Estimating the Analytical Performance of Raman Spectroscopy for Quantification of Active Ingredients in Human Stratum Corneum
Abstract
:1. Introduction
2. Material and Methods
2.1. Chemicals
2.2. Isolated Human Stratum Corneum
2.3. Preparation of Infused SC Samples
2.4. HPLC Analysis
2.5. Confocal Raman Microscopy Analysis
2.6. Data Analysis
2.7. Analytical Performance Figures of Merit
3. Results
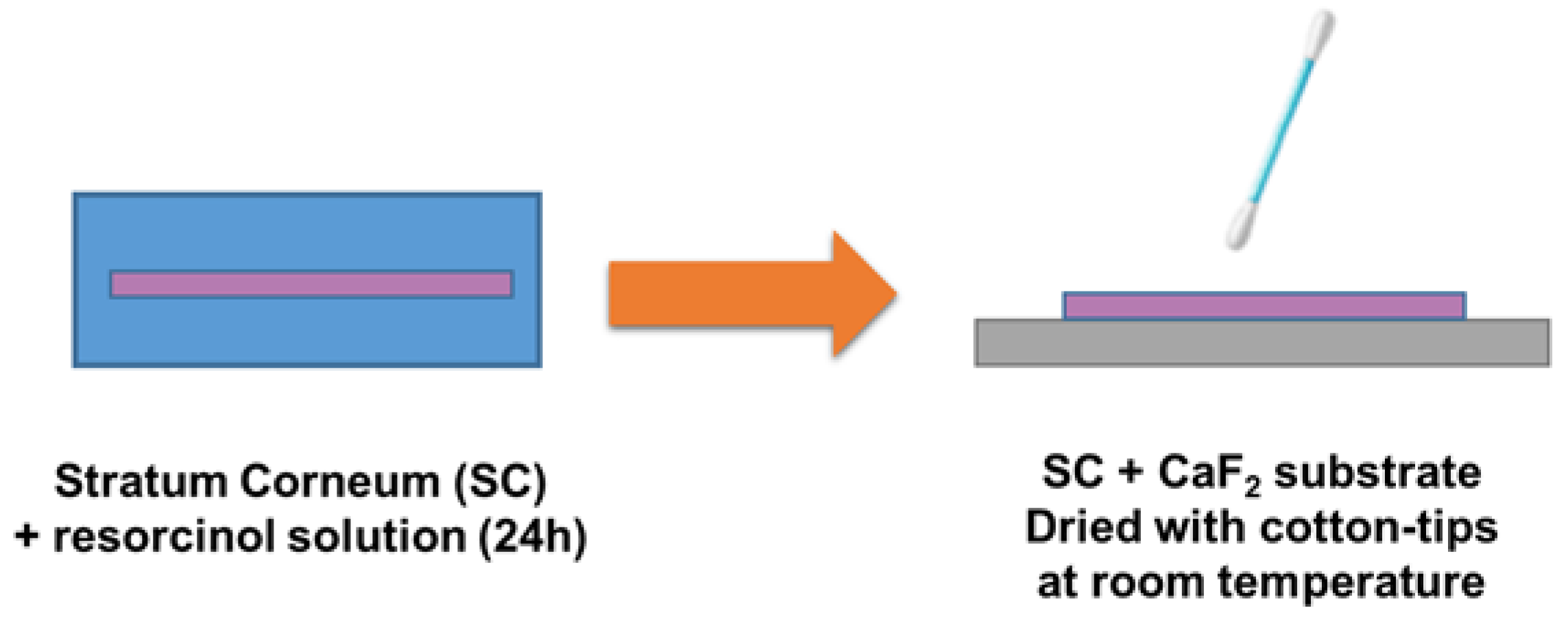
3.1. Validation of the Protocol for Sample Preparation
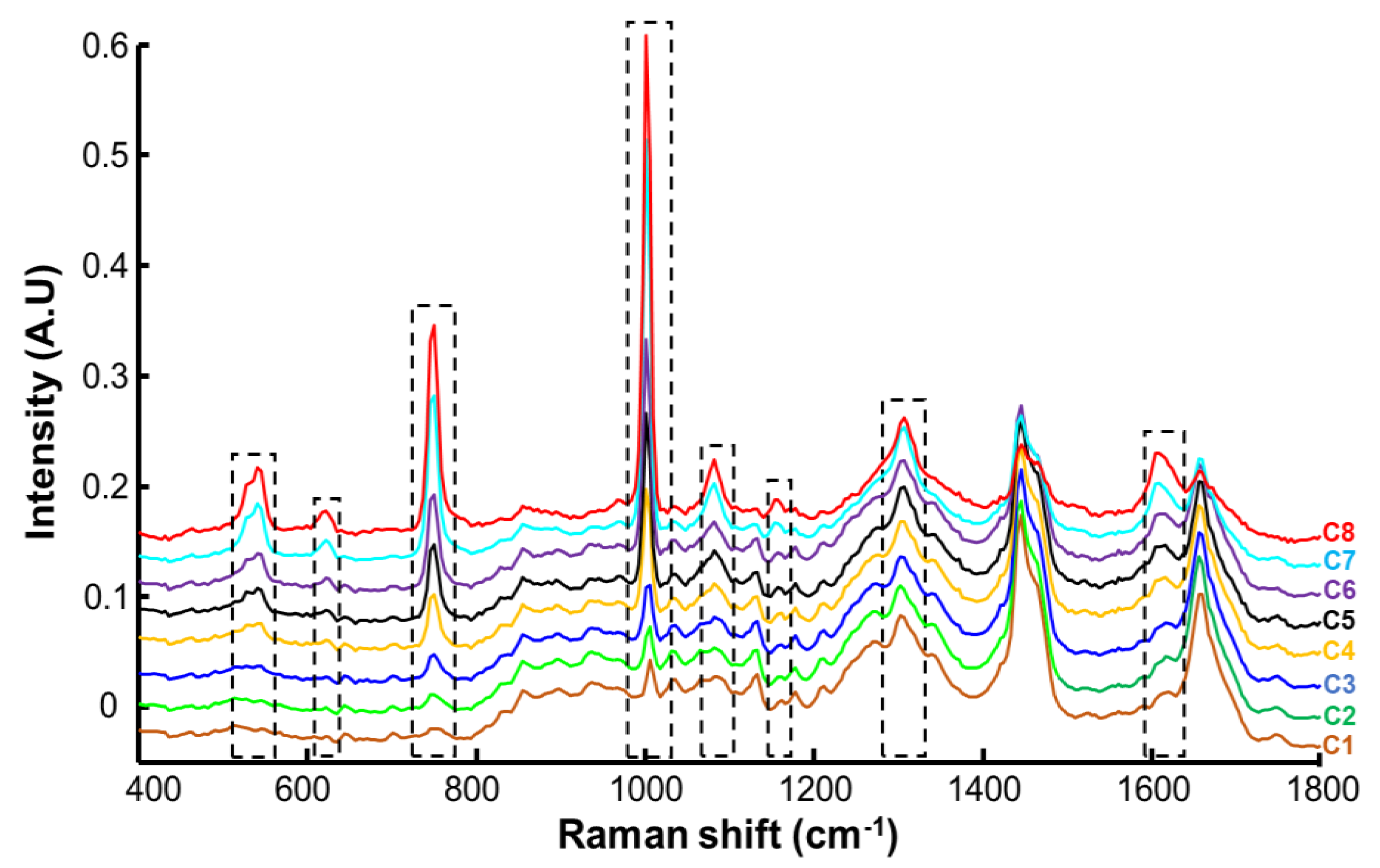
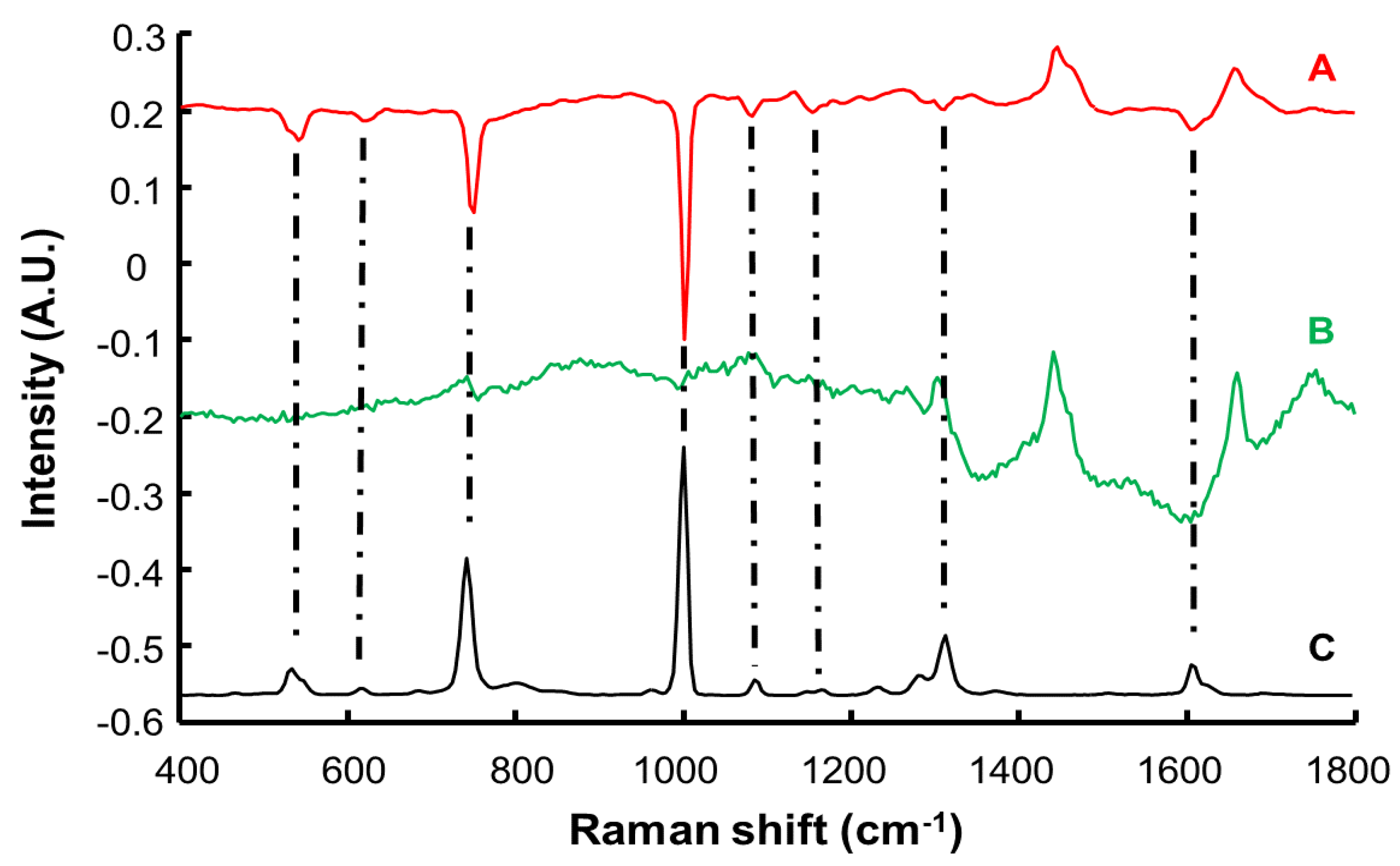
3.2. Spectral Characterisation of Human Stratum Corneum and Resorcinol by Confocal Raman Microscopy (CRM)
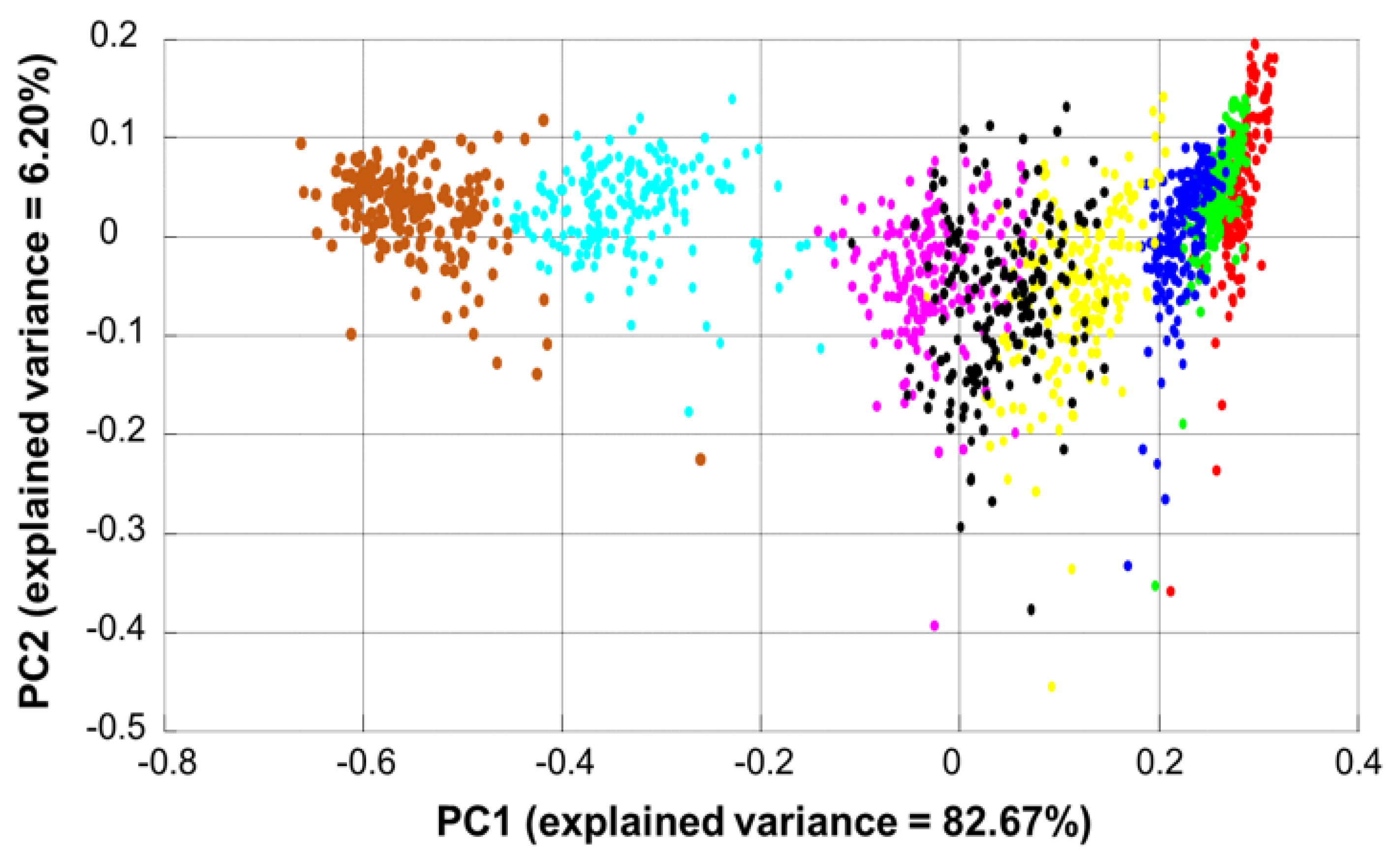
3.3. Data Exploration by Principal Component Analysis (PCA)
3.4. Data Analysis Using Area under the Curve
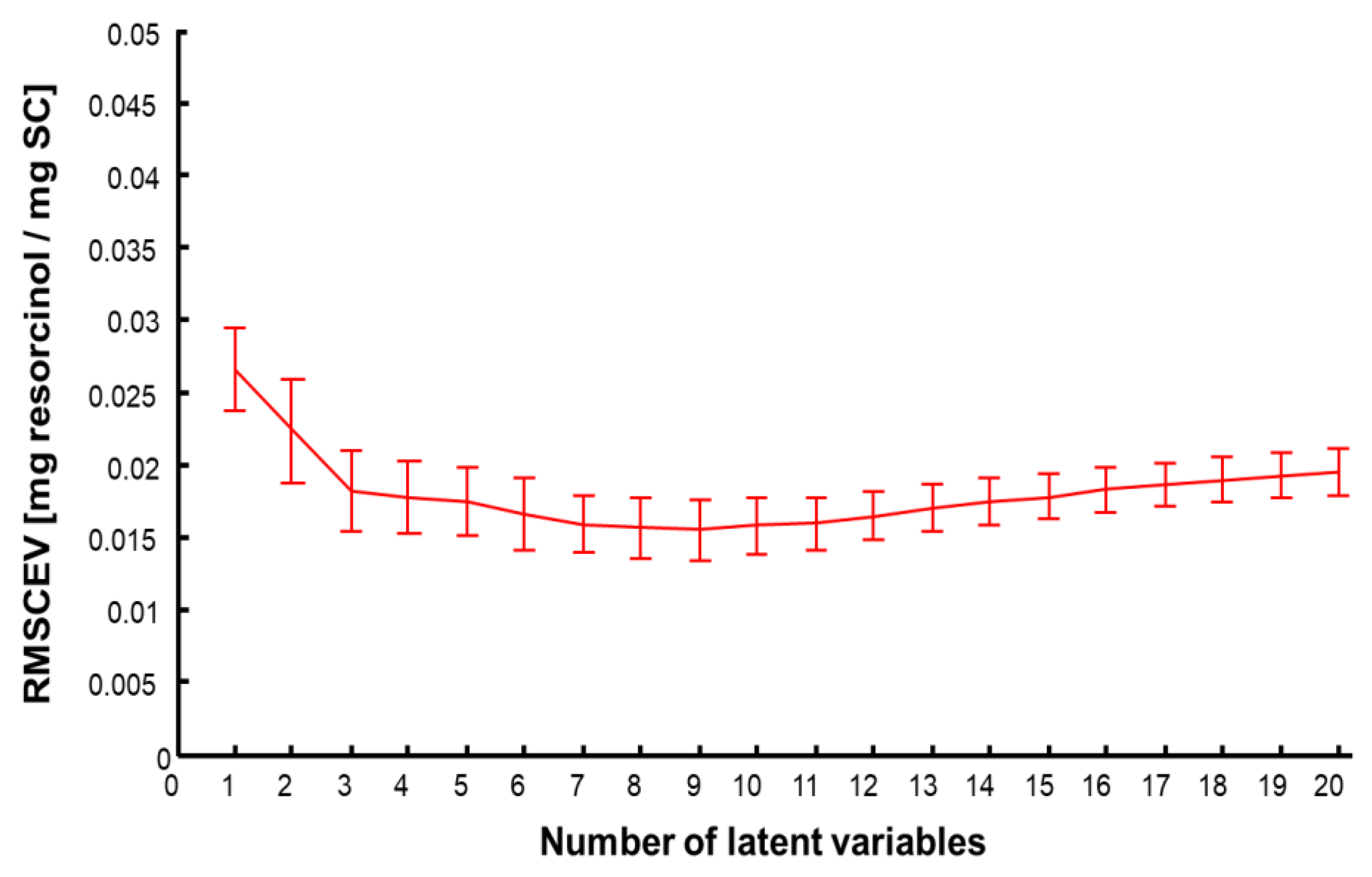
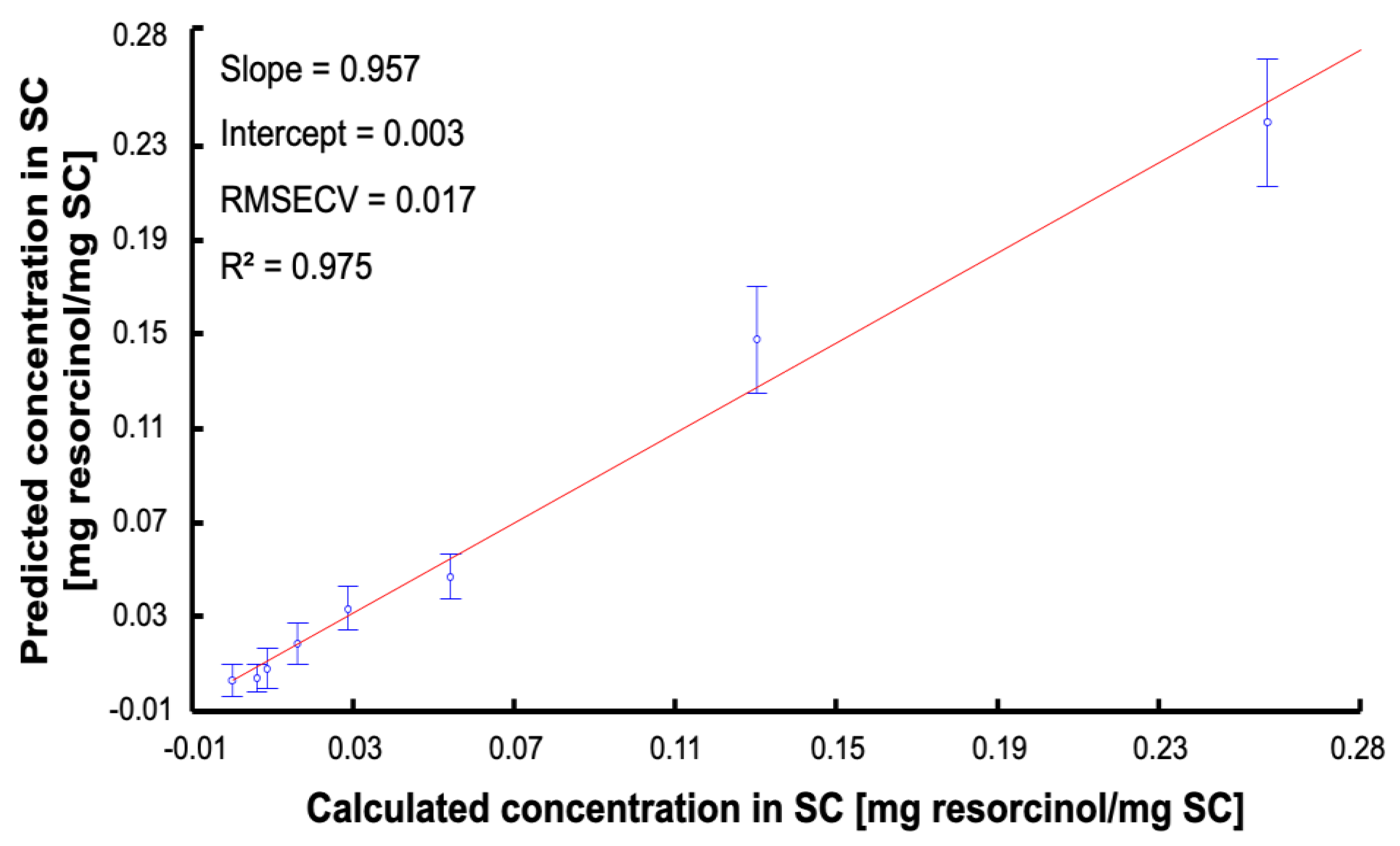
3.5. Data Analysis Using Partial Least Squares Regression (PLSR)
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Van Smeden, J.; Janssens, M.; Gooris, G.S.; Bouwstra, J.A. The important role of Stratum corneum lipids for the cutaneous barrier function. Biochim. Biophys. Acta BBA Mol. Cell Biol. Lipids 2014, 1841, 295–313. [Google Scholar] [CrossRef] [PubMed]
- Elias, P.M. Epidermal Lipids, Barrier Function, and Desquamation. J. Investig. Dermatol. 1983, 80, S44–S49. [Google Scholar] [CrossRef] [PubMed]
- Organisation for Economic Co-operation and Development (OECD). OECD Guidelines for the Testing of Chemicals, Section 4: Test No. 428: Skin Absorption: In Vitro Method; OECD Publishing: Paris, France, 2004. [Google Scholar] [CrossRef]
- Monteiro-Riviere, N.A. Toxicology of the Skin, 1st ed.; CRC Press: Boca Raton, FL, USA, 2010. [Google Scholar]
- Schmidberger, M.; Nikolic, I.; Pantelic, I.; Lunter, D. Optimization of Rheological Behaviour and Skin Penetration of Thermogelling Emulsions with Enhanced Substantivity for Potential Application in Treatment of Chronic Skin Diseases. Pharmaceutics 2019, 11, 361. [Google Scholar] [CrossRef] [Green Version]
- Okunade, O.A.; Yekeen, N.; Padmanabhan, E.; Al-Yaseri, A.; Idris, A.K.; Khan, J.A. Shale core wettability alteration, foam and emulsion stabilization by surfactant: Impact of surfactant concentration, rock surface roughness and nanoparticles. J. Pet. Sci. Eng. 2021, 207, 109139. [Google Scholar] [CrossRef]
- Franz, T.J. Percutaneous Absorption. On the Relevance of in Vitro Data. J. Investig. Dermatol. 1975, 64, 190–195. [Google Scholar] [CrossRef] [Green Version]
- Directorate-General for Health and Consumers—European Commission. Basic Criteria for the In Vitro Assessment of Dermal Absorption of Cosmetic Ingredients; European Commission: Brussels, Belgium, 2010. [Google Scholar]
- European Medicines Agency (EMA). Draft Guideline on Quality and Equivalence of Topical Products; European Medicines Agency: Amsterdam, The Netherlands, 2018. [Google Scholar]
- Supe, S.; Takudage, P. Methods for evaluating penetration of drug into the skin: A review. Skin Res. Technol. 2021, 27, 299–308. [Google Scholar] [CrossRef]
- Hughes, A.J.; Tawfik, S.S.; Baruah, K.P.; O’Toole, E.A.; O’Shaughnessy, R.F.L. Tape strips in dermatology research. Br. J. Dermatol. 2021, 185, 26–35. [Google Scholar] [CrossRef]
- Escobar-Chavez, J.J.; Merino-Sanjuán, V.; López-Cervantes, M.; Urban-Morlan, Z.; Piñón-Segundo, E.; Quintanar-Guerrero, D.; Ganem-Quintanar, A. The Tape-Stripping Technique as a Method for Drug Quantification in Skin. J. Pharm. Pharm. Sci. 2008, 11, 104. [Google Scholar] [CrossRef] [Green Version]
- Bashir, S.J.; Chew, A.-L.; Anigbogu, A.; Dreher, F.; Maibach, H.I. Physical and physiological effects of Stratum corneum tape stripping: Effects of SC tape stripping. Skin Res. Technol. 2001, 7, 40–48. [Google Scholar] [CrossRef]
- Baumann, K.Y.; Church, M.K.; Clough, G.F. Skin microdialysis: Methods, applications and future opportunities—An EAACI position paper. Clin. Transl. Allergy 2019, 9, 24. [Google Scholar] [CrossRef] [Green Version]
- Bodenlenz, M.; Augustin, T.; Birngruber, T.; Tiffner, K.I.; Boulgaropoulos, B.; Schwingenschuh, S.; Raney, S.G.; Rantou, E.; Sinner, F. Variability of Skin Pharmacokinetic Data: Insights from a Topical Bioequivalence Study Using Dermal Open Flow Microperfusion. Pharm. Res. 2020, 37, 204. [Google Scholar] [CrossRef] [PubMed]
- Caspers, P.J.; Lucassen, G.W.; Wolthuis, R.; Bruining, H.A.; Puppels, G.J. In vitro and in vivo Raman spectroscopy of human skin. Biospectroscopy 1998, 4, S31–S39. [Google Scholar] [CrossRef]
- Krafft, C.; Schie, I.W.; Meyer, T.; Schmitt, M.; Popp, J. Developments in spontaneous and coherent Raman scattering microscopic imaging for biomedical applications. Chem. Soc. Rev. 2016, 45, 1819–1849. [Google Scholar] [CrossRef] [PubMed]
- Krafft, C.; Schmitt, M.; Schie, I.W.; Cialla-May, D.; Matthäus, C.; Bocklitz, T.; Popp, J. Label-Free Molecular Imaging of Biological Cells and Tissues by Linear and Nonlinear Raman Spectroscopic Approaches. Angew. Chem. Int. Ed. 2017, 56, 4392–4430. [Google Scholar] [CrossRef]
- Silveira, L.; Pasqualucci, C.A.; Bodanese, B.; Pacheco, M.T.T.; Zângaro, R.A. Normal-subtracted preprocessing of Raman spectra aiming to discriminate skin actinic keratosis and neoplasias from benign lesions and normal skin tissues. Lasers Med. Sci. 2020, 35, 1141–1151. [Google Scholar] [CrossRef]
- Tfaili, S.; Josse, G.; Angiboust, J.F.; Manfait, M.; Piot, O. Monitoring caffeine and resveratrol cutaneous permeation by confocal Raman microspectroscopy. J. Biophotonics 2014, 7, 676–681. [Google Scholar] [CrossRef]
- Mateus, R.; Abdalghafor, H.; Oliveira, G.; Hadgraft, J.; Lane, M.E. A new paradigm in dermatopharmacokinetics—Confocal Raman spectroscopy. Int. J. Pharm. 2013, 444, 106–108. [Google Scholar] [CrossRef]
- Lunter, D.; Daniels, R. Confocal Raman microscopic investigation of the effectiveness of penetration enhancers for procaine delivery to the skin. J. Biomed. Opt. 2014, 19, 126015. [Google Scholar] [CrossRef]
- dos Santos, L.; Tippavajhala, V.K.; Mendes, T.O.; da Silva, M.G.P.; Fávero, P.P.; Téllez Soto, C.A.; Martin, A.A. Evaluation of penetration process into young and elderly skin using confocal Raman spectroscopy. Vib. Spectrosc. 2019, 100, 123–130. [Google Scholar] [CrossRef]
- de Azevedo, J.R.; Bolzinger, M.-A.; Briançon, S.; Chevalier, Y.; Dancik, Y. Confocal Raman Microspectroscopy in the Assessment of Skin Barrier Function and Drug Penetration. In Percutaneous Absorption; CRC Press: Boca Raton, FL, USA, 2021; pp. 799–836. [Google Scholar]
- Essendoubi, M.; Gobinet, C.; Reynaud, R.; Angiboust, J.F.; Manfait, M.; Piot, O. Human skin penetration of hyaluronic acid of different molecular weights as probed by Raman spectroscopy. Skin Res. Technol. 2016, 22, 55–62. [Google Scholar] [CrossRef]
- Pereira, A.F.M.; Rodrigues, B.V.M.; Neto, L.M.; Lopes, L.D.O.; da Costa, A.L.F.; Santos, A.S.; Viana, B.C.; Tosato, M.G.; da Silva, G.C.; Gusmao, G.O.M.; et al. Confocal Raman spectroscopy as a tool to assess advanced glycation end products on solar-exposed human skin. Vib. Spectrosc. 2021, 114, 103234. [Google Scholar] [CrossRef]
- Boncheva, M.; de Sterke, J.; Caspers, P.J.; Puppels, G.J. Depth profiling of Stratum corneum hydration in vivo: A comparison between conductance and confocal Raman spectroscopic measurements. Exp. Dermatol. 2009, 7, 870–876. [Google Scholar] [CrossRef] [PubMed]
- Dancik, Y.; Sriram, G.; Rout, B.; Zou, Y.; Bigliardi-Qi, M.; Bigliardi, P.L. Physical and compositional analysis of differently cultured 3D human skin equivalents by confocal Raman spectroscopy. Analyst 2018, 143, 1065–1076. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z. Confocal Raman microspectroscopy as an alternative method to investigate the extraction of lipids from Stratum corneum by emulsifiers and formulations. Eur. J. Pharm. Biopharm. 2018, 11, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Pudney, P.D.A.; Mélot, M.; Caspers, P.J.; van der Pol, A.; Puppels, G.J. An In Vivo Confocal Raman Study of the Delivery of Trans-Retinol to the Skin. Appl. Spectrosc. 2007, 61, 804–811. [Google Scholar] [CrossRef] [PubMed]
- Iliopoulos, F.; Caspers, P.J.; Puppels, G.J.; Lane, M.E. Franz Cell Diffusion Testing and Quantitative Confocal Raman Spectroscopy: In Vitro-In Vivo Correlation. Pharmaceutics 2020, 12, 887. [Google Scholar] [CrossRef]
- Caspers, P.J.; Nico, C.; Bakker Schut, T.C.; Sterke, J.; Pudney, P.D.A.; Curto, P.R.; Illand, A.; Puppels, G.J. Method to quantify the in vivo skin penetration of topically applied materials based on confocal Raman spectroscopy. Transl. Biophotonics 2019, 1, e201900004. [Google Scholar] [CrossRef] [Green Version]
- Food and Drug Administration (FDA); Center for Drug Evaluation and Research (CDER); Center for Veterinary Medicine (CVM). Bioanalytical Method Validation Guidance for Industry; U.S. Department of Health and Human Services, Food and Drug Administration: Silver Spring, MD, USA, 2018; 44p. [Google Scholar]
- European Medicines Agency (EMA). Guideline on Bioanalytical Method Validation; European Medicines Agency: Amsterdam, The Netherlands, 2011. [Google Scholar]
- Guideline, I.H. Bioanalytical Method Validation M10; European Medicines Agency: Amsterdam, The Netherlands, 2019. [Google Scholar]
- Franzen, L.; Anderski, J.; Windbergs, M. Quantitative detection of caffeine in human skin by confocal Raman spectroscopy—A systematic in vitro validation study. Eur. J. Pharm. Biopharm. 2015, 95, 110–116. [Google Scholar] [CrossRef]
- Alonso, C.; Carrer, V.; Barba, C.; Coderch, L. Caffeine delivery in porcine skin: A confocal Raman study. Arch. Dermatol. Res. 2018, 310, 657–664. [Google Scholar] [CrossRef]
- Dancik, Y.; Kichou, H.; Eklouh-Molinier, C.; Soucé, M.; Munnier, E.; Chourpa, I.; Bonnier, F. Freezing Weakens the Barrier Function of Reconstructed Human Epidermis as Evidenced by Raman Spectroscopy and Percutaneous Permeation. Pharmaceutics 2020, 12, 1041. [Google Scholar] [CrossRef]
- Lieber, C.A.; Mahadevan-Jansen, A. Automated Method for Subtraction of Fluorescence from Biological Raman Spectra. Appl. Spectrosc. 2003, 57, 1363–1367. [Google Scholar] [CrossRef] [PubMed]
- Miloudi, L.; Bonnier, F.; Tfayli, A.; Yvergnaux, F.; Byrne, H.J.; Chourpa, I.; Munnier, E. Confocal Raman spectroscopic imaging for in vitro monitoring of active ingredient penetration and distribution in reconstructed human epidermis model. J. Biophotonics 2018, 11, e201700221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Callao, M.P.; Rius, F.X. Multivariate standardization techniques using UV-Vis data. Chemom. Intell. Lab. Syst. 1997, 38, 63–73. [Google Scholar]
- ISO 11843-2; Capability of Detection–Part 2: Methodology in the Linear Calibration Case. International Organization for Standardization (ISO): Geneva, Switzerland, 2000.
- Li, M.; Ebel, B.; Chauchard, F.; Guédon, E.; Marc, A. Parallel comparison of in situ Raman and NIR spectroscopies to simultaneously measure multiple variables toward real-time monitoring of CHO cell bioreactor cultures. Biochem. Eng. J. 2018, 137, 205–213. [Google Scholar] [CrossRef]
- Lorber, A.; Faber, K.; Kowalski, B.R. Net Analyte Signal Calculation in Multivariate Calibration. Anal. Chem. 1997, 69, 1620–1626. [Google Scholar] [CrossRef]
- Schönbichler, S.A.; Bittner, L.K.H.; Weiss, A.K.H.; Griesser, U.J.; Pallua, J.D.; Huck, C.W. Comparison of NIR chemical imaging with conventional NIR, Raman and ATR-IR spectroscopy for quantification of furosemide crystal polymorphs in ternary powder mixtures. Eur. J. Pharm. Biopharm. 2013, 84, 616–625. [Google Scholar] [CrossRef]
- Tfayli, A.; Piot, O.; Draux, F.; Pitre, F.; Manfait, M. Molecular characterization of reconstructed skin model by Raman microspectroscopy: Comparison with excised human skin. Biopolymers 2007, 87, 261–274. [Google Scholar] [CrossRef]
- Tfaili, S.; Gobinet, C.; Josse, G.; Angiboust, J.-F.; Manfait, M.; Piot, O. Confocal Raman microspectroscopy for skin characterization: A comparative study between human skin and pig skin. Analyst 2012, 137, 3673–3682. [Google Scholar] [CrossRef]
- Barry, B.W.; Edwards, H.G.M.; Williams, A.C. Fourier transform Raman and infrared vibrational study of human skin: Assignment of spectral bands. J. Raman. Spectrosc. 1992, 23, 641–645. [Google Scholar] [CrossRef]
- Gniadecka, M.; Wulf, H.C.; Nielsen, O.F.; Christensen, D.H.; Hercogova, J. Distinctive Molecular Abnormalities in Benign and Malignant Skin Lesions: Studies by Raman Spectroscopy. Photochem. Photobiol. 1997, 66, 418–423. [Google Scholar] [CrossRef]
- Drużbicki, K.; Mikuli, E.; Pałka, N.; Zalewski, S.; Ossowska-Chruściel, M.D. Polymorphism of Resorcinol Explored by Complementary Vibrational Spectroscopy (FT-RS, THz-TDS, INS) and First-Principles Solid-State Computations (Plane-Wave DFT). J Phys. Chem. B 2015, 119, 1681–1695. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Wu, C.; Shi, P.; Wang, L.; Gao, S. General Toxicity Prediction Approach for Mixtures Containing Polar Narcotic Chemicals. Bull. Environ. Contam. Toxicol. 2003, 71, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Allegrini, F.; Olivieri, A.C. IUPAC-consistent approach to the limit of detection in partial least-squares calibration. Anal. Chem. 2014, 86, 7858–7866. [Google Scholar] [CrossRef] [PubMed]
- Oleneva, E.; Khaydukova, M.; Ashina, J.; Yaroshenko, I.; Jahatspanian, I.; Legin, A.; Kirsanov, D. A Simple Procedure to Assess Limit of Detection for Multisensor Systems. Sensors 2019, 19, 1359. [Google Scholar] [CrossRef] [Green Version]
- ISO 11843-1; Capability of Detection—Part 1: Terms and Definitions. International Organization for Standardization (ISO): Geneva, Switzerland, 1997.
- Makki, A.A.; Massot, V.; Byrne, H.J.; Respaud, R.; Bertrand, D.; Mohammed, E.; Chourpa, I.; Bonnier, F. Understanding the discrimination and quantification of monoclonal antibodies preparations using Raman spectroscopy. J. Pharm. Biomed. Anal. 2021, 194, 113734. [Google Scholar] [CrossRef]
- Makki, A.A.; Massot, V.; Byrne, H.J.; Respaud, R.; Bertrand, D.; Mohammed, E.; Chourpa, I.; Bonnier, F. Vibrational spectroscopy for discrimination and quantification of clinical chemotherapeutic preparations. Vib. Spectrosc. 2021, 113, 103200. [Google Scholar] [CrossRef]
- Burgués, J.; Marco, S. Multivariate estimation of the limit of detection by orthogonal partial least squares in temperature-modulated MOX sensors. Anal. Chim. Acta 2018, 1019, 49–64. [Google Scholar] [CrossRef]
- Ostra, M.; Ubide, C.; Vidal, M.; Zuriarrain, J. Detection limit estimator for multivariate calibration by an extension of the IUPAC recommendations for univariate methods. Analyst 2008, 133, 532. [Google Scholar] [CrossRef]
- Short, S.M.; Cogdill, R.P.; Anderson, C.A. Determination of figures of merit for near-infrared and raman spectrometry by net analyte signal analysis for a 4-component solid dosage system. Aaps Pharmscitech 2007, 8, 109–119. [Google Scholar] [CrossRef] [Green Version]
- Joshi, R.; Cho, B.-K.; Joshi, R.; Lohumi, S.; Faqeerzada, M.A.; Amanah, H.Z.; Lee, J.; Mo, C.; Lee, H. Raman spectroscopic analysis to detect olive oil mixtures in argan oil. Korean J. Agric. Sci. 2019, 46, 183–194. [Google Scholar] [CrossRef]
- Fan, Y.; Lai, K.; Rasco, B.A.; Huang, Y. Determination of carbaryl pesticide in Fuji apples using surface-enhanced Raman spectroscopy coupled with multivariate analysis. LWT-Food Sci. Technol. 2015, 60, 352–357. [Google Scholar] [CrossRef]
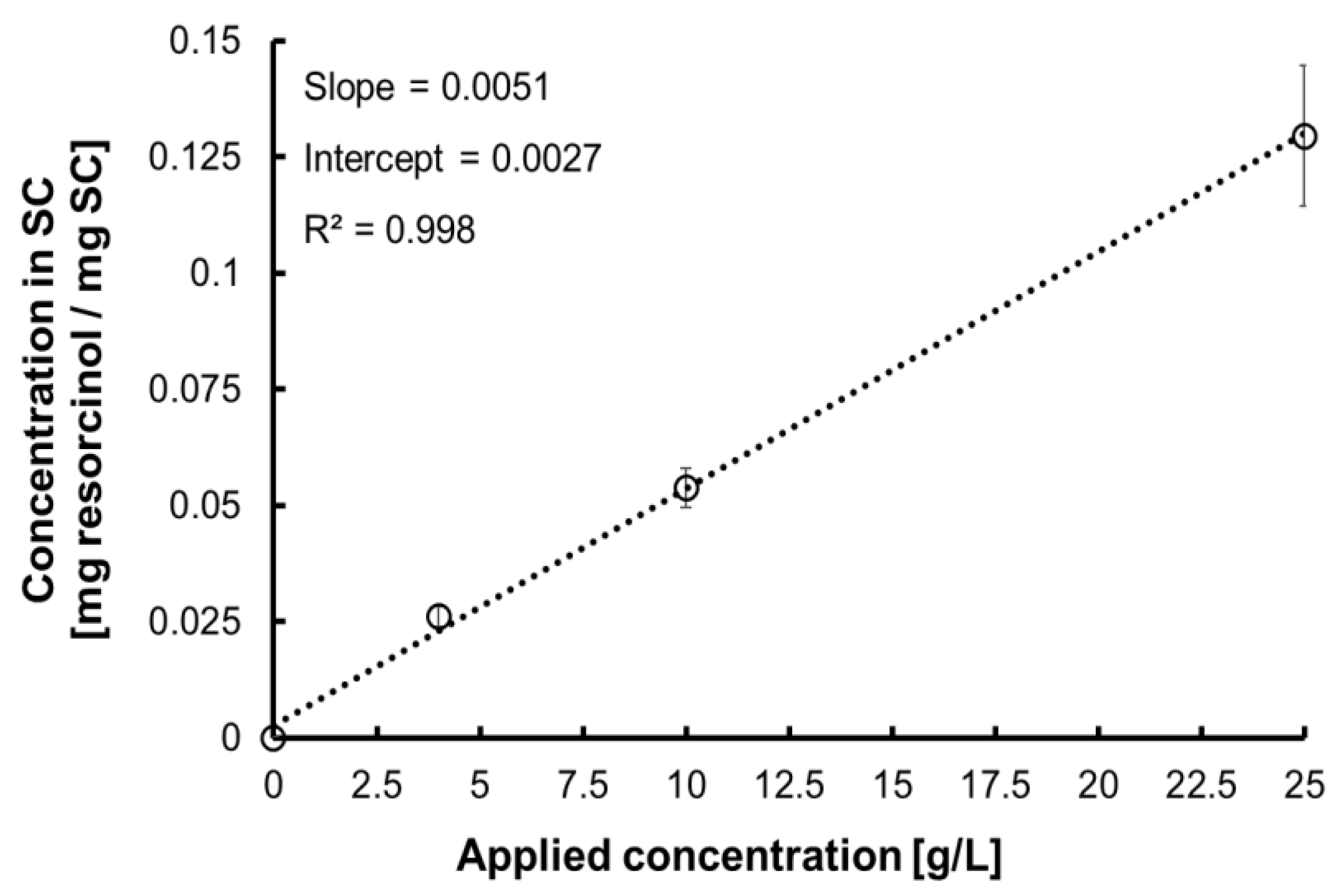

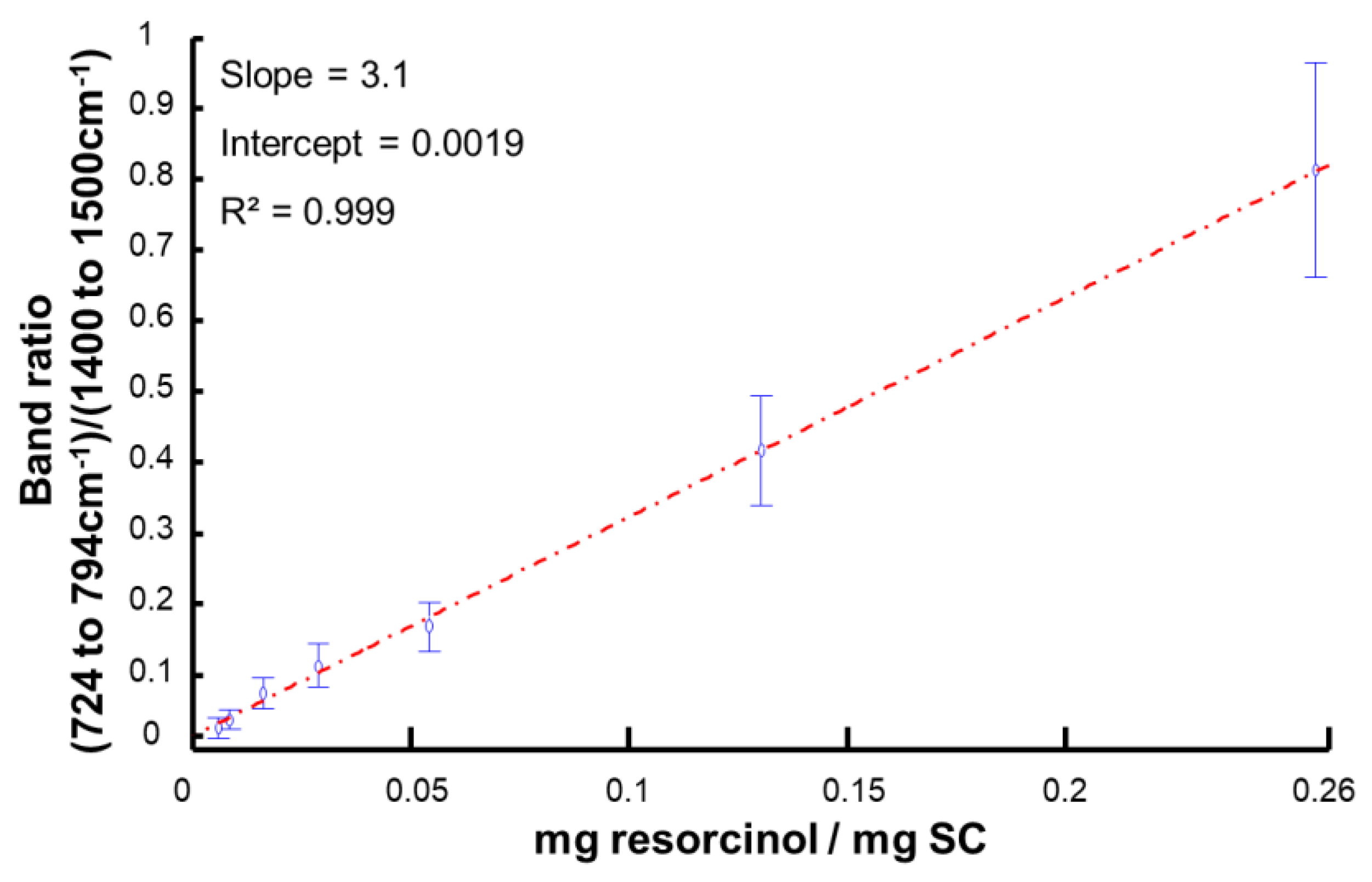

| Applied Concentration [g·L−1] | Concentration in SC [mg resorcinol/mg SC] | |
|---|---|---|
| C1 | 0 | 0 |
| C2 | 0.5 | 0.006 |
| C3 | 1 | 0.008 |
| C4 | 2.5 | 0.016 |
| C5 | 5 | 0.028 |
| C6 | 10 | 0.054 |
| C7 | 25 | 0.130 |
| C8 | 50 | 0.257 |
| Raman Shift (cm−1) | Assignment |
|---|---|
| 614 | Phenylalanine ring |
| 646 | Tyrosine ring, ν C–S |
| 702 | ν C–S gauche of cysteine cholesterol |
| 751 | Aromatic ring puckering |
| 864 | Proline, RNA |
| 898 | Tryptophan |
| 939 | ρ CH3 terminal, m CC a helix (secondary structure), phospholipids |
| 1005 | Symmetric ring breathing phenylalanine |
| 1033 | C–H Phenylalanine |
| 1067 | Lipids: skeletal trans conformation ceramides; ν |
| 1130 | Lipids: hydrocarbon chain, trans conformation ceramides |
| 1177 | CH Tyrosine, phenylalanine |
| 1303 | Amide III, CH2 phospholipids |
| 1444 | δ CH proteins and lipids |
| 1524 | –C=C– carotenoids |
| 1655 | Amide I |
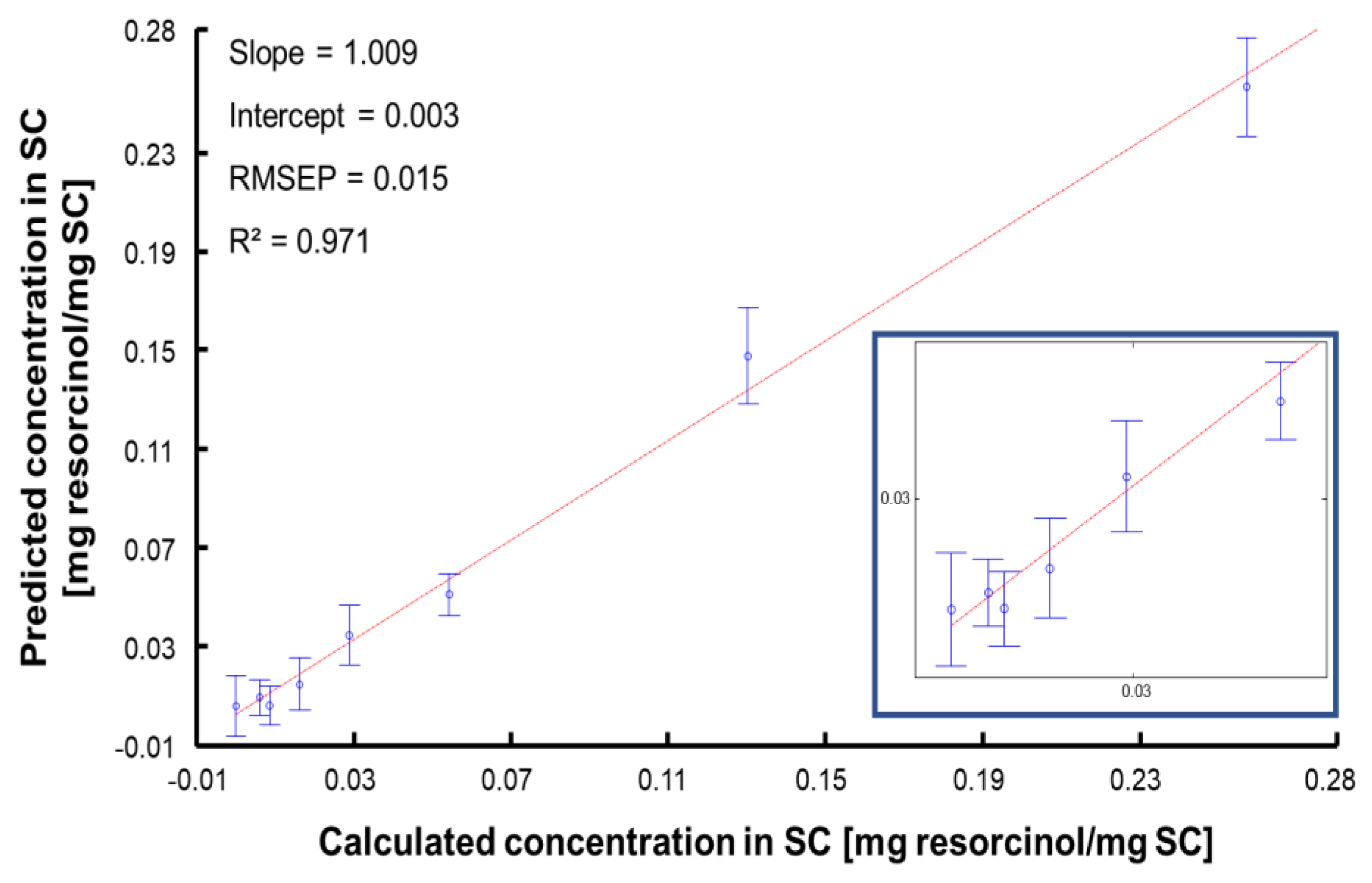
| RMSECV | 0.017 |
| RMSEP | 0.015 |
| Linearity (R2) | 0.971 |
| RPD | 8.112 |
| BCMSEP | 5.15 × 10−5 |
| Sensitivity (sen) | 18.994 |
| Selectivity (sel) | 0.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kichou, H.; Munnier, E.; Dancik, Y.; Kemel, K.; Byrne, H.J.; Tfayli, A.; Bertrand, D.; Soucé, M.; Chourpa, I.; Bonnier, F. Estimating the Analytical Performance of Raman Spectroscopy for Quantification of Active Ingredients in Human Stratum Corneum. Molecules 2022, 27, 2843. https://doi.org/10.3390/molecules27092843
Kichou H, Munnier E, Dancik Y, Kemel K, Byrne HJ, Tfayli A, Bertrand D, Soucé M, Chourpa I, Bonnier F. Estimating the Analytical Performance of Raman Spectroscopy for Quantification of Active Ingredients in Human Stratum Corneum. Molecules. 2022; 27(9):2843. https://doi.org/10.3390/molecules27092843
Chicago/Turabian StyleKichou, Hichem, Emilie Munnier, Yuri Dancik, Kamilia Kemel, Hugh J. Byrne, Ali Tfayli, Dominique Bertrand, Martin Soucé, Igor Chourpa, and Franck Bonnier. 2022. "Estimating the Analytical Performance of Raman Spectroscopy for Quantification of Active Ingredients in Human Stratum Corneum" Molecules 27, no. 9: 2843. https://doi.org/10.3390/molecules27092843







