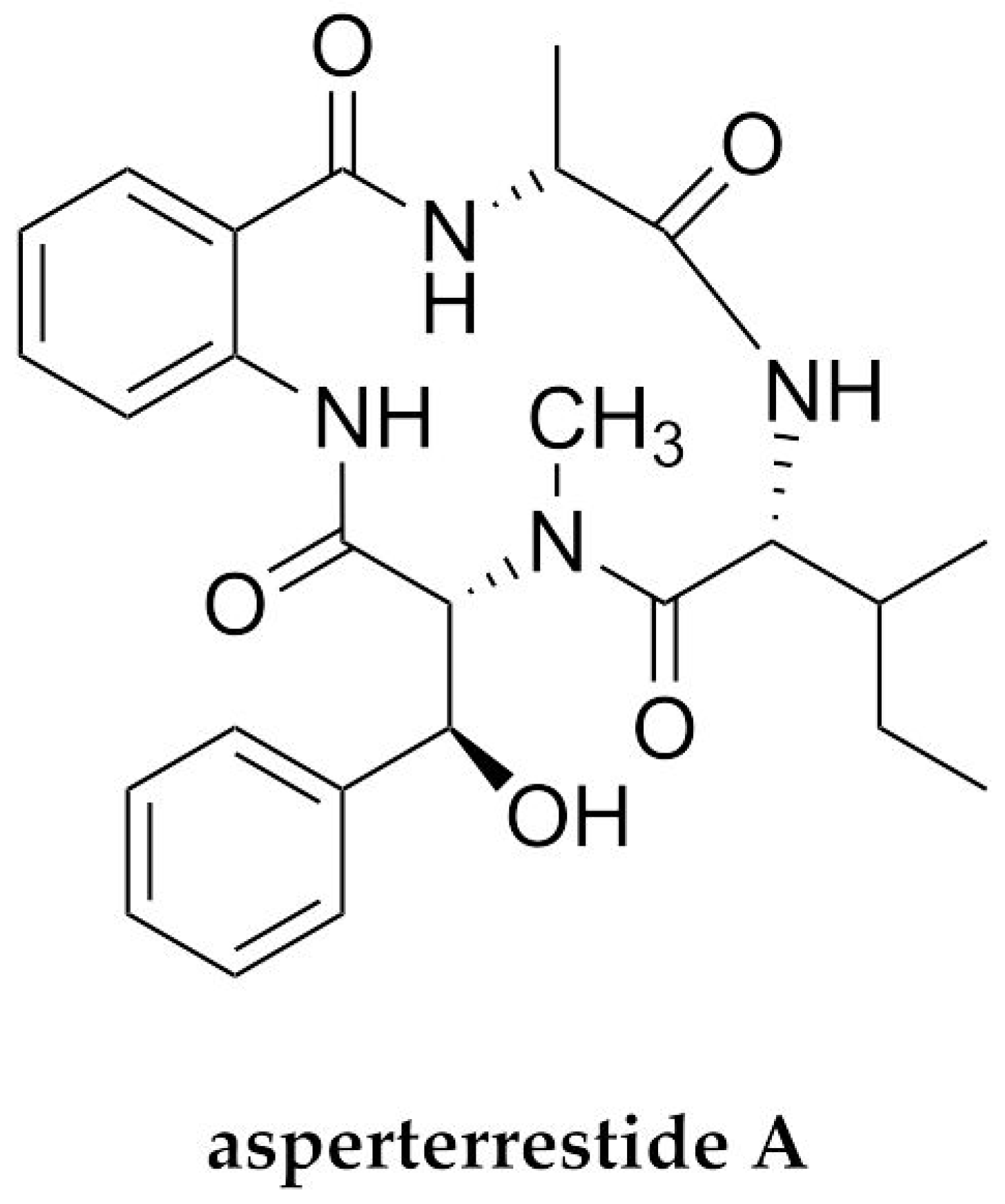
| H1N1/H3N2 | Asperterrestide A | Cyclopeptide/NRP | Endophytic fungus Aspergillus terreus | 20.2 and 0.41 μM | ND * | [73] |
| HSV-1 | Aspergillipeptide D | Cyclopeptide/NRP | Endophytic fungus Aspergillus sp. SCSIO 41501 | 9.5 μM (HSV-1) 12.5 M (ACV-HSV-1) | Viral intercellular spread (Viral glycoprotein gB) | [74] |
| HSV-1 | Aspergillipeptide E | Linear peptide/NRP | Endophytic fungus Aspergillus sp. SCSIO 41501 | 19.8 μM | ND * | [74] |
| HSV-1 | Simplicilliumtide J | Cyclodepsipeptide/NRP | Fungus Simplicillium obclavatum EIODSF 0210 | 14.1 μM | ND * | [75] |
| HSV-1 | Verlamelines A and B | Cyclodepsipeptide/NRPs | Fungus Simplicillium obclavatum EIODSF 0210 | 16.7 and 15.6 μM | ND * | [75] |
| HSV-1 | Acremonpeptides A and B | Cyclopeptide/NRPs | Fungus Acremonium persicinum SCSIO 115 | 16 and 8.7 μM | Viral replication | [76] |
| Al(III)-acremonpeptide D | Cyclopeptide/NRPs | Fungus Acremonium persicinum SCSIO 115 | 14 μM | Viral replication | [76] | |
| HSV-1/HSV-2 | Myticin C | Ribosomal peptide | Mollusk Mytilus galloprovincialis | 7.69–8.21/8.32–10.5 | The intracellular phase of viral replication | [77] |
| HSV-1/HSV-2 | Pa-MAP | Ribosomal peptide | polar fish Pleuronectes americanus | 82% (45 μM)/90% (23 μM) | Virucidal effect | [78] |
| HCV | Cyclo(l-Tyr-l-Pro) diketopiperazine | Cyclopeptide diketopiperazine/NRP | Endophytic fungus Aspergillus versicolor | 8.2 μg mL−1 | NS3-4A protease | [79] |
| HCV | Valinomycin; streptodepsipeptides P11A and SV21 | Cyclodepsipeptides/NRPs | Bacterial symbiont Streptomyces sp. SV21 | 0–5% | ND * | [80] |
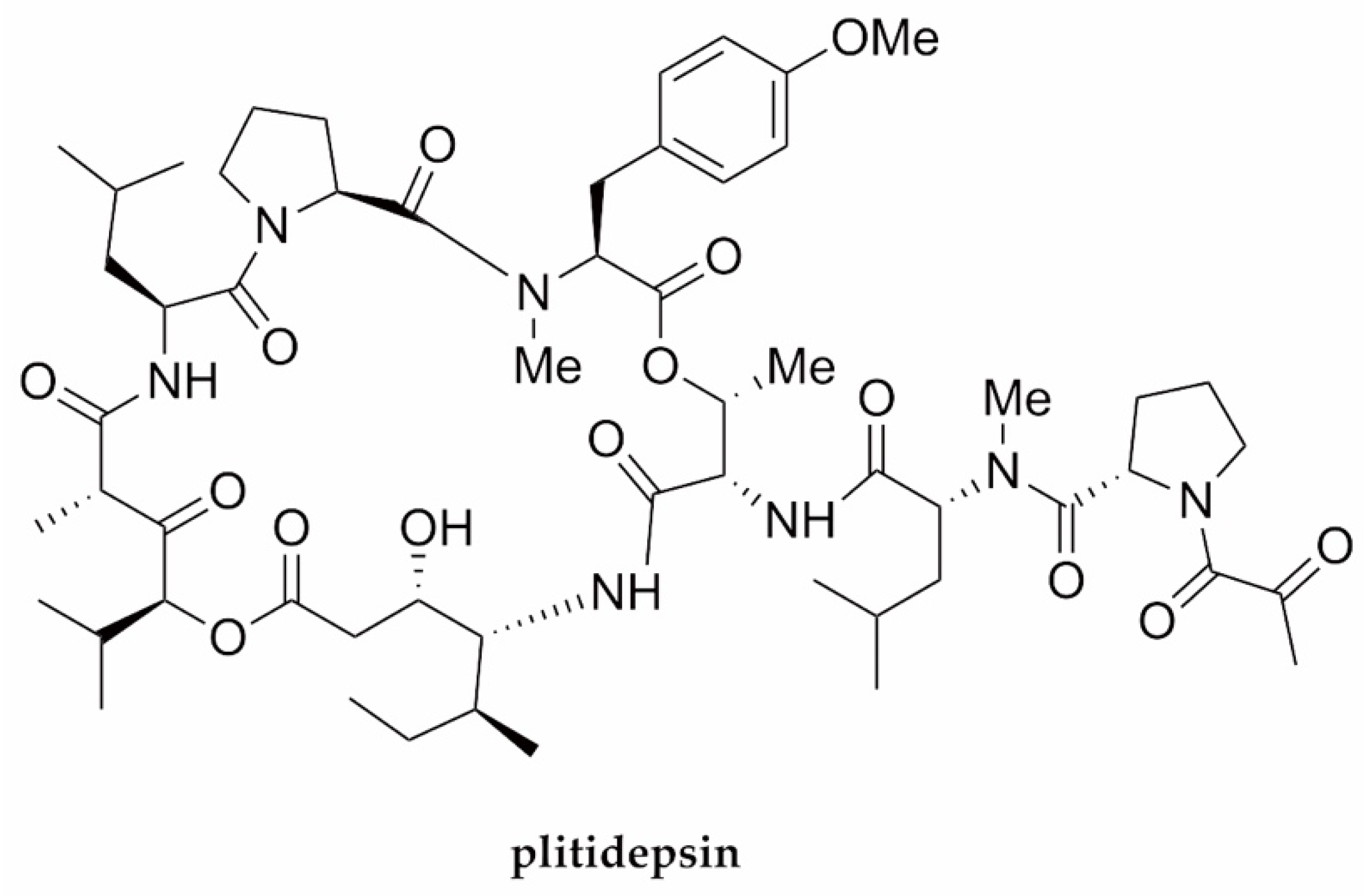
| SARS-CoV-2 | Plitidepsin | Cyclodepsipeptide/NRP | Tunicate Aplidium albicans | 0.88 nM | Viral replication (eEF1A) | [80] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sukmarini, L. Antiviral Peptides (AVPs) of Marine Origin as Propitious Therapeutic Drug Candidates for the Treatment of Human Viruses. Molecules 2022, 27, 2619. https://doi.org/10.3390/molecules27092619
Sukmarini L. Antiviral Peptides (AVPs) of Marine Origin as Propitious Therapeutic Drug Candidates for the Treatment of Human Viruses. Molecules. 2022; 27(9):2619. https://doi.org/10.3390/molecules27092619
Chicago/Turabian StyleSukmarini, Linda. 2022. "Antiviral Peptides (AVPs) of Marine Origin as Propitious Therapeutic Drug Candidates for the Treatment of Human Viruses" Molecules 27, no. 9: 2619. https://doi.org/10.3390/molecules27092619
APA StyleSukmarini, L. (2022). Antiviral Peptides (AVPs) of Marine Origin as Propitious Therapeutic Drug Candidates for the Treatment of Human Viruses. Molecules, 27(9), 2619. https://doi.org/10.3390/molecules27092619






